Micro RNAs in Regulation of Cellular Redox Homeostasis
Abstract
1. Introduction
2. ROS/RNS Production and Neutralization in Cells
3. Roles of ROS and RNS in Cellular Processes
4. Role of miRNAs in Regulation of ROS/RNS-Producing Enzymes
4.1. NADPH Oxidases
4.2. Nitric Oxide Synthases
5. MiRNAs Participating in Regulation of H2O2 Level
5.1. Regulation of Superoxide Dismutases by miRNAs
5.2. Regulation of Catalase, Glutathione Peroxidases, Peroxiredoxins and Thioredoxin System Enzymes by miRNAs
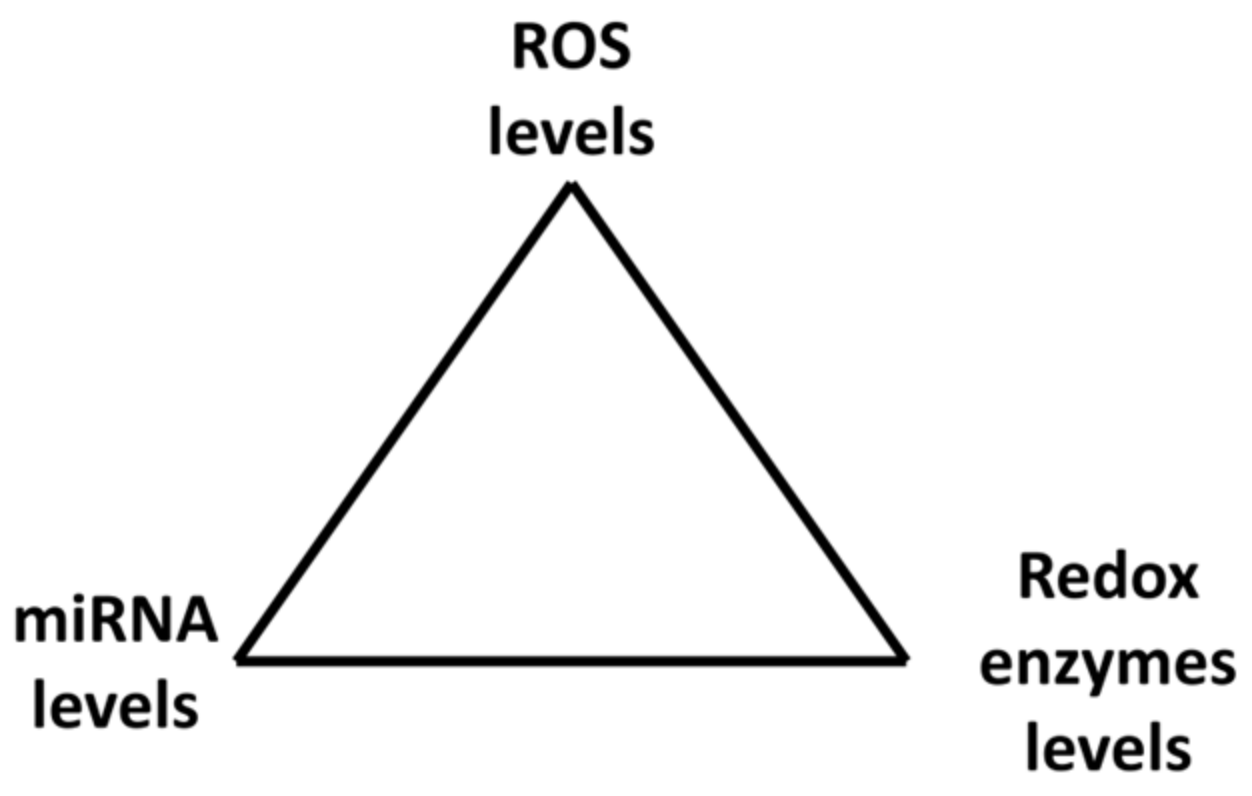
6. Mutual Regulation of Elements of the Redox System
6.1. Changes of Cellular H2O2 Levels Are Accompanied by Changes in Levels of miRNAs and Their Targets
6.2. Unexpected Effects of Changes in Levels of ROS Producers and Neutralizers
6.3. The Same miRNAs May Regulate the Expression of Both ROS-Producing and ROS-Neutralizing Enzymes
7. Role of miRNAs as Regulators of Redox Balance in Cancer Development
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Jonas, S.; Izaurralde, E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat. Rev. Genet. 2015, 16, 421–433. [Google Scholar] [CrossRef] [PubMed]
- Selbach, M.; Schwanhäusser, B.; Thierfelder, N.; Fang, Z.; Khanin, R.; Rajewsky, N. Widespread changes in protein synthesis induced by microRNAs. Nature 2008, 455, 58–63. [Google Scholar] [CrossRef] [PubMed]
- Friedman, R.C.; Farh, K.K.H.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef] [PubMed]
- Podralska, M.; Ciesielska, S.; Kluiver, J.; van den Berg, A.; Dzikiewicz-Krawczyk, A.; Slezak-Prochazka, I. Non-coding RNAs in cancer radiosensitivity: MicroRNAs and lncrnas as regulators of radiation-induced signaling pathways. Cancers 2020, 12, 1–27. [Google Scholar] [CrossRef]
- Qadir, M.M.F.; Klein, D.; Álvarez-Cubela, S.; Domínguez-Bendala, J.; Pastori, R.L. The role of microRNAs in diabetes-related oxidative stress. Int. J. Mol. Sci. 2019, 20, 5423. [Google Scholar] [CrossRef]
- Kura, B.; Bacova, B.S.; Kalocayova, B.; Sykora, M.; Slezak, J. Oxidative stress-responsive microRNAs in heart injury. Int. J. Mol. Sci. 2020, 21, 358. [Google Scholar] [CrossRef]
- Kurutas, E.B. The importance of antioxidants which play the role in cellular response against oxidative/nitrosative stress: Current state. Nutr. J. 2016, 15. [Google Scholar] [CrossRef]
- Tauler Riera, P.; Volterrani, M.; Iellamo, F.; Fallo, F.; Ermolao, A.; Kraemer, W.J.; Ratamess, N.A.; Faigenbaum, A.; Philp, A.; Baar, K. Redox Status. In Encyclopedia of Exercise Medicine in Health and Disease; Springer: Berlin/Heidelberg, Germany, 2012; pp. 751–753. [Google Scholar]
- Kohen, R.; Nyska, A. Oxidation of biological systems: Oxidative stress phenomena, antioxidants, redox reactions, and methods for their quantification. Toxicol. Pathol. 2002, 30, 620–650. [Google Scholar] [CrossRef]
- Marengo, B.; Nitti, M.; Furfaro, A.L.; Colla, R.; Ciucis, C.D.; Marinari, U.M.; Pronzato, M.A.; Traverso, N.; Domenicotti, C. Redox homeostasis and cellular antioxidant systems: Crucial players in cancer growth and therapy. Oxid. Med. Cell. Longev. 2016, 2016, 6235641. [Google Scholar] [CrossRef]
- Phaniendra, A.; Jestadi, D.B.; Periyasamy, L. Free Radicals: Properties, Sources, Targets, and Their Implication in Various Diseases. Indian J. Clin. Biochem. 2015, 30, 11–26. [Google Scholar] [CrossRef]
- Schrader, M.; Fahimi, H.D. Peroxisomes and oxidative stress. Biochim. Biophys. Acta Mol. Cell Res. 2006, 1763, 1755–1766. [Google Scholar] [CrossRef]
- Skonieczna, M.; Hejmo, T.; Poterala-Hejmo, A.; Cieslar-Pobuda, A.; Buldak, R.J. NADPH Oxidases: Insights into Selected Functions and Mechanisms of Action in Cancer and Stem Cells. Oxid. Med. Cell. Longev. 2017, 2017, 9420539. [Google Scholar] [CrossRef]
- Jadhav, V.S.; Krause, K.H.; Singh, S.K. HIV-1 Tat C modulates NOX2 and NOX4 expressions through miR-17 in a human microglial cell line. J. Neurochem. 2014, 131, 803–815. [Google Scholar] [CrossRef]
- Kim, H.-L.; Park, Y.-S. Maintenance of cellular tetrahydrobiopterin homeostasis. BMB Rep. 2010, 43, 584–592. [Google Scholar] [CrossRef]
- Förstermann, U.; Closs, E.I.; Pollock, J.S.; Nakane, M.; Schwarz, P.; Gath, I.; Kleinert, H. Nitric oxide synthase isozymes. Characterization, purification, molecular cloning, and functions. Hypertension 1994, 23, 1121–1131. [Google Scholar] [CrossRef]
- Andrew, P.J.; Mayer, B. Enzymatic function of nitric oxide synthases. Cardiovasc. Res. 1999, 43, 521–531. [Google Scholar] [CrossRef]
- Vasquez-Vivar, J.; Martasek, P.; Whitsett, J.; Joseph, J.; Kalyanaraman, B. The ratio between tetrahydrobiopterin and oxidized tetrahydrobiopterin analogues controls superoxide release from endothelial nitric oxide synthase: An EPR spin trapping study. Biochem. J. 2002, 362, 733–739. [Google Scholar] [CrossRef]
- Morris, S.M. Enzymes of arginine metabolism. J. Nutr. 2004, 134, 2743S–2747S. [Google Scholar] [CrossRef]
- Meister, A. Glutathione metabolism and its selective modification. J. Biol. Chem. 1988, 263, 17205–17208. [Google Scholar] [CrossRef]
- Bradshaw, P.C. Cytoplasmic and mitochondrial NADPH-coupled Redox systems in the regulation of aging. Nutrients 2019, 11, 504. [Google Scholar] [CrossRef]
- Candas, D.; Li, J.J. MnSOD in oxidative stress response-potential regulation via mitochondrial protein influx. Antioxid. Redox Signal. 2014, 20, 1599–1617. [Google Scholar] [CrossRef]
- Sarsour, E.H.; Kumar, M.G.; Chaudhuri, L.; Kalen, A.L.; Goswami, P.C. Redox control of the cell cycle in health and disease. Antioxid. Redox Signal. 2009, 11, 2985–3011. [Google Scholar] [CrossRef]
- Wong, H.S.; Dighe, P.A.; Mezera, V.; Monternier, P.A.; Brand, M.D. Production of superoxide and hydrogen peroxide from specific mitochondrial sites under different bioenergetic conditions. J. Biol. Chem. 2017, 292, 16804–16809. [Google Scholar] [CrossRef]
- Puppo, A.; Halliwellt, B. Formation of hydroxyl radicals from hydrogen peroxide in the presence of iron is haemoglobin a biological Fenton reagent? Biochem. J 1988, 249, 185–190. [Google Scholar] [CrossRef] [PubMed]
