Proangiogenic Effect of 2A-Peptide Based Multicistronic Recombinant Constructs Encoding VEGF and FGF2 Growth Factors
Abstract
1. Introduction
2. Materials and Methods
2.1. Recombinant Constructs, Isolation, and Restriction Analysis of Plasmid DNA
2.2. Genetic Modification of the Cells with Recombinant Plasmids
2.3. Quantitative Analysis of mRNA Expression
2.4. Immunofluorescent Assays for Transfected HEK293T Cells
2.5. Enzyme-Linked Immunosorbent Assay (ELISA) of Target Genes
2.6. Multiplex Analysis
2.7. Tube Formation Assay Using Matrigel Matrix
2.8. Statistical Analysis
3. Results
3.1. Characterization of Multigenic Constructs Containing Picornavirus 2A-Peptide Sequences
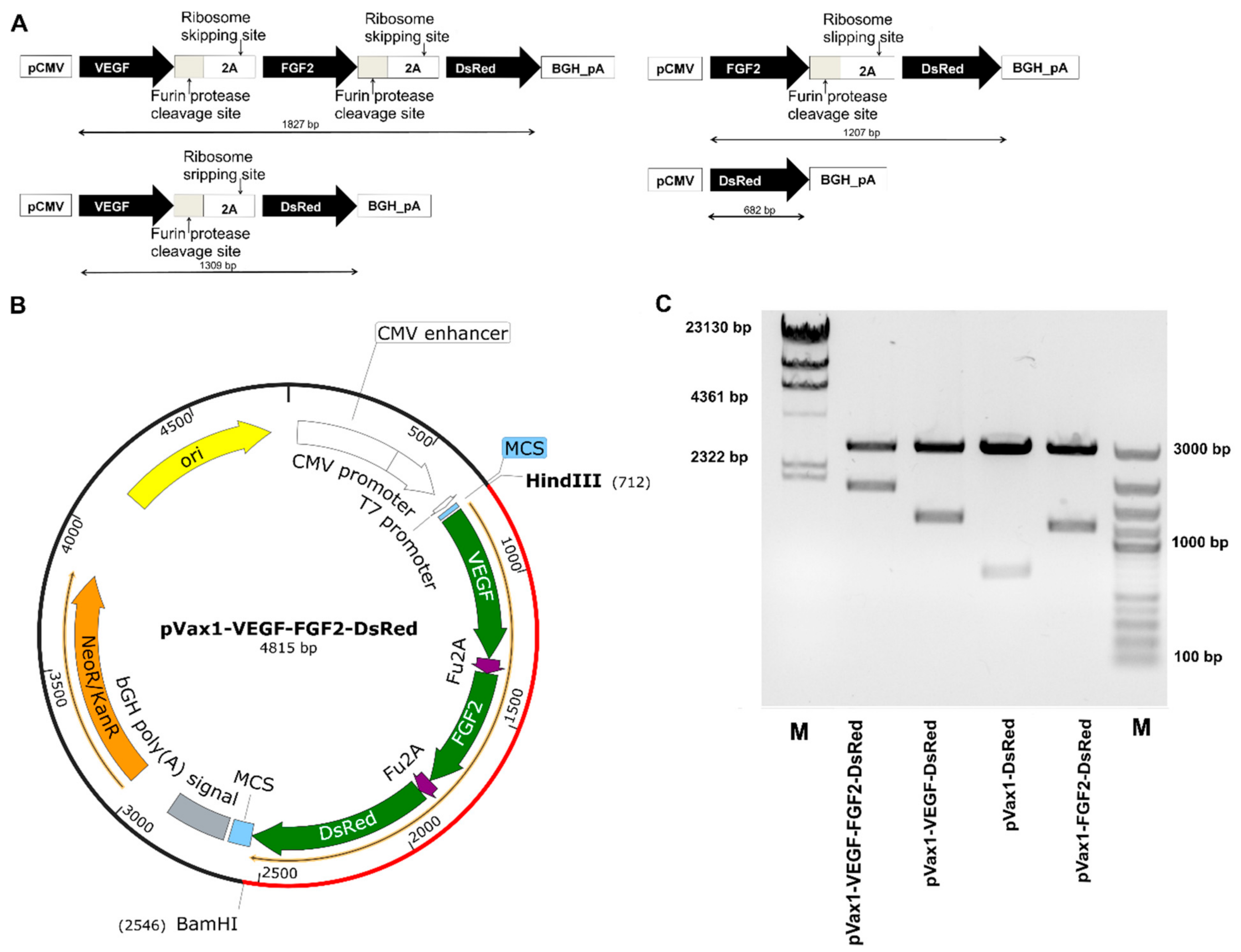
3.2. VEGF, FGF2 and DsRed Expression in Genetically Modified Cells In Vitro
3.3. Production of VEGF and FGF2 by Genetically Modified Cells
3.4. Cytokine Profile Study of Genetically Modified Cells
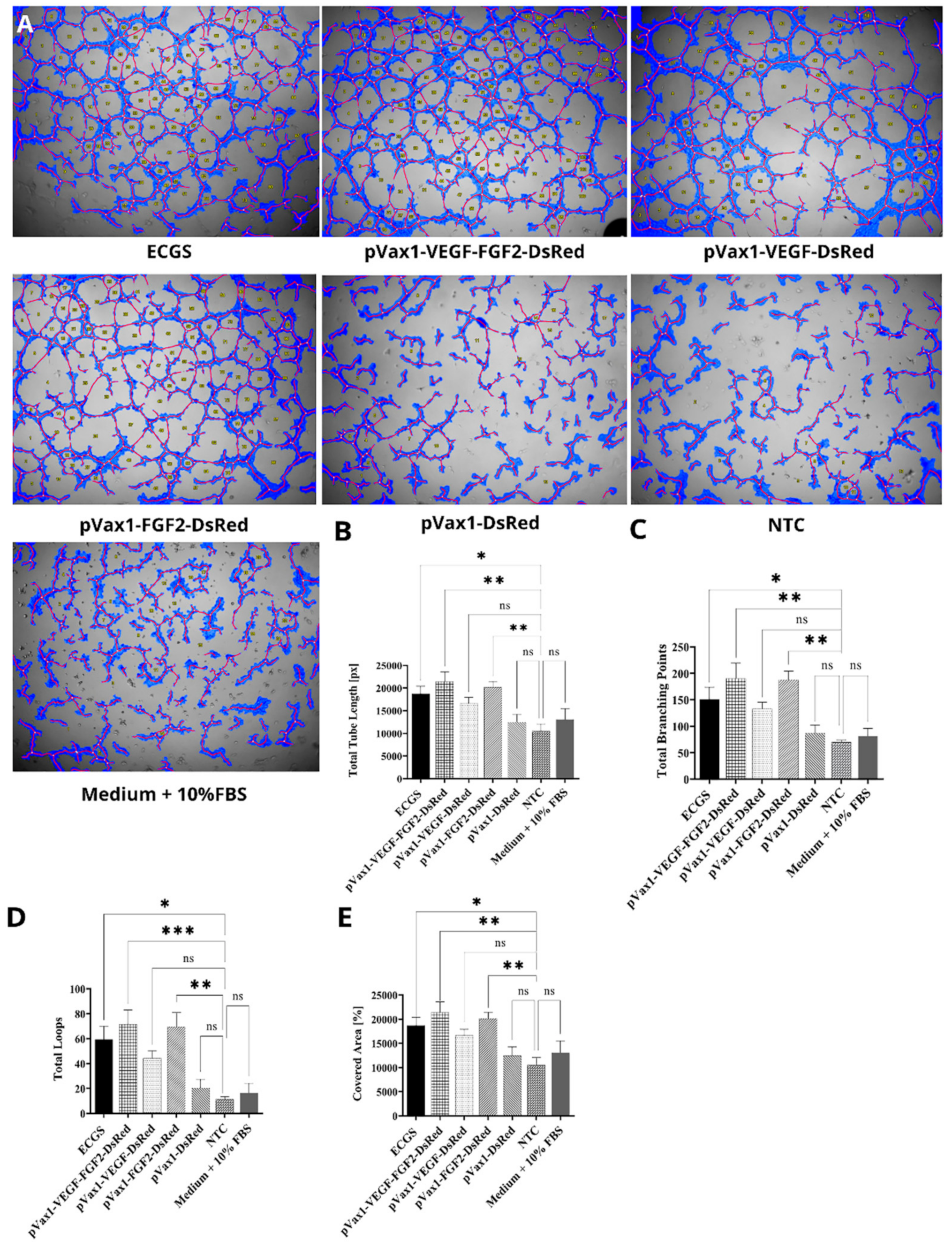
3.5. Effect of Increased VEGF and FGF2 Expression on Capillary-Like Structures Formation by HUVEC In Vitro
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- The Top 10 Causes of Death. 2020. Available online: www.who.int/news-room/fact-sheets/detail/the-top-10-causes-of-death (accessed on 23 April 2021).
