Peptide/β-Peptoid Hybrids with Activity against Vancomycin-Resistant Enterococci: Influence of Hydrophobicity and Structural Features on Antibacterial and Hemolytic Properties
Abstract
1. Introduction
2. Results and Discussion
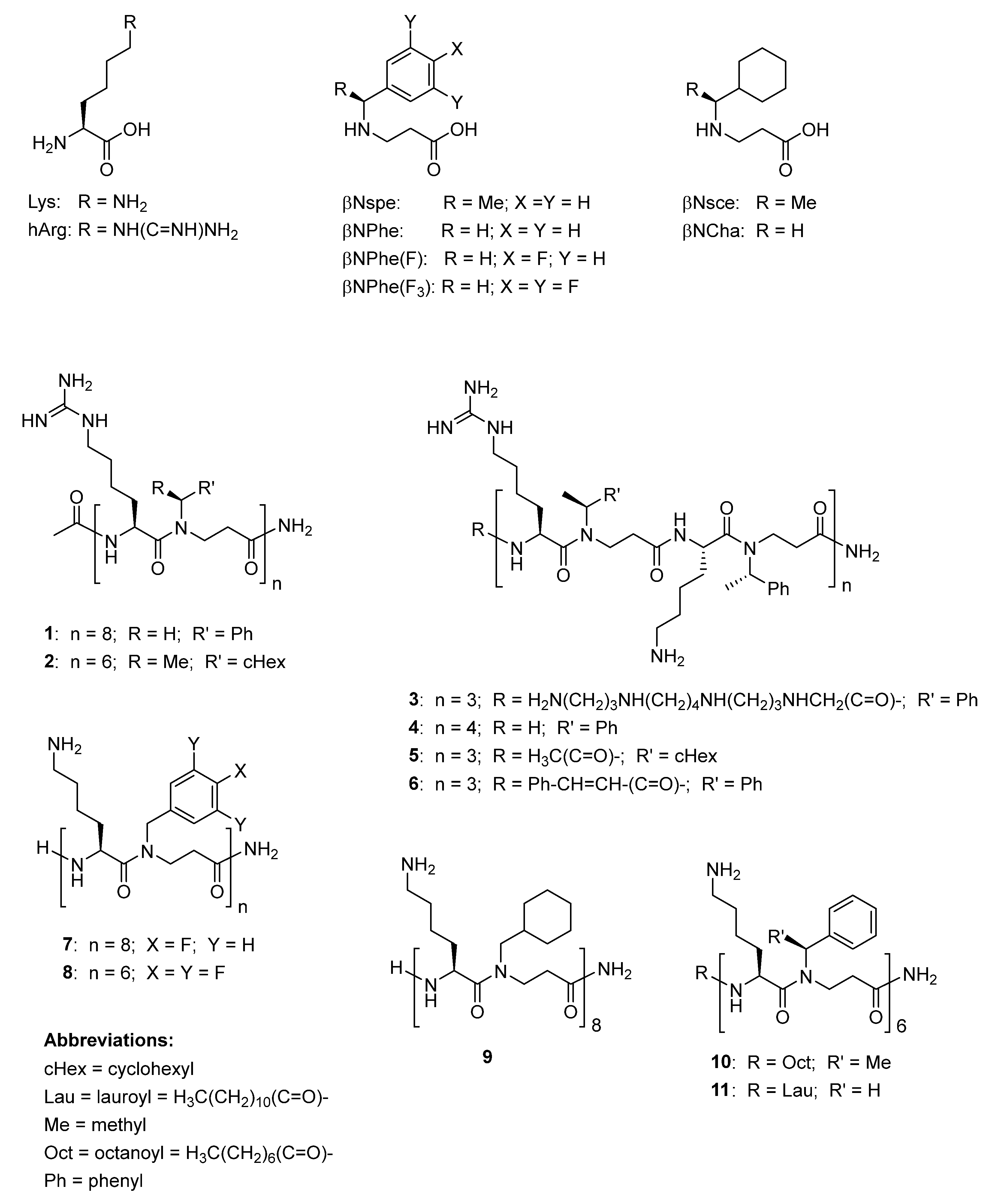
2.1. Structural Diversity of Studied Peptidomimetics
| No. | Peptidomimetic | Length (Residues) | MW 1 (g/mol) | Charge | Ref. |
|---|---|---|---|---|---|
| Subclass I | |||||
| 1 | Ac-[hArg-βNPhe]8-NH2 | 16 | 3622.56 | +8 | [20] |
| 2 | Ac-[hArg-βNsce]6-NH2 | 12 | 2852.13 | +6 | [20] |
| Subclass II | |||||
| 3 | SpermineAc-[hArg-βNspe-Lys-βNspe]3-NH2 | 13 | 3346.14 | +10 | [27] |
| 4 | H-[hArg-βNspe-Lys-βNspe]4-NH2 | 16 | 3638.59 | +9 | [27] |