- Florence, T.M. The production of hydroxyl radical from hydrogen peroxide. J. Inorg. Biochem. 1984, 22, 221–230. [Google Scholar] [CrossRef]
- Nandi, A.; Yan, L.-J.; Jana, C.K.; Das, N. Role of Catalase in Oxidative Stress- and Age-Associated Degenerative Diseases. Oxid. Med. Cell. Longev. 2019, 2019. [Google Scholar] [CrossRef] [PubMed]
- Adamiec, M.; Skonieczna, M. UV radiation in HCT 116 cells influences intracellular H2O2 and glutathione levels, antioxidant expression, and protein glutathionylation. Acta Biochim. Pol. 2019, 66, 605–610. [Google Scholar] [CrossRef]
- Immenschuh, S.; Baumgart-Vogt, E. Peroxiredoxins, oxidative stress, and cell proliferation. Antioxid. Redox Signal. 2005, 7, 768–777. [Google Scholar] [CrossRef]
- Sue, G.R.; Ho, Z.C.; Kim, K. Peroxiredoxins: A historical overview and speculative preview of novel mechanisms and emerging concepts in cell signaling. Free Radic. Biol. Med. 2005, 38, 1543–1552. [Google Scholar]
- Glorieux, C.; Calderon, P.B. Catalase, a remarkable enzyme: Targeting the oldest antioxidant enzyme to find a new cancer treatment approach. Biol. Chem. 2017, 398, 1095–1108. [Google Scholar] [CrossRef]
- Bai, J.; Cederbaum, A.I. Mitochondrial catalase and oxidative injury. NeuroSignals 2001, 10, 189–199. [Google Scholar] [CrossRef]
- Ciesielska, S.; Bil, P.; Gajda, K.; Poterala-Hejmo, A.; Hudy, D.; Rzeszowska-Wolny, J. Cell type-specific differences in redox regulation and proliferation after low UVA doses. PLoS ONE 2019, 14, e0205215. [Google Scholar] [CrossRef]
- Bil, P.; Ciesielska, S.; Jaksik, R.; Rzeszowska-Wolny, J. Circuits Regulating Superoxide and Nitric Oxide Production and Neutralization in Different Cell Types: Expression of Participating Genes and Changes Induced by Ionizing Radiation. Antioxidants 2020, 9, 701. [Google Scholar] [CrossRef]
- Powis, G.; Montfort, W.R. Properties and biological activities of thioredoxins. Annu. Rev. Biophys. Biomol. Struct. 2001, 30, 421–455. [Google Scholar] [CrossRef]
- Arnér, E.S.J.; Holmgren, A. Physiological functions of thioredoxin and thioredoxin reductase. Eur. J. Biochem. 2000, 267, 6102–6109. [Google Scholar] [CrossRef]
- Dreyer, J.; Schleicher, M.; Tappe, A.; Schilling, K.; Kuner, T.; Kusumawidijaja, G.; Müller-Esterl, W.; Oess, S.; Kuner, R. Nitric oxide synthase (NOS)-interacting protein interacts with neuronal NOS and regulates its distribution and activity. J. Neurosci. 2004, 24, 10454–10465. [Google Scholar] [CrossRef]
- Thannickal, V.J.; Fanburg, B.L. Reactive oxygen species in cell signaling. Am. J. Physiol. Lung Cell. Mol. Physiol. 2000, 279, L1005–L1028. [Google Scholar] [CrossRef]
- Feinendegen, L.E. The role of adaptive responses following exposure to ionizing radiation. Hum. Exp. Toxicol. 1999, 18, 426–432. [Google Scholar] [CrossRef]
- Zhang, J.; Xing, D.; Gao, X. Low-power laser irradiation activates Src tyrosine kinase through reactive oxygen species-mediated signaling pathway. J. Cell. Physiol. 2008, 217, 518–528. [Google Scholar] [CrossRef]
- Wang, X.; Liu, J.Z.; Hu, J.X.; Wu, H.; Li, Y.L.; Chen, H.L.; Bai, H.; Hai, C.X. ROS-activated p38 MAPK/ERK-Akt cascade plays a central role in palmitic acid-stimulated hepatocyte proliferation. Free Radic. Biol. Med. 2011, 51, 539–551. [Google Scholar] [CrossRef]
- Yasuda, M.; Ohzeki, Y.; Shimizu, S.; Naito, S.; Ohtsuru, A.; Yamamoto, T.; Kuroiwa, Y. Stimulation of in vitro angiogenesis by hydrogen peroxide and the relation with ets-1 in endothelial cells. Life Sci. 1998, 64, 249–258. [Google Scholar] [CrossRef]
- Day, R.M.; Suzuki, Y.J. Cell Proliferation, Reactive Oxygen and Cellular Glutathione. Dose-Response 2005, 3. [Google Scholar] [CrossRef] [PubMed]
- Brown, M.R.; Miller, F.J.; Li, W.-G.; Ellingson, A.N.; Mozena, J.D.; Chatterjee, P.; Engelhardt, J.F.; Zwacka, R.M.; Oberley, L.W.; Fang, X.; et al. Overexpression of Human Catalase Inhibits Proliferation and Promotes Apoptosis in Vascular Smooth Muscle. Circ. Res. 1999, 85, 524–533. [Google Scholar] [CrossRef] [PubMed]
- Murrell, G.A.C.; Francis, M.J.; Bromley, L. Modulation of fibroblast proliferation by oxygen free radicals. Biochem. J. 1990, 265, 659–665. [Google Scholar] [CrossRef] [PubMed]
- Davies, K. The Broad Spectrum of Responses to Oxidants in Proliferating Cells: A New Paradigm for Oxidative Stress. IUBMB Life 1999, 48, 41–47. [Google Scholar] [CrossRef] [PubMed]
- Davies, J.M.S.; Lowry, C.V.; Davies, K.J.A. Transient Adaptation to Oxidative Stress in Yeast. Arch. Biochem. Biophys. 1995, 317, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Christman, M.F.; Morgan, R.W.; Jacobson, F.S.; Ames, B.N. Positive control of a regulon for defenses against oxidative stress and some heat-shock proteins in Salmonella typhimurium. Cell 1985, 41, 753–762. [Google Scholar] [CrossRef]
- Trachootham, D.; Lu, W.; Ogasawara, M.A.; Valle, N.R.; Del, H.P. Redox regulation of cell survival. Antioxid. Redox Signal. 2008, 10, 1343–1374. [Google Scholar] [CrossRef]
- He, F.; Ru, X.; Wen, T. NRF2, a transcription factor for stress response and beyond. Int. J. Mol. Sci. 2020, 21, 4777. [Google Scholar] [CrossRef]
- Zhang, C.; Shu, L.; Kong, A.N.T. MicroRNAs: New Players in Cancer Prevention Targeting Nrf2, Oxidative Stress and Inflammatory Pathways. Curr. Pharmacol. Rep. 2015, 1, 21–30. [Google Scholar] [CrossRef]
- Kitamoto, K.; Miura, Y.; Karnan, S.; Ota, A.; Konishi, H.; Hosokawa, Y.; Sato, K. Inhibition of NADPH oxidase 2 induces apoptosis in osteosarcoma: The role of reactive oxygen species in cell proliferation. Oncol. Lett. 2018, 15, 7955–7962. [Google Scholar] [CrossRef]
- Raffel, J.; Bhattacharyya, A.K.; Gallegos, A.; Cui, H.; Einspahr, J.G.; Alberts, D.S.; Powis, G. Increased expression of thioredoxin-1 in human colorectal cancer is associated with decreased patient survival. J. Lab. Clin. Med. 2003, 142, 46–51. [Google Scholar] [CrossRef]