- Gianni-Barrera, R.; Di Maggio, N.; Melly, L.; Burger, M.G.; Mujagic, E.; Gürke, L.; Schaefer, D.J.; Banfi, A. Therapeutic vascularization in regenerative medicine. Stem Cells Transl. Med. 2020, 9, 433–444. [Google Scholar] [CrossRef] [PubMed]
- Bian, X.; Ma, K.; Zhang, C.; Fu, X. Therapeutic angiogenesis using stem cell-derived extracellular vesicles: An emerging approach for treatment of ischemic diseases. Stem Cell Res. Ther. 2019, 10, 158. [Google Scholar] [CrossRef] [PubMed]
- Makarevich, P.I.; Parfyonova, Y.V. Therapeutic Angiogenesis: Foundations and Practical Application. In Physiologic and Pathologic Angiogenesis: Signaling Mechanisms and Targeted Therapy; Simionescu, D., Simionescu, A., Eds.; InTech: Rijeka, Croatia, 2017; ISBN 978-953-51-3023-9. [Google Scholar]
- Ferrara, N.; Carver-Moore, K.; Chen, H.; Dowd, M.; Lu, L.; O’Shea, K.S.; Powell-Braxton, L.; Hillan, K.J.; Moore, M.W. Heterozygous embryonic lethality induced by targeted inactivation of the VEGF gene. Nat. Cell Biol. 1996, 380, 439–442. [Google Scholar] [CrossRef] [PubMed]
- Giacca, M.; Zacchigna, S. VEGF gene therapy: Therapeutic angiogenesis in the clinic and beyond. Gene Ther. 2012, 19, 622–629. [Google Scholar] [CrossRef]
- Uccelli, A.; Wolff, T.; Valente, P.; Di Maggio, N.; Pellegrino, M.; Gürke, L.; Banfi, A.; Gianni-Barrera, R. Vascular endothelial growth factor biology for regenerative angiogenesis. Swiss Med. Wkly. 2019, 149, 20011. [Google Scholar] [CrossRef]
- Sanada, F.; Taniyama, Y.; Muratsu, J.; Otsu, R.; Shimizu, H.; Rakugi, H.; Morishita, R. Gene-Therapeutic Strategies Targeting Angiogenesis in Peripheral Artery Disease. Medicines 2018, 5, 31. [Google Scholar] [CrossRef]
- Kim, J.; Mirando, A.C.; Popel, A.; Green, J.J. Gene delivery nanoparticles to modulate angiogenesis. Adv. Drug Deliv. Rev. 2017, 119, 20–43. [Google Scholar] [CrossRef]
- Jazwa, A.; Florczyk, U.; Grochot-Przeczek, A.; Krist, B.; Loboda, A.; Jozkowicz, A.; Dulak, J. Limb ischemia and vessel regeneration: Is there a role for VEGF? Vasc. Pharmacol. 2016, 86, 18–30. [Google Scholar] [CrossRef]
- Gerber, H.-P.; McMurtrey, A.; Kowalski, J.; Yan, M.; Keyt, B.A.; Dixit, V.; Ferrara, N. Vascular Endothelial Growth Factor Regulates Endothelial Cell Survival through the Phosphatidylinositol 3′-Kinase/Akt Signal Transduction Pathway. J. Biol. Chem. 1998, 273, 30336–30343. [Google Scholar] [CrossRef]
- Wu, J.-B.; Tang, Y.-L.; Liang, X.-H. Targeting VEGF pathway to normalize the vasculature: An emerging insight in cancer therapy. OncoTargets Ther. 2018, 11, 6901–6909. [Google Scholar] [CrossRef]
- Gerber, H.-P.; Dixit, V.; Ferrara, N. Vascular Endothelial Growth Factor Induces Expression of the Antiapoptotic Proteins Bcl-2 and A1 in Vascular Endothelial Cells. J. Biol. Chem. 1998, 273, 13313–13316. [Google Scholar] [CrossRef]
- Carmeliet, P.; Ferreira, V.; Breier, G.; Pollefeyt, S.; Kieckens, L.; Gertsenstein, M.; Fahrig, M.; Vandenhoeck, A.; Harpal, K.; Eberhardt, C.; et al. Abnormal blood vessel development and lethality in embryos lacking a single VEGF allele. Nat. Cell Biol. 1996, 380, 435–439. [Google Scholar] [CrossRef]
- Riccardi, C.; Napolitano, E.; Platella, C.; Musumeci, D.; Melone, M.A.B.; Montesarchio, D. Anti-VEGF DNA-based aptamers in cancer therapeutics and diagnostics. Med. Res. Rev. 2021, 41, 464–506. [Google Scholar] [CrossRef]
- Gan, L.-M.; Lagerström-Fermér, M.; Carlsson, L.G.; Arfvidsson, C.; Egnell, A.-C.; Rudvik, A.; Kjaer, M.; Collén, A.; Thompson, J.D.; Joyal, J.; et al. Intradermal delivery of modified mRNA encoding VEGF-A in patients with type 2 diabetes. Nat. Commun. 2019, 10, 871. [Google Scholar] [CrossRef]
- Demidova, O.A.; Bokeria, L.A.; Bokeria, O.L.; Arakelyan, V.S.; Deev, R.V. Neovasculgen” in the treatment of patients with chronic ischemia of the lower limbs, clinical study. Byulleten’ NTSSSKH im. A.N. Bakuleva RAMN. Serdechno Sosud. Zabol. 2017, 18, 210. [Google Scholar]
- Shvalb, P.; Gavrilenko, A.; Kalinin, R.; Chervyakov, Y.; Voronov, D.; Staroverov, I.; Gryaznov, S.; Mzhavanadze, N.; Nersesyan, E.; Kiselev, S.; et al. Efficacy and safety of application Neovasculgen in the complex treatment patients with chronic lower limb ischemia (IIb-III phase of clinical trials). Cell. Transplant. Tissue Eng. 2011, 6, 76–83. [Google Scholar]
- Kattoor, A.J.; Mathur, P.; Mehta, J.L. Trials of Angiogenesis Therapy in Patients with Ischemic Heart Disease. In Biochemical Basis and Therapeutic Implications of Angiogenesis; Springer Science and Business Media LLC: Berlin/Heidelberg, Germany, 2017; pp. 393–421. [Google Scholar]
- Ribatti, D.; Presta, M. The role of fibroblast growth factor-2 in the vascularization of the chick embryo chorioallantoic membrane. J. Cell. Mol. Med. 2002, 6, 439–446. [Google Scholar] [CrossRef]
- Trafermin—Kaken Pharmaceutical. 2001. Available online: https://adisinsight.springer.com/drugs/800009962 (accessed on 23 April 2021).
- Nie, K.; Li, P.; Zeng, X.; Sun, G.; Jin, W.; Wei, Z.; Wang, B.; Qi, J.; Wang, Y.; Wang, D. Clinical observation of basic fibroblast growth factor combined with topical oxygen therapy in enhancing burn wound healing. Chin. J. Rep. Rec. Surg. 2010, 24, 643–646. (In Chinese) [Google Scholar]
- Walicke, P.A. Basic and acidic fibroblast growth factors have trophic effects on neurons from multiple CNS regions. J. Neurosci. 1988, 8, 2618–2627. [Google Scholar] [CrossRef]
- Del Corral, R.D.; Morales, A.V. The Multiple Roles of FGF Signaling in the Developing Spinal Cord. Front. Cell Dev. Biol. 2017, 5, 58. [Google Scholar] [CrossRef]
- Bikfalvi, A.; Klein, S.; Pintucci, G.; Rifkin, D.B. Biological Roles of Fibroblast Growth Factor-2. Endocr. Rev. 1997, 18, 26–45. [Google Scholar] [CrossRef] [PubMed]
- Kano, M.R.; Morishita, Y.; Iwata, C.; Iwasaka, S.; Watabe, T.; Ouchi, Y.; Miyazono, K.; Miyazawa, K. VEGF-A and FGF-2 synergistically promote neoangiogenesis through enhancement of endogenous PDGF-B–PDGFRβ signaling. J. Cell Sci. 2005, 118, 3759–3768. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-S.; Kim, J.-M.; Kim, K.L.; Jang, H.-S.; Shin, I.-S.; Jeon, E.-S.; Suh, W.; Byun, J.; Kim, D.-K. Combined administration of naked DNA vectors encoding VEGF and bFGF enhances tissue perfusion and arteriogenesis in ischemic hindlimb. Biochem. Biophys. Res. Commun. 2007, 360, 752–758. [Google Scholar] [CrossRef] [PubMed]
- Spanholtz, T.A.; Theodorou, P.; Holzbach, T.; Wutzler, S.; Giunta, R.E.; Machens, H.-G. Vascular Endothelial Growth Factor (VEGF165) Plus Basic Fibroblast Growth Factor (bFGF) Producing Cells induce a Mature and Stable Vascular Network—a Future Therapy for Ischemically Challenged Tissue. J. Surg. Res. 2011, 171, 329–338. [Google Scholar] [CrossRef]
- Tille, J.C.; Wood, J.; Mandriota, S.J.; Schnell, C.; Ferrari, S.; Mestan, J.; Zhu, Z.; Witte, L.; Pepper, M.S. Vascular endothelial growth factor (VEGF) receptor-2 antagonists inhibit VEGF- and basic fibroblast growth factor-induced angiogenesis in vivo and in vitro. J. Pharmacol. Exp. Ther. 2001, 299, 1073–1085. [Google Scholar]
- Bouïs, D.; Kusumanto, Y.; Meijer, C.; Mulder, N.H.; Hospers, G.A. A review on pro- and anti-angiogenic factors as targets of clinical intervention. Pharmacol. Res. 2006, 53, 89–103. [Google Scholar] [CrossRef]
- Fujita, M.; Ishihara, M.; Simizu, M.; Obara, K.; Ishizuka, T.; Saito, Y.; Yura, H.; Morimoto, Y.; Takase, B.; Matsui, T.; et al. Vascularization in vivo caused by the controlled release of fibroblast growth factor-2 from an injectable chitosan/non-anticoagulant heparin hydrogel. Biomaterials 2004, 25, 699–706. [Google Scholar] [CrossRef]
- Cross, M.J.; Claesson-Welsh, L. FGF and VEGF function in angiogenesis: Signalling pathways, biological responses and therapeutic inhibition. Trends Pharmacol. Sci. 2001, 22, 201–207. [Google Scholar] [CrossRef]
- Carano, R.A.; Filvaroff, E.H. Angiogenesis and bone repair. Drug Discov. Today 2003, 8, 980–989. [Google Scholar] [CrossRef]
- Boilly, B.; Vercoutter-Edouart, A.-S.; Hondermarck, H.; Nurcombe, V.; Le Bourhis, X. FGF signals for cell proliferation and migration through different pathways. Cytokine Growth Factor Rev. 2000, 11, 295–302. [Google Scholar] [CrossRef]
- Martino, M.M.; Ebrkic, S.; Ebovo, E.; Eburger, M.; Schaefer, D.J.; Ewolff, T.; Gãrke, L.; Briquez, P.S.; Larsson, H.M.; Barrera, R.E.; et al. Extracellular Matrix and Growth Factor Engineering for Controlled Angiogenesis in Regenerative Medicine. Front. Bioeng. Biotechnol. 2015, 3, 45. [Google Scholar] [CrossRef]