- Chiu, J.; Dawes, I.W. Redox control of cell proliferation. Trends Cell Biol. 2012, 22, 592–601. [Google Scholar] [CrossRef]
- Sun, J.; Steenbergen, C.; Murphy, E. S-nitrosylation: NO-related redox signaling to protect against oxidative stress. Antioxid. Redox Signal. 2006, 8, 1693–1705. [Google Scholar] [CrossRef]
- Cho, S.H.; Lee, C.H.; Ahn, Y.; Kim, H.; Kim, H.; Ahn, C.Y.; Yang, K.S.; Lee, S.R. Redox regulation of PTEN and protein tyrosine phosphatases in H2O2-mediated cell signaling. FEBS Lett. 2004, 560, 7–13. [Google Scholar] [CrossRef]
- Sha, Y.; Marshall, H.E. S-nitrosylation in the regulation of gene transcription. Biochim. Biophys. Acta Gen. Subj. 2012, 1820, 701–711. [Google Scholar] [CrossRef]
- Yakovlev, V.A.; Mikkelsen, R.B. Protein tyrosine nitration in cellular signal transduction pathways. J. Recept. Signal Transduct. 2010, 30, 420–429. [Google Scholar] [CrossRef]
- Shen, W.; Lu, Y.; Hu, J.; Le, H.; Yu, W.; Xu, W.; Yu, W.; Zheng, J. Mechanism of miR-320 in Regulating Biological Characteristics of Ischemic Cerebral Neuron by Mediating Nox2/ROS Pathway. J. Mol. Neurosci. 2020, 70, 449–457. [Google Scholar] [CrossRef]
- Zuo, M.-L.; Wang, A.-P.; Song, G.-L.; Yang, Z.-B. miR-652 protects rats from cerebral ischemia/reperfusion oxidative stress injury by directly targeting NOX2. Biomed. Pharmacother. 2020, 124, 109860. [Google Scholar] [CrossRef]
- Li, S.Z.; Hu, Y.Y.; Zhao, J.; Zhao, Y.B.; Sun, J.D.; Yang, Y.F.; Ji, C.C.; Liu, Z.B.; Cao, W.D.; Qu, Y.; et al. MicroRNA-34a induces apoptosis in the human glioma cell line, A172, through enhanced ROS production and NOX2 expression. Biochem. Biophys. Res. Commun. 2014, 444, 6–12. [Google Scholar] [CrossRef]
- Im, Y.B.; Jee, M.K.; Choi, J.I.; Cho, H.T.; Kwon, O.H.; Kang, S.K. Molecular targeting of NOX4 for neuropathic pain after traumatic injury of the spinal cord. Cell Death Dis. 2012, 3, e426. [Google Scholar] [CrossRef]
- Xu, Y.; Zhang, J.; Fan, L.; He, X. miR-423-5p suppresses high-glucose-induced podocyte injury by targeting Nox4. Biochem. Biophys. Res. Commun. 2018, 505, 339–345. [Google Scholar] [CrossRef]
- Sun, M.; Hong, S.; Li, W.; Wang, P.; You, J.; Zhang, X.; Tang, F.; Wang, P.; Zhang, C. MIR-99a regulates ROS-mediated invasion and migration of lung adenocarcinoma cells by targeting NOX4. Oncol. Rep. 2016, 35, 2755–2766. [Google Scholar] [CrossRef]
- Wu, Q.; Zheng, B.; Weng, G.; Yang, H.; Ren, Y.; Weng, X.; Zhang, S.; Zhu, W. Downregulated NOX4 underlies a novel inhibitory role of microRNA-137 in prostate cancer. J. Cell. Biochem. 2019, 120, 10215–10227. [Google Scholar] [CrossRef]
- Li, Z.-N.; Ge, M.-X.; Yuan, Z.-F. MicroRNA-182-5p protects human lens epithelial cells against oxidative stress-induced apoptosis by inhibiting NOX4 and p38 MAPK signalling. BMC Ophthalmol. 2020, 20, 233. [Google Scholar] [CrossRef]
- Gu, C.; Draga, D.; Zhou, C.; Su, T.; Zou, C.; Gu, Q.; Lahm, T.; Zheng, Z.; Qiu, Q. MiR-590-3p inhibits pyroptosis in diabetic retinopathy by targeting NLRP1 and inactivating the NOX4 signaling pathway. Investig. Ophthalmol. Vis. Sci. 2019, 60, 4215–4223. [Google Scholar] [CrossRef]
- Fierro-Fernández, M.; Busnadiego, Ó.; Sandoval, P.; Espinosa-Díez, C.; Blanco-Ruiz, E.; Rodríguez, M.; Pian, H.; Ramos, R.; López-Cabrera, M.; García-Bermejo, M.L.; et al. miR-9-5p suppresses pro-fibrogenic transformation of fibroblasts and prevents organ fibrosis by targeting NOX 4 and TGFBR 2. EMBO Rep. 2015, 16, 1358–1377. [Google Scholar] [CrossRef]
- Gordillo, G.M.; Biswas, A.; Khanna, S.; Pan, X.; Sinha, M.; Roy, S.; Sen, C.K. Dicer knockdown inhibits Endothelial cell tumor growth via microRNA 21a-3p targeting of nox-4. J. Biol. Chem. 2014, 289, 9027–9038. [Google Scholar] [CrossRef]
- Zhao, L.; Jiang, X.; Shi, J.; Gao, S.; Zhu, Y.; Gu, T.; Shi, E. Exosomes derived from bone marrow mesenchymal stem cells overexpressing microRNA-25 protect spinal cords against transient ischemia. J. Thorac. Cardiovasc. Surg. 2019, 157, 508–517. [Google Scholar] [CrossRef]
- Varga, Z.V.; Kupai, K.; Szucs, G.; Gáspár, R.; Pálóczi, J.; Faragó, N.; Zvara, Á.; Puskás, L.G.; Rázga, Z.; Tiszlavicz, L.; et al. MicroRNA-25-dependent up-regulation of NADPH oxidase 4 (NOX4) mediates hypercholesterolemia-induced oxidative/nitrative stress and subsequent dysfunction in the heart. J. Mol. Cell Cardiol. 2013, 62, 111–121. [Google Scholar] [CrossRef]
- Li, P.; Fan, C.; Cai, Y.; Fang, S.; Zeng, Y.; Zhang, Y.; Lin, X.; Zhang, H.; Xue, Y.; Guan, M. Transplantation of brown adipose tissue up-regulates miR-99a to ameliorate liver metabolic disorders in diabetic mice by targeting NOX4. Adipocyte 2020, 9, 57–67. [Google Scholar] [CrossRef]
- Yang, J.; Brown, M.E.; Zhang, H.; Martinez, M.; Zhao, Z.; Bhutani, S.; Yin, S.; Trac, D.; Xi, J.J.; Davis, M.E. High-throughput screening identifies microRNAs that target Nox2 and improve function after acute myocardial infarction. Am. J. Physiol. Heart Circ. Physiol. 2017, 312, H1002–H1012. [Google Scholar] [CrossRef]
- Wang, H.-J.; Huang, Y.-L.; Shih, Y.-Y.; Wu, H.-Y.; Peng, C.-T.; Lo, W.-Y. MicroRNA-146a Decreases High Glucose/Thrombin-Induced Endothelial Inflammation by Inhibiting NAPDH Oxidase 4 Expression. Mediat. Inflamm. 2014, 2014. [Google Scholar] [CrossRef]
- Geng, Y.; Zhao, X.; Xu, J.; Zhang, X.; Hu, G.; Fu, S.C.; Dai, K.; Chen, X.; Patrick, Y.S.; Zhang, X. Overexpression of mechanical sensitive miR-337-3p alleviates ectopic ossification in rat tendinopathy model via targeting IRS1 and Nox4 of tendon-derived stem cells. J. Mol. Cell Biol. 2020, 12, 305–317. [Google Scholar] [CrossRef]
- Zhang, J.; Zhao, F.; Yu, X.; Lu, X.; Zheng, G. MicroRNA-155 modulates the proliferation of vascular smooth muscle cells by targeting endothelial nitric oxide synthase. Int. J. Mol. Med. 2015, 35, 1708–1714. [Google Scholar] [CrossRef]