- Springer, M.L.; Chen, A.S.; Kraft, P.E.; Bednarski, M.; Blau, H.M. VEGF Gene Delivery to Muscle. Mol. Cell 1998, 2, 549–558. [Google Scholar] [CrossRef]
- Lukashev, A.N.; Zamyatnin, A.A. Viral vectors for gene therapy: Current state and clinical perspectives. Biochemistry 2016, 81, 700–708. [Google Scholar] [CrossRef]
- Merten, O.-W.; Gaillet, B. Viral vectors for gene therapy and gene modification approaches. Biochem. Eng. J. 2016, 108, 98–115. [Google Scholar] [CrossRef]
- Hardee, C.L.; Arévalo-Soliz, L.M.; Hornstein, B.D.; Zechiedrich, L. Advances in non-viral DNA vectors for gene therapy. Genes 2017, 8, 65. [Google Scholar] [CrossRef]
- Giacca, M.; Zacchigna, S. Virus-mediated gene delivery for human gene therapy. J. Control. Release 2012, 161, 377–388. [Google Scholar] [CrossRef]
- Lundstrom, K. Gene Therapy Today and Tomorrow. Diseases 2019, 7, 37. [Google Scholar] [CrossRef]
- Shaimardanova, A.A.; Chulpanova, D.S.; Kitaeva, K.V.; Abdrakhmanova, I.I.; Chernov, V.M.; Rutland, C.S.; Rizvanov, A.A.; Solovyeva, V.V. Production and Application of Multicistronic Constructs for Various Human Disease Therapies. Pharmaceutics 2019, 11, 580. [Google Scholar] [CrossRef]
- Solovyeva, V.V.; Chulpanova, D.S.; Tazetdinova, L.G.; Salafutdinov, I.I.; Bozo, I.Y.; Isaev, A.; Deev, R.V.; Rizvanov, A.A. In Vitro Angiogenic Properties of Plasmid DNA Encoding SDF-1α and VEGF165 Genes. Appl. Biochem. Biotechnol. 2019, 190, 773–788. [Google Scholar] [CrossRef]
- Salafutdinov, I.I.; Gazizov, I.M.; Gatina, D.K.; Mullin, R.I.; Bogov, A.A.; Islamov, R.R.; Kiassov, A.P.; Masgutov, R.F.; Rizvanov, A.A. Influence of Recombinant Codon-Optimized Plasmid DNA Encoding VEGF and FGF2 on Co-Induction of Angiogenesis. Cells 2021, 10, 432. [Google Scholar] [CrossRef]
- Yu, X.; Zhan, X.; D’Costa, J.; Tanavde, V.M.; Ye, Z.; Peng, T.; Malehorn, M.T.; Yang, X.; Civin, C.I.; Cheng, L. Lentiviral vectors with two independent internal promoters transfer high-level expression of multiple transgenes to human hematopoietic stem-progenitor cells. Mol. Ther. 2003, 7, 827–838. [Google Scholar] [CrossRef]
- Berger, A.; Maire, S.; Gaillard, M.-C.; Sahel, J.-A.; Hantraye, P.; Bemelmans, A.-P. mRNAtrans-splicing in gene therapy for genetic diseases. Wiley Interdiscip. Rev. RNA 2016, 7, 487–498. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Wang, Y.; Liu, M.; Lan, X. Multimodality reporter gene imaging: Construction strategies and application. Theranostics 2018, 8, 2954–2973. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Kim, J.-A.; Kim, H.-D.; Chung, S.; Kim, K.; Choe, H.K. Real-Time Temporal Dynamics of Bicistronic Expression Mediated by Internal Ribosome Entry Site and 2A Cleaving Sequence. Mol. Cells 2019, 42, 418–425. [Google Scholar] [CrossRef]
- Hadpech, S.; Jinathep, W.; Saoin, S.; Thongkum, W.; Chupradit, K.; Yasamut, U.; Moonmuang, S.; Tayapiwatana, C. Impairment of a membrane-targeting protein translated from a downstream gene of a “self-cleaving” T2A peptide conjunction. Protein Expr. Purif. 2018, 150, 17–25. [Google Scholar] [CrossRef]
- Al-Allaf, F.A.; Abduljaleel, Z.; Athar, M.; Taher, M.M.; Khan, W.; Mehmet, H.; Colakogullari, M.; Apostolidou, S.; Bigger, B.; Waddington, S.; et al. Modifying inter-cistronic sequence significantly enhances IRES dependent second gene expression in bicistronic vector: Construction of optimised cassette for gene therapy of familial hypercholesterolemia. Non Coding RNA Res. 2019, 4, 1–14. [Google Scholar] [CrossRef]
- Kim, J.H.; Lee, S.-R.; Li, L.-H.; Park, H.-J.; Park, J.-H.; Lee, K.Y.; Kim, M.-K.; Shin, B.A.; Choi, S.-Y. High Cleavage Efficiency of a 2A Peptide Derived from Porcine Teschovirus-1 in Human Cell Lines, Zebrafish and Mice. PLoS ONE 2011, 6, e18556. [Google Scholar] [CrossRef]
- Lee, K.; Kim, S.Y.; Seo, Y.; Kwon, H.; Kwon, Y.J.; Lee, H. Multicistronic IVT mRNA for simultaneous expression of multiple fluorescent proteins. J. Ind. Eng. Chem. 2019, 80, 770–777. [Google Scholar] [CrossRef]
- Mizuguchi, H.; Xu, Z.; Ishii-Watabe, A.; Uchida, E.; Hayakawa, T. IRES-Dependent Second Gene Expression Is Significantly Lower Than Cap-Dependent First Gene Expression in a Bicistronic Vector. Mol. Ther. 2000, 1, 376–382. [Google Scholar] [CrossRef]
- Minskaia, E.; Nicholson, J.; Ryan, M.D. Optimisation of the foot-and-mouth disease virus 2A co-expression system for biomedical applications. BMC Biotechnol. 2013, 13, 67. [Google Scholar] [CrossRef]
- Doronina, V.A.; Wu, C.; De Felipe, P.; Sachs, M.; Ryan, M.D.; Brown, J.D. Site-Specific Release of Nascent Chains from Ribosomes at a Sense Codon. Mol. Cell. Biol. 2008, 28, 4227–4239. [Google Scholar] [CrossRef]
- Donnelly, M.L.L.; Luke, G.; Mehrotra, A.; Li, X.; Hughes, L.E.; Gani, D.; Ryan, M.D. Analysis of the aphthovirus 2A/2B polyprotein ‘cleavage’ mechanism indicates not a proteolytic reaction, but a novel translational effect: A putative ribosomal skip. J. Gen. Virol. 2001, 82, 1013–1025. [Google Scholar] [CrossRef]
- De Felipe, P.; Luke, G.A.; Hughes, L.E.; Gani, D.; Halpin, C.; Ryan, M.D. E unum pluribus: Multiple proteins from a self-processing polyprotein. Trends Biotechnol. 2006, 24, 68–75. [Google Scholar] [CrossRef]
- Szymczak, A.L.; Workman, C.J.; Wang, Y.; Vignali, K.M.; Dilioglou, S.; Vanin, E.F.; Vignali, D.A.A. Correction of multi-gene deficiency in vivo using a single ‘self-cleaving’ 2A peptide–based retroviral vector. Nat. Biotechnol. 2004, 22, 589–594. [Google Scholar] [CrossRef]
- Thomas, G. Furin at the cutting edge: From protein traffic to embryogenesis and disease. Nat. Rev. Mol. Cell Biol. 2002, 3, 753–766. [Google Scholar] [CrossRef]
- Rothwell, D.G.; Crossley, R.; Bridgeman, J.S.; Sheard, V.; Zhang, Y.; Sharp, T.V.; Hawkins, R.E.; Gilham, D.E.; McKay, T.R. Functional Expression of Secreted Proteins from a Bicistronic Retroviral Cassette Based on Foot-and-Mouth Disease Virus 2A Can Be Position Dependent. Hum. Gene Ther. 2010, 21, 1631–1637. [Google Scholar] [CrossRef]
- Fisicaro, N.; Londrigan, S.L.; Brady, J.L.; Salvaris, E.; Nottle, M.B.; O’Connell, P.J.; Robson, S.C.; D’Apice, A.J.F.; Lew, A.M.; Cowan, P.J. Versatile co-expression of graft-protective proteins using 2A-linked cassettes. Xenotransplantation 2011, 18, 121–130. [Google Scholar] [CrossRef]
- Yang, Y.-P.; Li, Y.-H.; Zhang, A.-H.; Bi, L.; Fan, M.-W. Good Manufacturing Practices production and analysis of a DNA vaccine against dental caries. Acta Pharmacol. Sin. 2009, 30, 1513–1521. [Google Scholar] [CrossRef][Green Version]
- Garanina, E.E.; Mukhamedshina, Y.O.; Salafutdinov, I.I.; Kiyasov, A.P.; Lima, L.M.; Reis, H.J.; Palotás, A.; Islamov, R.R.; Rizvanov, A.A. Construction of recombinant adenovirus containing picorna-viral 2A-peptide sequence for the co-expression of neuro-protective growth factors in human umbilical cord blood cells. Spinal Cord 2015, 54, 423–430. [Google Scholar] [CrossRef][Green Version]
- Xu, Y.; Zhang, N.-Z.; Tan, Q.-D.; Chen, J.; Lu, J.; Xu, Q.-M.; Zhu, X.-Q. Evaluation of immuno-efficacy of a novel DNA vaccine encoding Toxoplasma gondiirhoptry protein 38 (TgROP38) against chronic toxoplasmosis in a murine model. BMC Infect. Dis. 2014, 14, 525. [Google Scholar] [CrossRef]