- Sun, H.X.; Zeng, D.Y.; Li, R.T.; Pang, R.P.; Yang, H.; Hu, Y.L.; Zhang, Q.; Jiang, Y.; Huang, L.Y.; Tang, Y.B.; et al. Essential role of microRNA-155 in regulating endothelium-dependent vasorelaxation by targeting endothelial nitric oxide synthase. Hypertension 2012, 60, 1407–1414. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, X.; Li, Y.; Lai, J.; Zhang, N.; Ming, J.; Ma, X.; Ji, Q.; Xing, Y. Downregulation of microRNA-155 ameliorates high glucose-induced endothelial injury by inhibiting NF-κB activation and promoting HO-1 and NO production. Biomed. Pharmacother. 2017, 88, 1227–1234. [Google Scholar] [CrossRef]
- Wang, L.; Wu, W.; Chen, J.; Li, Y.; Xu, M.; Cai, Y. Microrna microarray-based identification of involvement of mir-155 and mir-19a in development of oral lichen planus (OLP) by modulating th1/th2 balance via targeting enos and toll-like receptor 2 (TLR2). Med. Sci. Monit. 2018, 24, 3591–3603. [Google Scholar] [CrossRef]
- Jiang, F.; Li, J.; Wu, G.; Miao, Z.; Lu, L.; Ren, G.; Wang, X. Upregulation of microRNA-335 and microRNA-584 contributes to the pathogenesis of severe preeclampsia through downregulation of endothelial nitric oxide synthase. Mol. Med. Rep. 2015, 12, 5383–5390. [Google Scholar] [CrossRef]
- Fu, Q.; Liu, X.; Liu, Y.; Yang, J.; Lv, G.; Dong, S. MicroRNA-335 and -543 suppress bone metastasis in prostate cancer via targeting endothelial nitric oxide synthase. Int. J. Mol. Med. 2015, 36, 1417–1425. [Google Scholar] [CrossRef]
- Meloni, M.; Marchetti, M.; Garner, K.; Littlejohns, B.; Sala-Newby, G.; Xenophontos, N.; Floris, I.; Suleiman, M.S.; Madeddu, P.; Caporali, A.; et al. Local inhibition of microRNA-24 improves reparative angiogenesis and left ventricle remodeling and function in mice with myocardial infarction. Mol. Ther. 2013, 21, 1390–1402. [Google Scholar] [CrossRef]
- Carlomosti, F.; D’Agostino, M.; Beji, S.; Torcinaro, A.; Rizzi, R.; Zaccagnini, G.; Maimone, B.; Di Stefano, V.; De Santa, F.; Cordisco, S.; et al. Oxidative Stress-Induced miR-200c Disrupts the Regulatory Loop among SIRT1, FOXO1, and eNOS. Antioxid. Redox Signal. 2017, 27, 328–344. [Google Scholar] [CrossRef]
- Kim, S.; Lee, K.S.; Choi, S.; Kim, J.; Lee, D.K.; Park, M.; Park, W.; Kim, T.H.; Hwang, J.Y.; Won, M.H.; et al. NF-B–responsive miRNA-31-5p elicits endothelial dysfunction associated with preeclampsia via downregulation of endothelial nitric-oxide synthase. J. Biol. Chem. 2018, 293, 18989–19000. [Google Scholar] [CrossRef]
- Qin, J.Z.; Wang, S.J.; Xia, C. microRNAs regulate nitric oxide release from endothelial cells by targeting NOS3. J. Thromb. Thrombolysis 2018, 46, 275–282. [Google Scholar] [CrossRef]
- Sun, X.; Lv, H.; Zhao, P.; He, J.; Cui, Q.; Wei, M.; Feng, S.; Zhu, Y. Commutative regulation between endothelial NO synthase and insulin receptor substrate 2 by microRNAs. J. Mol. Cell Biol. 2019, 11, 509–520. [Google Scholar] [CrossRef]
- Janaszak-Jasiecka, A.; Siekierzycka, A.; Bartoszewska, S.; Serocki, M.; Dobrucki, L.W.; Collawn, J.F.; Kalinowski, L.; Bartoszewski, R. eNOS expression and NO release during hypoxia is inhibited by miR-200b in human endothelial cells. Angiogenesis 2018, 21, 711–724. [Google Scholar] [CrossRef]
- Liu, Y.; Usa, K.; Wang, F.; Liu, P.; Geurts, A.M.; Li, J.; Williams, A.M.; Regner, K.R.; Kong, Y.; Liu, H.; et al. MicroRNA-214-3p in the kidney contributes to the development of hypertension. J. Am. Soc. Nephrol. 2018, 29, 2518–2528. [Google Scholar] [CrossRef]
- Reilly, S.N.; Liu, X.; Carnicer, R.; Recalde, A.; Muszkiewicz, A.; Jayaram, R.; Carena, C.M.; Wijesurendra, R.; Stefanini, M.; Surdo, N.C.; et al. Up-regulation of MIR-31 in human atrial fibrillation begets the arrhythmia by depleting dystrophin and neuronal nitric oxide synthase. Sci. Transl. Med. 2016, 8, 340ra74. [Google Scholar] [CrossRef]
- Guilbaud, M.; Gentil, C.; Peccate, C.; Gargaun, E.; Holtzmann, I.; Gruszczynski, C.; Falcone, S.; Mamchaoui, K.; Ben Yaou, R.; Leturcq, F.; et al. miR-708-5p and miR-34c-5p are involved in nNOS regulation in dystrophic context. Skelet. Muscle 2018, 8, 15. [Google Scholar] [CrossRef]
- Ding, J.; Tang, Y.; Tang, Z.; Zhang, X.; Wang, G. A variant in the precursor of microRNA-146a is responsible for development of erectile dysfunction in patients with chronic prostatitis via targeting NOS1. Med. Sci. Monit. 2017, 23, 929–937. [Google Scholar] [CrossRef]
- Zhang, X.; Huo, Q.; Sun, W.; Zhang, C.; Wu, Z.; Xing, B.; Li, Q. Rs2910164 in microRNA-146a confers an elevated risk of depression in patients with coronary artery disease by modulating the expression of NOS1. Mol. Med. Rep. 2018, 18, 603–609. [Google Scholar] [CrossRef]
- Guo, Z.; Shao, L.; Zheng, L.; Du, Q.; Li, P.; John, B.; Geller, D.A. miRNA-939 regulates human inducible nitric oxide synthase posttranscriptional gene expression in human hepatocytes. Proc. Natl. Acad. Sci. USA 2012, 109, 5826–5831. [Google Scholar] [CrossRef]
- Duan, Y.F.; Sun, D.L.; Chen, J.; Zhu, F.; An, Y. MicroRNA-29a/b/c targets iNOS and is involved in protective remote ischemic preconditioning in an ischemia-reperfusion rat model of non-alcoholic fatty liver disease. Oncol. Lett. 2017, 13, 1775–1782. [Google Scholar] [CrossRef]
- Rasheed, Z.; Al-Shobaili, H.A.; Rasheed, N.; Mahmood, A.; Khan, M.I. MicroRNA-26a-5p regulates the expression of inducible nitric oxide synthase via activation of NF-κB pathway in human osteoarthritis chondrocytes. Arch. Biochem. Biophys. 2016, 594, 61–67. [Google Scholar] [CrossRef]
- Zhou, W.; Yuan, T.; Gao, Y.; Yin, P.; Liu, W.; Pan, C.; Liu, Y.; Yu, X. IL-1β-induces NF-κB and upregulates microRNA-372 to inhibit spinal cord injury recovery. J. Neurophysiol. 2017, 117, 2282–2291. [Google Scholar] [CrossRef]
- Schröder, K.; Zhang, M.; Benkhoff, S.; Mieth, A.; Pliquett, R.; Kosowski, J.; Kruse, C.; Luedike, P.; Michaelis, U.R.; Weissmann, N.; et al. Nox4 Is a protective reactive oxygen species generating vascular NADPH oxidase. Circ. Res. 2012, 110, 1217–1225. [Google Scholar] [CrossRef]
- Xu, Y.; Fang, F.; Zhang, J.; Josson, S.; Clair, W.H.S.; Clair, D.K.S. Mir-17* suppresses tumorigenicity of prostate cancer by inhibiting mitochondrial antioxidant enzymes. PLoS ONE 2010, 5, e14356. [Google Scholar] [CrossRef]
- Xu, Z.; Zhang, Y.; Ding, J.; Hu, W.; Tan, C.; Wang, M.; Tang, J.; Xu, Y. miR-17-3p Downregulates Mitochondrial Antioxidant Enzymes and Enhances the Radiosensitivity of Prostate Cancer Cells. Mol. Ther. Nucleic Acids 2018, 13, 64–77. [Google Scholar] [CrossRef]
- Tian, B.; Maidana, D.E.; Dib, B.; Miller, J.B.; Bouzika, P.; Miller, J.W.; Vavvas, D.G.; Lin, H. miR-17-3p Exacerbates Oxidative Damage in Human Retinal Pigment Epithelial Cells. PLoS ONE 2016, 11, e0160887. [Google Scholar] [CrossRef]
- Long, B.; Gan, T.Y.; Zhang, R.C.; Zhang, Y.H. miR-23a regulates cardiomyocyte apoptosis by targeting manganese superoxide dismutase. Mol. Cells 2017, 40, 542–549. [Google Scholar] [CrossRef]
- Xia, S.; Zhen, Y.; Ma, H.; Wang, A. Abnormal expression of microRNA-575 leads to missed abortion through regulating apoptosis and angiogenesis. Exp. Ther. Med. 2017, 14, 3993–4000. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Wu, J.; Pan, C.; Wang, H.; Ying, X.; Zhou, Y.; Yu, H.; Zuo, Y.; Pan, Z.; Liu, R.Y.; et al. Genetic and Epigenetic Down-regulation of MicroRNA-212 Promotes Colorectal Tumor Metastasis via Dysregulation of MnSOD. Gastroenterology 2013, 145, 426.e6–436.e6. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Yu, J.; Jiang, L.; Wang, A.; Shi, F.; Ye, H.; Zhou, X. MicroRNA-222 regulates cell invasion by targeting matrix metalloproteinase 1 (MMP1) and manganese superoxide dismutase 2 (SOD2) in tongue squamous cell carcinoma cell lines. Cancer Genom. Proteom. 2009, 6, 131–139. [Google Scholar]
- Dubois-Deruy, E.; Cuvelliez, M.; Fiedler, J.; Charrier, H.; Mulder, P.; Hebbar, E.; Pfanne, A.; Beseme, O.; Chwastyniak, M.; Amouyel, P.; et al. MicroRNAs regulating superoxide dismutase 2 are new circulating biomarkers of heart failure. Sci. Rep. 2017, 7, 1–10. [Google Scholar] [CrossRef]
- Rossi, C.; Zini, R.; Rontauroli, S.; Ruberti, S.; Prudente, Z.; Barbieri, G.; Bianchi, E.; Salati, S.; Genovese, E.; Bartalucci, N.; et al. Role of TGF-β1/miR-382-5p/SOD2 axis in the induction of oxidative stress in CD34+ cells from primary myelofibrosis. Mol. Oncol. 2018, 12, 2102–2123. [Google Scholar] [CrossRef]
- Bai, X.Y.; Ma, Y.; Ding, R.; Fu, B.; Shi, S.; Chen, X.M. miR-335 and miR-34a promote renal senescence by suppressing mitochondrial antioxidative enzymes. J. Am. Soc. Nephrol. 2011, 22, 1252–1261. [Google Scholar] [CrossRef]
- Wang, Q.; Wang, Y.; Minto, A.W.; Wang, J.; Shi, Q.; Li, X.; Quigg, R.J. MicroRNA-377 is up-regulated and can lead to increased fibronectin production in diabetic nephropathy. FASEB J. 2008, 22, 4126–4135. [Google Scholar] [CrossRef]
- Zhang, X.; Ng, W.L.; Wang, P.; Tian, L.L.; Werner, E.; Wang, H.; Doetsch, P.; Wang, Y. MicroRNA-21 modulates the levels of reactive oxygen species by targeting SOD3 and TNFα. Cancer Res. 2012, 72, 4707–4713. [Google Scholar] [CrossRef]
- Papaioannou, M.D.; Lagarrigue, M.; Vejnar, C.E.; Rolland, A.D.; Kühne, F.; Aubry, F.; Schaad, O.; Fort, A.; Descombes, P.; Neerman-Arbez, M.; et al. Loss of dicer in sertoli cells has a major impact on the testicular proteome of mice. Mol. Cell. Proteom. 2011, 10. [Google Scholar] [CrossRef]
- Wang, L.; Xu, J.; Liu, H.; Li, J.; Hao, H. PM2.5 inhibits SOD1 expression by up-regulating microRNA-206 and promotes ROS accumulation and disease progression in asthmatic mice. Int. Immunopharmacol. 2019, 76, 105871. [Google Scholar] [CrossRef]
- Ji, G.; Lv, K.; Chen, H.; Wang, T.; Wang, Y.; Zhao, D.; Qu, L.; Li, Y. MiR-146a Regulates SOD2 Expression in H2O2 Stimulated PC12 Cells. PLoS ONE 2013, 8, e69351. [Google Scholar] [CrossRef]
- Haque, R.; Chun, E.; Howell, J.C.; Sengupta, T.; Chen, D.; Kim, H. MicroRNA-30b-mediated regulation of catalase expression in human ARPE-19 cells. PLoS ONE 2012, 7, e42542. [Google Scholar] [CrossRef]
- Haque, R.; Chen, D.; Sengupta, T.; Chun, E.; Howell, J.; Kim, H. Role of Mir-30b On The Regulation Of The Catalase Gene In Human ARPE-19 Cells. Invest. Ophthalmol. Vis. Sci. 2011, 52, 907. [Google Scholar]
- Xu, X.; Wells, A.; Padilla, M.T.; Kato, K.; Kim, K.C.; Lin, Y. A signaling pathway consisting of miR-551b, catalase and MUC1 contributes to acquired apoptosis resistance and chemoresistance. Carcinogenesis 2014, 35, 2457–2466. [Google Scholar] [CrossRef]
- Wang, L.; Huang, H.; Fan, Y.; Kong, B.; Hu, H.; Hu, K.; Guo, J.; Mei, Y.; Liu, W.-L. Effects of downregulation of microRNA-181a on H2O2-induced H9c2 cell apoptosis via the mitochondrial apoptotic pathway. Oxid. Med. Cell. Longev. 2014, 2014, 960362. [Google Scholar] [CrossRef]
- La Sala, L.; Cattaneo, M.; De Nigris, V.; Pujadas, G.; Testa, R.; Bonfigli, A.R.; Genovese, S.; Ceriello, A. Oscillating glucose induces microRNA-185 and impairs an efficient antioxidant response in human endothelial cells. Cardiovasc. Diabetol. 2016, 15, 71. [Google Scholar] [CrossRef]
- Dong, X.; Liu, H.; Chen, F.; Li, D.; Zhao, Y. MiR-214 Promotes the Alcohol-Induced Oxidative Stress via Down-Regulation of Glutathione Reductase and Cytochrome P450 Oxidoreductase in Liver Cells. Alcohol. Clin. Exp. Res. 2014, 38, 68–77. [Google Scholar] [CrossRef]
- Liu, Q.; Bai, W.; Huang, F.; Tang, J.; Lin, X. Downregulation of microRNA-196a inhibits stem cell self-renewal ability and stemness in non-small-cell lung cancer through upregulating GPX3 expression. Int. J. Biochem. Cell Biol. 2019, 115, 105571. [Google Scholar] [CrossRef]
- Guo, Q.J.; Mills, J.N.; Bandurraga, S.G.; Nogueira, L.M.; Mason, N.J.; Camp, E.R.; Larue, A.C.; Turner, D.P.; Findlay, V.J. MicroRNA-510 promotes cell and tumor growth by targeting peroxiredoxin1 in breast cancer. Breast Cancer Res. 2013, 15, R70. [Google Scholar] [CrossRef]
- Zhang, Z.; Dai, D.Q. MicroRNA-596 acts as a tumor suppressor in gastric cancer and is upregulated by promotor demethylation. World J. Gastroenterol. 2019, 25, 1224–1237. [Google Scholar] [CrossRef]