- Smith, T.R.F.; Patel, A.; Ramos, S.; Elwood, D.; Zhu, X.; Yan, J.; Gary, E.N.; Walker, S.N.; Schultheis, K.; Purwar, M.; et al. Immunogenicity of a DNA vaccine candidate for COVID. Nat. Commun. 2020, 11, 2601. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Chen, O.; Wall, J.B.J.; Zheng, M.; Zhou, Y.; Wang, L.; Vaseghi, H.R.; Qian, L.; Liu, J. Systematic comparison of 2A peptides for cloning multi-genes in a polycistronic vector. Sci. Rep. 2017, 7, 2193. [Google Scholar] [CrossRef] [PubMed]
- Luke, G.A.; Ryan, M.D. Therapeutic applications of the ‘NPGP’ family of viral 2As. Rev. Med. Virol. 2018, 28, e2001. [Google Scholar] [CrossRef] [PubMed]
- Daniels, R.W.; Rossano, A.; MacLeod, G.T.; Ganetzky, B. Expression of Multiple Transgenes from a Single Construct Using Viral 2A Peptides in Drosophila. PLoS ONE 2014, 9, e100637. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, F.; Xu, S.; Wang, R.; Chen, W.; Hou, K.; Tian, C.; Wang, F.; Zhao, P.; Xia, Q. Optimization of a 2A self-cleaving peptide-based multigene expression system for efficient expression of upstream and downstream genes in silkworm. Mol. Genet. Genom. 2019, 294, 849–859. [Google Scholar] [CrossRef]
- Geisse, S.; Fux, C. Chapter 15 Recombinant Protein Production by Transient Gene Transfer into Mammalian Cells. In Methods in Enzymology; Elsevier BV: Amsterdam, The Netherlands, 2009; Volume 463, pp. 223–238. [Google Scholar]
- Vink, T.; Oudshoorn-Dickmann, M.; Roza, M.; Reitsma, J.-J.; de Jong, R.N. A simple, robust and highly efficient transient expression system for producing antibodies. Methods 2014, 65, 5–10. [Google Scholar] [CrossRef]
- Halabian, R.; Roudkenar, M.H.; Jahanian-Najafabadi, A.; Hosseini, K.M.; Tehrani, H.A. Co-culture of bone marrow-derived mesenchymal stem cells overexpressing lipocalin 2 with HK-2 and HEK293 cells protects the kidney cells against cisplatin-induced injury. Cell Biol. Int. 2014, 39, 152–163. [Google Scholar] [CrossRef]
- Serban, A.I.; Stanca, L.; Geicu, O.I.; Dinischiotu, A. AGEs-Induced IL-6 Synthesis Precedes RAGE Up-Regulation in HEK 293 Cells: An Alternative Inflammatory Mechanism? Int. J. Mol. Sci. 2015, 16, 20100–20117. [Google Scholar] [CrossRef]
- Solovyeva, V.; Salafutdinov, I.; Tazetdinova, L.; Khaiboullina, S.; Masgutov, R.; Rizvanov, A. Genetic modification of adipose derived stem cells with recombinant plasmid DNA pBud-VEGF-FGF2 results in increased of IL-8 and MCP-1 secretion. J. Pure Appl. Microbiol. 2014, 8, 523–528. [Google Scholar]
- Jin, S.; Yang, C.; Huang, J.; Liu, L.; Zhang, Y.; Li, S.; Zhang, L.; Sun, Q.; Yang, P. Conditioned medium derived from FGF-2-modified GMSCs enhances migration and angiogenesis of human umbilical vein endothelial cells. Stem Cell Res. Ther. 2020, 11, 68. [Google Scholar] [CrossRef]
- Yukita, A.; Hara, M.; Hosoya, A.; Nakamura, H. Relationship between localization of proteoglycans and induction of neurotrophic factors in mouse dental pulp. J. Oral Biosci. 2017, 59, 31–37. [Google Scholar] [CrossRef]
- Bauters, C.; Asahara, T.; Zheng, L.P.; Takeshita, S.; Bunting, S.; Ferrara, N.; Symes, J.F.; Isner, J.M. Site-specific therapeutic angiogenesis after systemic administration of vascular endothelial growth factor. J. Vasc. Surg. 1995, 21, 314–325. [Google Scholar] [CrossRef]