- Diao, S.; Zhang, J.; Wang, H.; He, M.; Lin, M.C.; Chen, Y.; Kung, H. Proteomic identification of microRNA-122a target proteins in hepatocellular carcinoma. Proteomics 2010, 10, 3723–3731. [Google Scholar] [CrossRef]
- Cortez, M.A.; Valdecanas, D.; Zhang, X.; Zhan, Y.; Bhardwaj, V.; Calin, G.A.; Komaki, R.; Giri, D.K.; Quini, C.C.; Wolfe, T.; et al. Therapeutic delivery of mir-200c enhances radiosensitivity in lung cancer. Mol. Ther. 2014, 22, 1494–1503. [Google Scholar] [CrossRef]
- Lv, Z.; Wei, J.; You, W.; Wang, R.; Shang, J.; Xiong, Y.; Yang, H.; Yang, X.; Fu, Z. Disruption of the c-Myc/miR-200b-3p/PRDX2 regulatory loop enhances tumor metastasis and chemotherapeutic resistance in colorectal cancer. J. Transl. Med. 2017, 15, 257. [Google Scholar] [CrossRef]
- Jiang, W.; Min, J.; Sui, X.; Qian, Y.; Liu, Y.; Liu, Z.; Zhou, H.; Li, X.; Gong, Y. MicroRNA-26a-5p and microRNA-23b-3p up-regulate peroxiredoxin III in acute myeloid leukemia. Leuk. Lymphoma 2015, 56, 460–471. [Google Scholar] [CrossRef]
- He, H.C.; Zhu, J.G.; Chen, X.B.; Chen, S.M.; Han, Z.D.; Dai, Q.S.; Ling, X.H.; Fu, X.; Lin, Z.Y.; Deng, Y.H.; et al. MicroRNA-23b downregulates peroxiredoxin III in human prostate cancer. FEBS Lett. 2012, 586, 2451–2458. [Google Scholar] [CrossRef]
- Li, K.K.W.; Pang, J.C.S.; Lau, K.M.; Zhou, L.; Mao, Y.; Wang, Y.; Poon, W.S.; Ng, H.K. MiR-383 is downregulated in medulloblastoma and targets peroxiredoxin 3 (PRDX3). Brain Pathol. 2013, 23, 413–425. [Google Scholar] [CrossRef]
- Li, Q.; Wang, N.; Wei, H.; Li, C.; Wu, J.; Yang, G. miR-24-3p Regulates Progression of Gastric Mucosal Lesions and Suppresses Proliferation and Invasiveness of N87 Via Peroxiredoxin 6. Dig. Dis. Sci. 2016, 61, 3486–3497. [Google Scholar] [CrossRef]
- Alemdehy, M.F.; Haanstra, J.R.; De Looper, H.W.J.; Van Strien, P.M.H.; Verhagen-Oldenampsen, J.; Caljouw, Y.; Sanders, M.A.; Hoogenboezem, R.; De Ru, A.H.; Janssen, G.M.C.; et al. ICL-induced miR139-3p and miR199a-3p have opposite roles in hematopoietic cell expansion and leukemic transformation. Blood 2015, 125, 3937–3948. [Google Scholar] [CrossRef]
- Zhang, S.; Wang, W.; Gu, Q.; Xue, J.; Cao, H.; Tang, Y.; Xu, X.; Cao, J.; Zhou, J.; Wu, J.; et al. Protein and miRNA profiling of radiation-induced skin injury in rats: The protective role of peroxiredoxin-6 against ionizing radiation. Free Radic. Biol. Med. 2014, 69, 96–107. [Google Scholar] [CrossRef]
- Sahu, N.; Stephan, J.P.; Cruz, D.D.; Merchant, M.; Haley, B.; Bourgon, R.; Classon, M.; Settleman, J. Functional screening implicates miR-371-3p and peroxiredoxin 6 in reversible tolerance to cancer drugs. Nat. Commun. 2016, 7, 12351. [Google Scholar] [CrossRef]
- Kraemer, A.; Barjaktarovic, Z.; Sarioglu, H.; Winkler, K.; Eckardt-Schupp, F.; Tapio, S.; Atkinson, M.J.; Moertl, S. Cell Survival Following Radiation Exposure Requires miR-525-3p Mediated Suppression of ARRB1 and TXN1. PLoS ONE 2013, 8, e77484. [Google Scholar] [CrossRef]
- Machitani, M.; Sakurai, F.; Wakabayashi, K.; Nakatani, K.; Tachibana, M.; Mizuguchi, H. MicroRNA miR-27 Inhibits Adenovirus Infection by Suppressing the Expression of SNAP25 and TXN2. J. Virol. 2017, 91. [Google Scholar] [CrossRef] [PubMed]
- Mercatelli, N.; Fittipaldi, S.; De Paola, E.; Dimauro, I.; Paronetto, M.P.; Jackson, M.J.; Caporossi, D. MiR-23-TrxR1 as a novel molecular axis in skeletal muscle differentiation. Sci. Rep. 2017, 7, 7219. [Google Scholar] [CrossRef]
- Hao, C.; Xu, X.; Ma, J.; Xia, J.; Dai, B.; Liu, L.; Ma, Y. MicroRNA-124 regulates the radiosensitivity of non-small cell lung cancer cells by targeting TXNRD1. Oncol. Lett. 2017, 13, 2071–2078. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Liu, H.; Wu, J.; Zhao, Y. miR-125a Suppresses TrxR1 Expression and Is Involved in H2O2-Induced Oxidative Stress in Endothelial Cells. J. Immunol. Res. 2018, 2018. [Google Scholar] [CrossRef]
- Vo, D.T.; Karanam, N.K.; Ding, L.; Saha, D.; Yordy, J.S.; Giri, U.; Heymach, J.V.; Story, M.D. miR-125a-5p Functions as Tumor Suppressor microRNA and is a Marker of Locoregional Recurrence and Poor Prognosis in Head and Neck Cancer. Neoplasia 2019, 21, 849–862. [Google Scholar] [CrossRef] [PubMed]
- Hua, S.; Quan, Y.; Zhan, M.; Liao, H.; Li, Y.; Lu, L. MiR-125b-5p inhibits cell proliferation, migration, and invasion in hepatocellular carcinoma via targeting TXNRD1. Cancer Cell Int. 2019, 19, 203. [Google Scholar] [CrossRef]
- Esposti, D.D.; Aushev, V.N.; Lee, E.; Cros, M.P.; Zhu, J.; Herceg, Z.; Chen, J.; Hernandez-Vargas, H. MiR-500a-5p regulates oxidative stress response genes in breast cancer and predicts cancer survival. Sci. Rep. 2017, 7, 1–10. [Google Scholar] [CrossRef]
- Thounaojam, M.C.; Jadeja, R.N.; Warren, M.; Powell, F.L.; Raju, R.; Gutsaeva, D.; Khurana, S.; Martin, P.M.; Bartoli, M. MicroRNA-34a (miR-34a) mediates retinal endothelial cell premature senescence through mitochondrial dysfunction and loss of antioxidant activities. Antioxidants 2019, 8, 328. [Google Scholar] [CrossRef]
- Dong, D.; Fu, N.; Yang, P. MiR-17 Downregulation by High Glucose Stabilizes Thioredoxin-Interacting Protein and Removes Thioredoxin Inhibition on ASK1 Leading to Apoptosis. Toxicol. Sci. 2016, 150, 84–96. [Google Scholar] [CrossRef]
- Li, X.F.; Shen, W.W.; Sun, Y.Y.; Li, W.X.; Sun, Z.H.; Liu, Y.H.; Zhang, L.; Huang, C.; Meng, X.M.; Li, J. MicroRNA-20a negatively regulates expression of NLRP3-inflammasome by targeting TXNIP in adjuvant-induced arthritis fibroblast-like synoviocytes. Jt. Bone Spine 2016, 83, 695–700. [Google Scholar] [CrossRef]
- Dong, F.; Dong, S.; Liang, Y.; Wang, K.; Qin, Y.; Zhao, X. MiR-20b inhibits the senescence of human umbilical vein endothelial cells through regulating the Wnt/β-catenin pathway via the TXNIP/NLRP3 axis. Int. J. Mol. Med. 2020, 45, 847–857. [Google Scholar] [CrossRef]
- Hu, L.; Zhang, H.; Wang, B.; Ao, Q.; He, Z. MicroRNA-152 attenuates neuroinflammation in intracerebral hemorrhage by inhibiting thioredoxin interacting protein (TXNIP)-mediated NLRP3 inflammasome activation. Int. Immunopharmacol. 2020, 80, 106141. [Google Scholar] [CrossRef]
- Jing, G.; Chen, J.; Xu, G.; Shalev, A. Mir-128 Targets Thioredoxin-Interacting Protein 3′UTR and Downregulates Its Expression in Pancreatic Beta Cells. Diabetes 2018, 67, 80-OR. [Google Scholar] [CrossRef]
- Zhu, H.J.; Wang, D.G.; Yan, J.; Xu, J. Up-regulation of microrna-135a protects against myocardial ischemia/reperfusion injury by decreasing txnip expression in diabetic mice. Am. J. Transl. Res. 2015, 7, 2661–2671. [Google Scholar]
- Heo, M.J.; Kim, T.H.; You, J.S.; Blaya, D.; Sancho-Bru, P.; Kim, S.G. Alcohol dysregulates miR-148a in hepatocytes through FoxO1, facilitating pyroptosis via TXNIP overexpression. Gut 2019, 68, 708–720. [Google Scholar] [CrossRef]
- Yan, G.R.; Xu, S.H.; Tan, Z.L.; Liu, L.; He, Q.Y. Global identification of miR-373-regulated genes in breast cancer by quantitative proteomics. Proteomics 2011, 11, 912–920. [Google Scholar] [CrossRef]
- Zhu, G.; Zhou, L.; Liu, H.; Shan, Y.; Zhang, X. MicroRNA-224 promotes pancreatic cancer cell proliferation and migration by targeting the TXNIP-mediated HIF1α pathway. Cell. Physiol. Biochem. 2018, 48, 1735–1746. [Google Scholar] [CrossRef] [PubMed]
- Knoll, S.; Fürst, K.; Kowtharapu, B.; Schmitz, U.; Marquardt, S.; Wolkenhauer, O.; Martin, H.; Pützer, B.M. E2F1 induces miR-224/452 expression to drive EMT through TXNIP downregulation. EMBO Rep. 2014, 15, 1315–1329. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Wang, H.; Liu, X.; Hu, Y.; Ding, L.; Zhang, X.; Sun, Q.; Li, Y. Oncogenic microRNA-411 promotes lung carcinogenesis by directly targeting suppressor genes SPRY4 and TXNIP. Oncogene 2019, 38, 1892–1904. [Google Scholar] [CrossRef]
- Simone, N.L.; Soule, B.P.; Ly, D.; Saleh, A.D.; Savage, J.E.; DeGraff, W.; Cook, J.; Harris, C.C.; Gius, D.; Mitchell, J.B. Ionizing Radiation-Induced Oxidative Stress Alters miRNA Expression. PLoS ONE 2009, 4, e6377. [Google Scholar] [CrossRef]
- Lin, H.; Qian, J.; Castillo, A.C.; Long, B.; Keyes, K.T.; Chen, G.; Ye, Y. Effect of miR-23 on Oxidant-induced injury in human retinal pigment epithelial cells. Investig. Ophthalmol. Vis. Sci. 2011, 52, 6308–6314. [Google Scholar] [CrossRef] [PubMed]
- Lv, G.; Shao, S.; Dong, H.; Bian, X.; Yang, X.; Dong, S. MicroRNA-214 protects cardiac myocytes against H2O2-induced injury. J. Cell. Biochem. 2014, 115, 93–101. [Google Scholar] [CrossRef]
- Chen, W.; Zhang, Y.N.; Jia, Q.Q.; Ji, A.; Shao, S.X.; Zhang, L.; Gong, M.; Yin, Q.; Huang, X.L. MicroRNA-214 protects L6 skeletal myoblasts against hydrogen peroxide-induced apoptosis. Free Radic. Res. 2020, 54, 162–172. [Google Scholar] [CrossRef]
- Lin, Y.; Liu, X.; Cheng, Y.; Yang, J.; Huo, Y.; Zhang, C. Involvement of MicroRNAs in hydrogen peroxide-mediated gene regulation and cellular injury response in vascular smooth muscle cells. J. Biol. Chem. 2009, 284, 7903–7913. [Google Scholar] [CrossRef]
- Lu, B.; Christensen, I.T.; Ma, L.W.; Wang, X.L.; Jiang, L.F.; Wang, C.X.; Feng, L.; Zhang, J.S.; Yan, Q.C. MiR-24-p53 pathway evoked by oxidative stress promotes lens epithelial cell apoptosis in age-related cataracts. Mol. Med. Rep. 2018, 17, 5021–5028. [Google Scholar] [CrossRef]
- Oltra, M.; Vidal-Gil, L.; Maisto, R.; Oltra, S.S.; Romero, F.J.; Sancho-Pelluz, J.; Barcia, J.M. miR302a and 122 are deregulated in small extracellular vesicles from ARPE-19 cells cultured with H2O2. Sci. Rep. 2019, 9, 17954. [Google Scholar] [CrossRef]
- Liu, N.; Shi, Y.F.; Diao, H.Y.; Li, Y.X.; Cui, Y.; Song, X.J.; Tian, X.; Li, T.Y.; Liu, B. MicroRNA-135a regulates apoptosis induced by hydrogen peroxide in rat cardiomyoblast cells. Int. J. Biol. Sci. 2017, 13, 13–21. [Google Scholar] [CrossRef]
- Yang, X.; Mao, X.; Ding, X.; Guan, F.; Jia, Y.; Luo, L.; Li, B.; Tan, H.; Cao, C. MiR-146a down-regulation alleviates H2O2-induced cytotoxicity of PC12 cells by regulating MCL1/JAK/STAT pathway. Cell Biol. Toxicol. 2018, 34, 479–489. [Google Scholar] [CrossRef]
- Zhao, L.; Ouyang, Y.; Bai, Y.; Gong, J.; Liao, H. miR-155-5p inhibits the viability of vascular smooth muscle cell via targeting FOS and ZIC3 to promote aneurysm formation. Eur. J. Pharmacol. 2019, 853, 145–152. [Google Scholar] [CrossRef]
- Xiao, Y.; Yan, W.; Lu, L.; Wang, Y.; Lu, W.; Cao, Y.; Cai, W. p38/p53/miR-200a-3p feedback loop promotes oxidative stress-mediated liver cell death. Cell Cycle 2015, 14, 1548–1558. [Google Scholar] [CrossRef]
- Xie, J.; Hong, E.; Ding, B.; Jiang, W.; Zheng, S.; Xie, Z.; Tian, D.; Chen, Y. Inhibition of NOX4/ROS Suppresses Neuronal and Blood-Brain Barrier Injury by Attenuating Oxidative Stress After Intracerebral Hemorrhage. Front. Cell. Neurosci. 2020, 14, 578060. [Google Scholar] [CrossRef]
- Michaeloudes, C.; Sukkar, M.B.; Khorasani, N.M.; Bhavsar, P.K.; Chung, K.F. TGF-β regulates Nox4, MnSOD and catalase expression, and IL-6 release in airway smooth muscle cells. Am. J. Physiol. Lung Cell. Mol. Physiol. 2011, 300, L295. [Google Scholar] [CrossRef]
- Bendall, J.K.; Rinze, R.; Adlam, D.; Tatham, A.L.; De Bono, J.; Channon, K.M. Endothelial Nox2 overexpression potentiates vascular oxidative stress and hemodynamic response to angiotensin II: Studies in endothelial-targeted Nox2 transgenic mice. Circ. Res. 2007, 100, 1016–1025. [Google Scholar] [CrossRef]
- Gregg, J.L.; Ii, R.M.T.; Chang, G.; Joshi, D.; Zhan, Y.; Chen, L.; Maranchie, J.K. Molecular and Cellular Pathobiology NADPH Oxidase NOX4 Supports Renal Tumorigenesis by Promoting the Expression and Nuclear Accumulation of HIF2a. Cancer Res. 2014, 74, 3501–3511. [Google Scholar] [CrossRef]
- Jeong, B.Y.; Lee, H.Y.; Park, C.G.; Kang, J.; Yu, S.-L.; Choi, D.; Han, S.-Y.; Park, M.H.; Cho, S.; Lee, S.Y.; et al. Oxidative stress caused by activation of NADPH oxidase 4 promotes contrast-induced acute kidney injury. PLoS ONE 2018, 13, e0191034. [Google Scholar] [CrossRef]