- Takeshita, S.; Weir, L.; Chen, D.; Zheng, L.P.; Riessen, R.; Bauters, C.; Symes, J.F.; Ferrara, N.; Isner, J.M. Therapeutic Angiogenesis Following Arterial Gene Transfer of Vascular Endothelial Growth Factor in a Rabbit Model of Hindlimb Ischemia. Biochem. Biophys. Res. Commun. 1996, 227, 628–635. [Google Scholar] [CrossRef]
- Cao, R.; Eriksson, A.; Kubo, H.; Alitalo, K.; Cao, Y.; Thyberg, J. Comparative Evaluation of FGF-2–, VEGF-A–, and VEGF-C–Induced Angiogenesis, Lymphangiogenesis, Vascular Fenestrations, and Permeability. Circ. Res. 2004, 94, 664–670. [Google Scholar] [CrossRef]
- Cartland, S.P.; Genner, S.W.; Zahoor, A.; Kavurma, M.M. Comparative Evaluation of TRAIL, FGF-2 and VEGF-A-Induced Angiogenesis In Vitro and In Vivo. Int. J. Mol. Sci. 2016, 17, 2025. [Google Scholar] [CrossRef]
- Song, M.; Finley, S.D. ERK and Akt exhibit distinct signaling responses following stimulation by pro-angiogenic factors. Cell Commun. Signal. 2020, 18, 114. [Google Scholar] [CrossRef]
- Lanza, A.M.; Curran, K.A.; Rey, L.G.; Alper, H.S. A condition-specific codon optimization approach for improved heterologous gene expression in Saccharomyces cerevisiae. BMC Syst. Biol. 2014, 8, 33. [Google Scholar] [CrossRef]
- Alexaki, A.; Hettiarachchi, G.K.; Athey, J.C.; Katneni, U.; Simhadri, V.; Hamasaki-Katagiri, N.; Nanavaty, P.; Lin, B.; Takeda, K.; Freedberg, D.; et al. Effects of codon optimization on coagulation factor IX translation and structure: Implications for protein and gene therapies. Sci. Rep. 2019, 9, 15449. [Google Scholar] [CrossRef]



| Primer Name | Nucleotide Sequence |
|---|---|
| rmh-18s-TMF | GCCGCTAGAGGTGAAATTCTTG |
| rmh-18s- TMR | CATTCTTGGCAAATGGTTTCG |
| rmh-18s-prode | (HEK)-ACCGGCGCAAGACGGACCA-(BHQ) |
| co-VEGF165-F | CAGATCATGGGGATCAAGCC |
| co-VEGF165-R | CATGGATTCTCCTGCCTTGC |
| co-VEGF165-Probe | (6-FAM)-CCAGGGCCAGCACATCGGCG -(BHQ1) |
| co-FGF2-F | GAGGCTGTACTGCAAGAACG |
| co-FGF2-R | TGATAGACACCAACGCCTCTC |
| co-FGF2-R | (6-FAM)-CCTCGGCCTGCAGCTGCTGCAGC -(BHQ1) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gatina, D.Z.; Garanina, E.E.; Zhuravleva, M.N.; Synbulatova, G.E.; Mullakhmetova, A.F.; Solovyeva, V.V.; Kiyasov, A.P.; Rutland, C.S.; Rizvanov, A.A.; Salafutdinov, I.I. Proangiogenic Effect of 2A-Peptide Based Multicistronic Recombinant Constructs Encoding VEGF and FGF2 Growth Factors. Int. J. Mol. Sci. 2021, 22, 5922. https://doi.org/10.3390/ijms22115922
Gatina DZ, Garanina EE, Zhuravleva MN, Synbulatova GE, Mullakhmetova AF, Solovyeva VV, Kiyasov AP, Rutland CS, Rizvanov AA, Salafutdinov II. Proangiogenic Effect of 2A-Peptide Based Multicistronic Recombinant Constructs Encoding VEGF and FGF2 Growth Factors. International Journal of Molecular Sciences. 2021; 22(11):5922. https://doi.org/10.3390/ijms22115922
Chicago/Turabian StyleGatina, Dilara Z., Ekaterina E. Garanina, Margarita N. Zhuravleva, Gulnaz E. Synbulatova, Adelya F. Mullakhmetova, Valeriya V. Solovyeva, Andrey P. Kiyasov, Catrin S. Rutland, Albert A. Rizvanov, and Ilnur I. Salafutdinov. 2021. "Proangiogenic Effect of 2A-Peptide Based Multicistronic Recombinant Constructs Encoding VEGF and FGF2 Growth Factors" International Journal of Molecular Sciences 22, no. 11: 5922. https://doi.org/10.3390/ijms22115922
APA StyleGatina, D. Z., Garanina, E. E., Zhuravleva, M. N., Synbulatova, G. E., Mullakhmetova, A. F., Solovyeva, V. V., Kiyasov, A. P., Rutland, C. S., Rizvanov, A. A., & Salafutdinov, I. I. (2021). Proangiogenic Effect of 2A-Peptide Based Multicistronic Recombinant Constructs Encoding VEGF and FGF2 Growth Factors. International Journal of Molecular Sciences, 22(11), 5922. https://doi.org/10.3390/ijms22115922