- Kumari, S.; Badana, A.K.; Murali Mohan, G.; Shailender, G.; Malla, R.R. Reactive Oxygen Species: A Key Constituent in Cancer Survival. Biomark. Insights 2018, 13, 1177271918755391. [Google Scholar] [CrossRef]
- Piskounova, E.; Agathocleous, M.; Murphy, M.M.; Hu, Z.; Huddlestun, S.E.; Zhao, Z.; Leitch, A.M.; Johnson, T.M.; DeBerardinis, R.J.; Morrison, S.J. Oxidative stress inhibits distant metastasis by human melanoma cells. Nature 2015, 527, 186–191. [Google Scholar] [CrossRef]
- Sayin, V.I.; Ibrahim, M.X.; Larsson, E.; Nilsson, J.A.; Lindahl, P.; Bergo, M.O. Cancer: Antioxidants accelerate lung cancer progression in mice. Sci. Transl. Med. 2014, 6, 221ra15. [Google Scholar] [CrossRef]
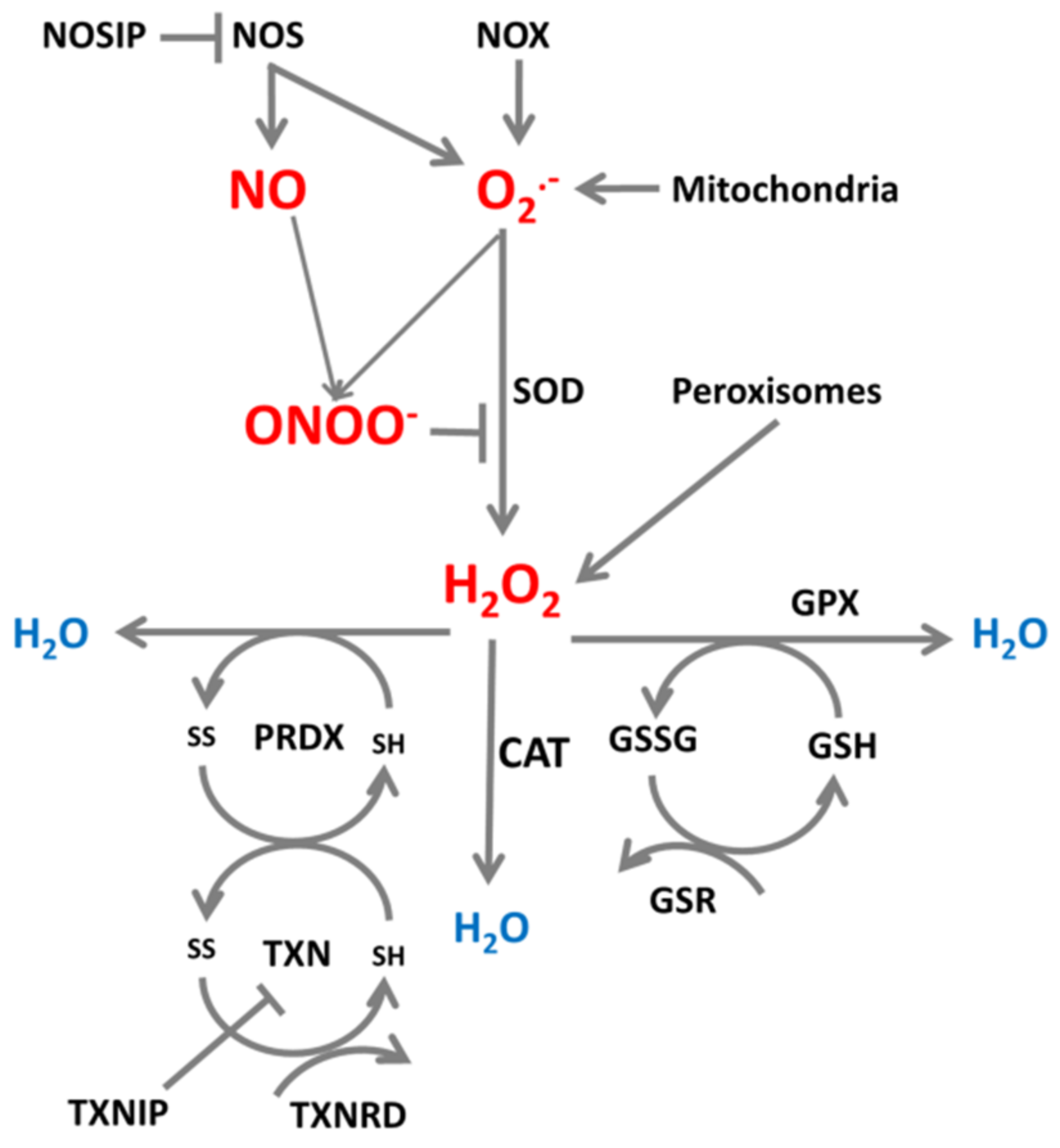
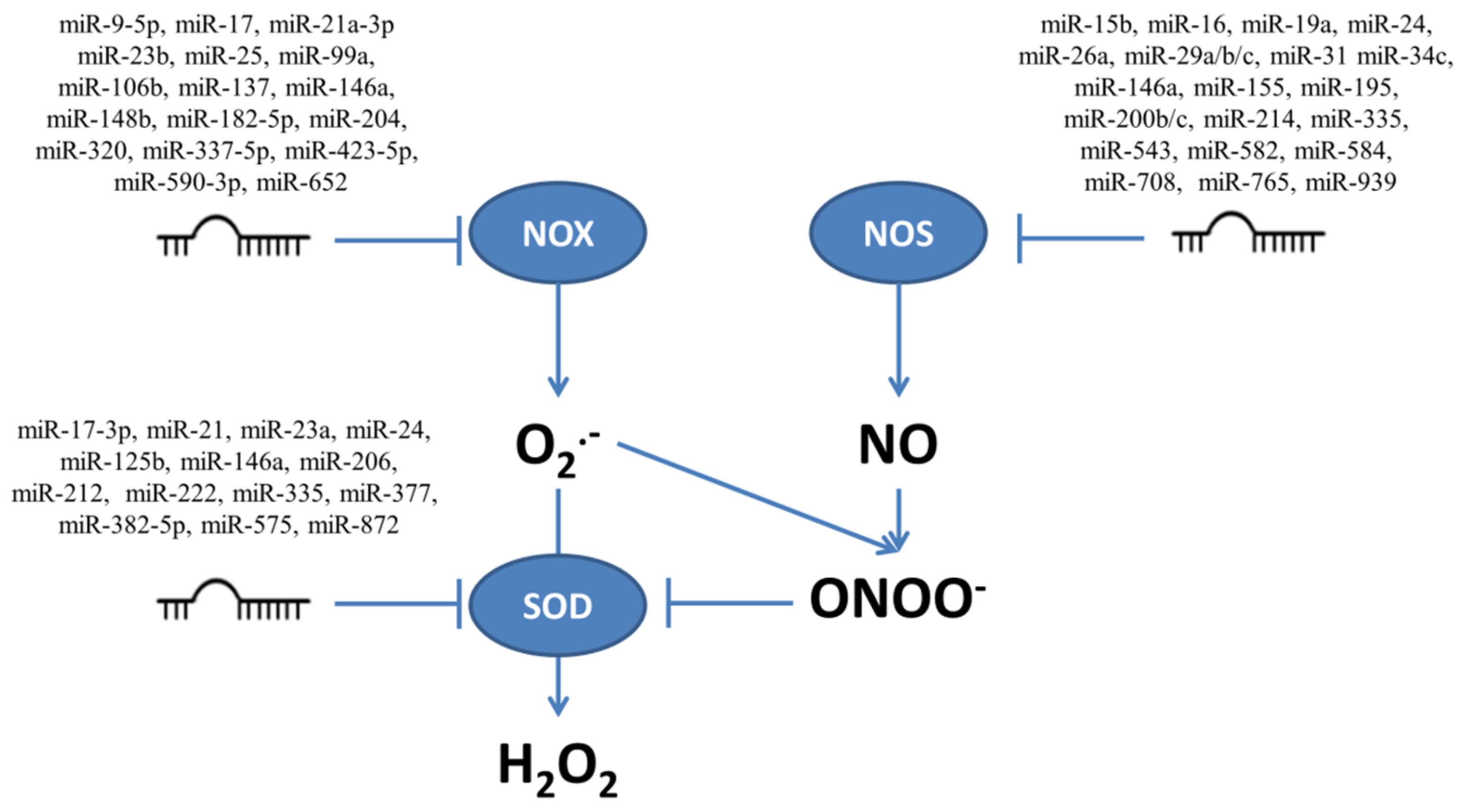
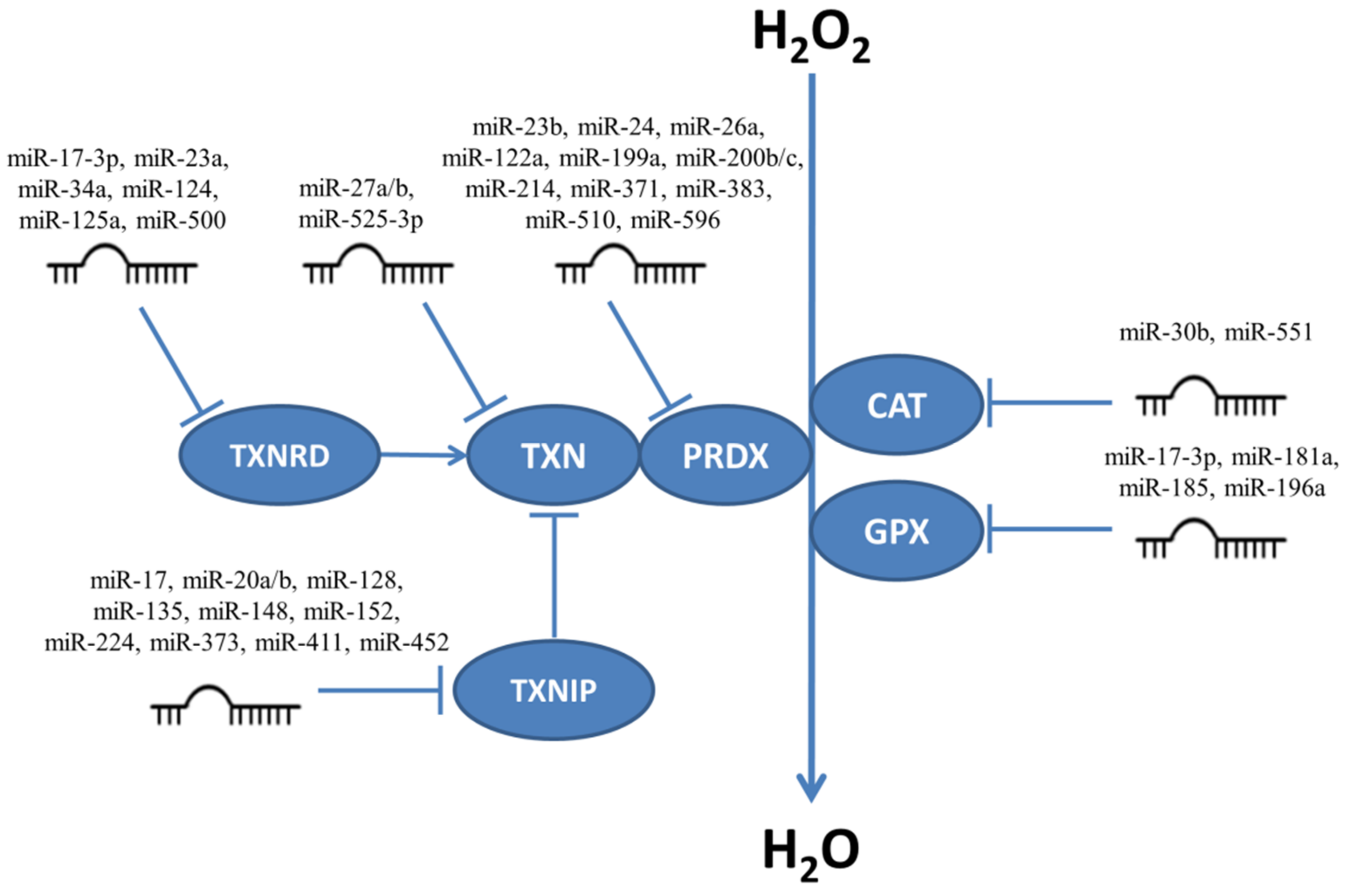

| Targeted Transcript and miRNAs | Cellular Effect | Cell Types | References |
|---|---|---|---|
| NOX2 ┣ miR-320, miR-652 | Pro-survival | Cerebral neurons, brain tissues of rats | [59,60] |
| NOX4 ┣ mir-23b, miR-182-5p, miR-423-5p, miR-590-3p | Pro-survival | Mouse spinal cord, human lens epithelial cells, mouse podocytes, human retinal microvascular endothelial cells | [62,63,66,67] |
| NOX4 ┣ miR-99a, miR-137 | Pro-apoptotic | Lung adenocarcinoma, prostate cancer | [64,65] |
| NOS3 ┣ miR-24, miR-155 | Pro-survival | VSMCs, cardiomyocytes, cardiac fibroblasts | [76,82,84] |
| NOS3 ┣ miR-24, miR-31-5p, miR-155, miR-200c, miR-335, miR-543, miR-584 | Pro-apoptotic | HUVECs, prostate cancer, trophoblasts, human microvascular endothelial cells | [77,78,80,81,82,83,84] |
| NOSIP ┣ miR-372 | Pro-apoptotic | Human neural stem cells | [96] |
| Targeted Transcript and miRNAs | Cellular Effect | Cell Types | References |
|---|---|---|---|
| SOD2 ┣ miR-17-3p, miR-23a, mir-146a, miR-212, miR-575 | Pro-apoptotic | Rat PC12 adrenal gland cells, villi cells, prostate cancer, ARPE-19 cells, cardiomyocytes, colorectal cancer | [98,99,100,101,102,103,112] |
| SOD2 ┣ miR-222, miR-382-5p | Pro-survival | Oral tongue squamous cell carcinoma, primary myelofibrosis CD34+ cells | [104,106] |
| SOD3┣ miR-21 | Pro-survival | Human bronchial epithelial cells | [109] |
| Targeted Transcript and Increased miRNAs | Cellular Effect | Cell Types | Reference |
|---|---|---|---|
| GPX1 ┣ miR-181a | Pro-apoptotic | Cardiomiocytes | [116] |
| GPX2 ┣ miR-17-3p | Pro-apoptotic | Prostate cancer | [98,99] |
| GPX3 ┣ miR-196a | Pro-survival | NSCLC | [119] |
| PRDX1 ┣ miR-510 | Pro-survival | Breast cancer | [120] |
| PRDX1 ┣ miR-596 | Pro-apoptotic | Gastric cancer | [121] |
| PRDX2 ┣ miR-122a, miR-200b, miR-200c | Pro-apoptotic | Hepatocellular carcinoma, colorectal cancer, lung cancer | [122,123,124] |
| PRDX3 ┣ miR-383 | Pro-apoptotic | Medulloblastoma | [127] |
| PRDX6 ┣ miR-199a-3p | Pro-survival | Leukemia | [129] |
| PRDX6 ┣ miR-24-3p, miR-371 | Pro-apoptotic | Gastric cancer PC9 xenograft mouse model | [128,131] |
| TXN1 ┣ miR-525-3p | Pro-survival | EA, HeLa, RPE, U2-OS | [132] |
| TXNRD1 ┣ miR-23a/b, miR124, miR-125a/b-5p | Pro-apoptotic | Skeletal muscle, lung cancer, head and neck cancer, hepatocellular carcinoma, human pigment epithelial cells, prostate cancer | [98,99,100,134,135,137,138] |
| TXNRD2┣ miR-17-3p | Pro-apoptotic | Mesangial cells | [107] |
| TXNIP ┣ miR-20b, miR-135a, miR-152, miR-224, miR-224/452, miR-373, miR-411-5p | Pro-survival | HUVECs, microglial BV2 cells, pancreatic cancer, melanoma, breast cancer, NSCLC | [143,144,146,148,149,150,151] |
| ** TXNIP ┣ miR-17 miR-148a | Pro-apoptotic | Myocardial cells of diabetic mice, hepatocytes | [141,147] |
| ** miR-17 and miR-148a levels were decreased, and thus a pro-apoptotic effect is expected. | |||
| miRNA and Targeted Transcripts | H2O2 Effect on miRNA Level | H2O2 Dose | Cell Type | Ref. |
|---|---|---|---|---|
| miR-17-3p ┫ SOD2, GPX2, TXNRD2 | Up Down | 0–100 µM 200 µM | ARPE-19 cells | [100] |
| miR-21 ┫ SOD3 | Up | 0–200 µM | VSMCs | [156] |
| miR-23a ┫ SOD2, TXNRD1 | Up Down | 100–200 µM 300–500 µM | ARPE-19 cells | [153] |
| miR-24 ┫ NOS3, SOD1, PRDX6 | Up | 400 µM | Human lens epithelial cells | [157] |
| miR-30b ┫ NOS3, CAT | Up | 200 µM | ARPE-19 cells | [113] |
| miR-122 ┫ PRDX2 | Down | 600 µM | ARPE-19 cells | [158] |
| miR-135a ┫ TXNIP | Up | 0–1 mM | Rat cardiomyoblasts | [159] |
| miR-146a ┫ NOX4, NOS1, SOD2 | Up | 0–200 µM | Rat PC12 cell from adrenal gland | [112,160] |
| miR-155 ┫ NOS3 | Up | 0–500 µM | VSMCs | [161] |
| miR-181a ┫ GPX1 | Down Up | 0–100 µM 200–600 µM | Rat cardiomyocytes H9c2 | [116] |
| miR-200c ┫ NOS3, PRDX2 | Up | 400 µM | Normal human liver LO2 cells | [162] |
| Up | 200 µM | HUVECs | [83] | |
| miR-214 ┫ NOS3, PRDX6, GSR | Up | 0–100 µM/L | Cardiomyocytes | [154] |
| Up | 0–600 µM/L | Skeletal myoblasts | [155] | |
| miR-500a ┫ TXNRD1 | Up | 0–10 µM | Breast cancer MCF-7 cells | [139] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ciesielska, S.; Slezak-Prochazka, I.; Bil, P.; Rzeszowska-Wolny, J. Micro RNAs in Regulation of Cellular Redox Homeostasis. Int. J. Mol. Sci. 2021, 22, 6022. https://doi.org/10.3390/ijms22116022
Ciesielska S, Slezak-Prochazka I, Bil P, Rzeszowska-Wolny J. Micro RNAs in Regulation of Cellular Redox Homeostasis. International Journal of Molecular Sciences. 2021; 22(11):6022. https://doi.org/10.3390/ijms22116022
Chicago/Turabian StyleCiesielska, Sylwia, Izabella Slezak-Prochazka, Patryk Bil, and Joanna Rzeszowska-Wolny. 2021. "Micro RNAs in Regulation of Cellular Redox Homeostasis" International Journal of Molecular Sciences 22, no. 11: 6022. https://doi.org/10.3390/ijms22116022
APA StyleCiesielska, S., Slezak-Prochazka, I., Bil, P., & Rzeszowska-Wolny, J. (2021). Micro RNAs in Regulation of Cellular Redox Homeostasis. International Journal of Molecular Sciences, 22(11), 6022. https://doi.org/10.3390/ijms22116022