| 5 | Ac-[hArg-βNsce-Lys-βNspe]3-NH2 | 12 | 2707.87 | +6 | [20] |
| 6 | Cinn-[hArg-βNspe-Lys-βNspe]3-NH2 | 13 | 2777.83 | +6 | [27] |
| Subclass III | |||||
| 7 | H-[Lys-βNPhe(F)]8-NH2 | 16 | 3502.20 | +9 | [28] |
| 8 | H-[Lys-βNPhe(F3)]6-NH2 | 12 | 2875.29 | +7 | [28] |
| 9 | H-[Lys-βNCha]8-NH2 | 16 | 2671.92 | +9 | [28] |
| 10 | Oct-[Lys-βNspe]6-NH2 | 13 | 2647.76 | +6 | [29] |
| 11 | Lau-[Lys-βNPhe]6-NH2 | 13 | 2619.71 | +6 | [30] |
2.2. Peptidomimetics Inhibit Growth of Enterococci, Including Vancomycin-Resistant E. faecium
2.3. No Synergy between Peptidomimetics and Conventional Antibiotics
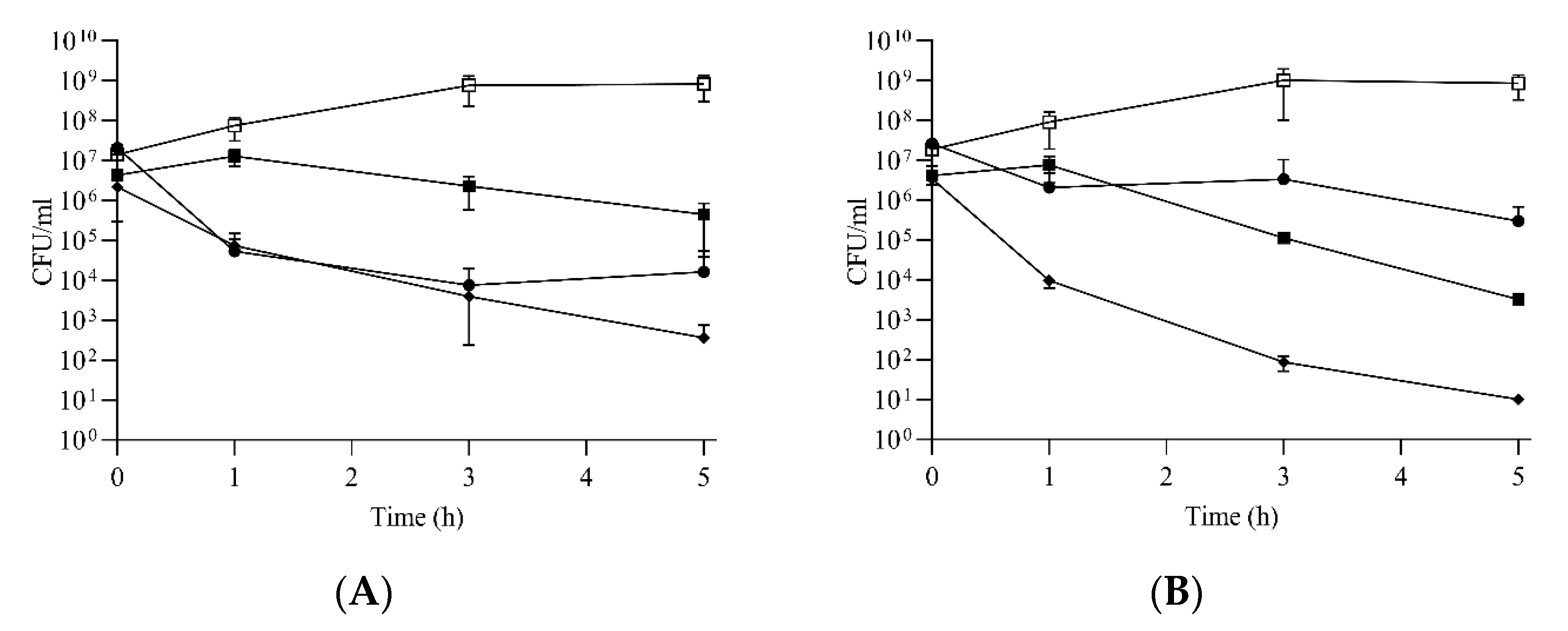
2.4. Compounds 2, 5 and 10 Kill Enterococci
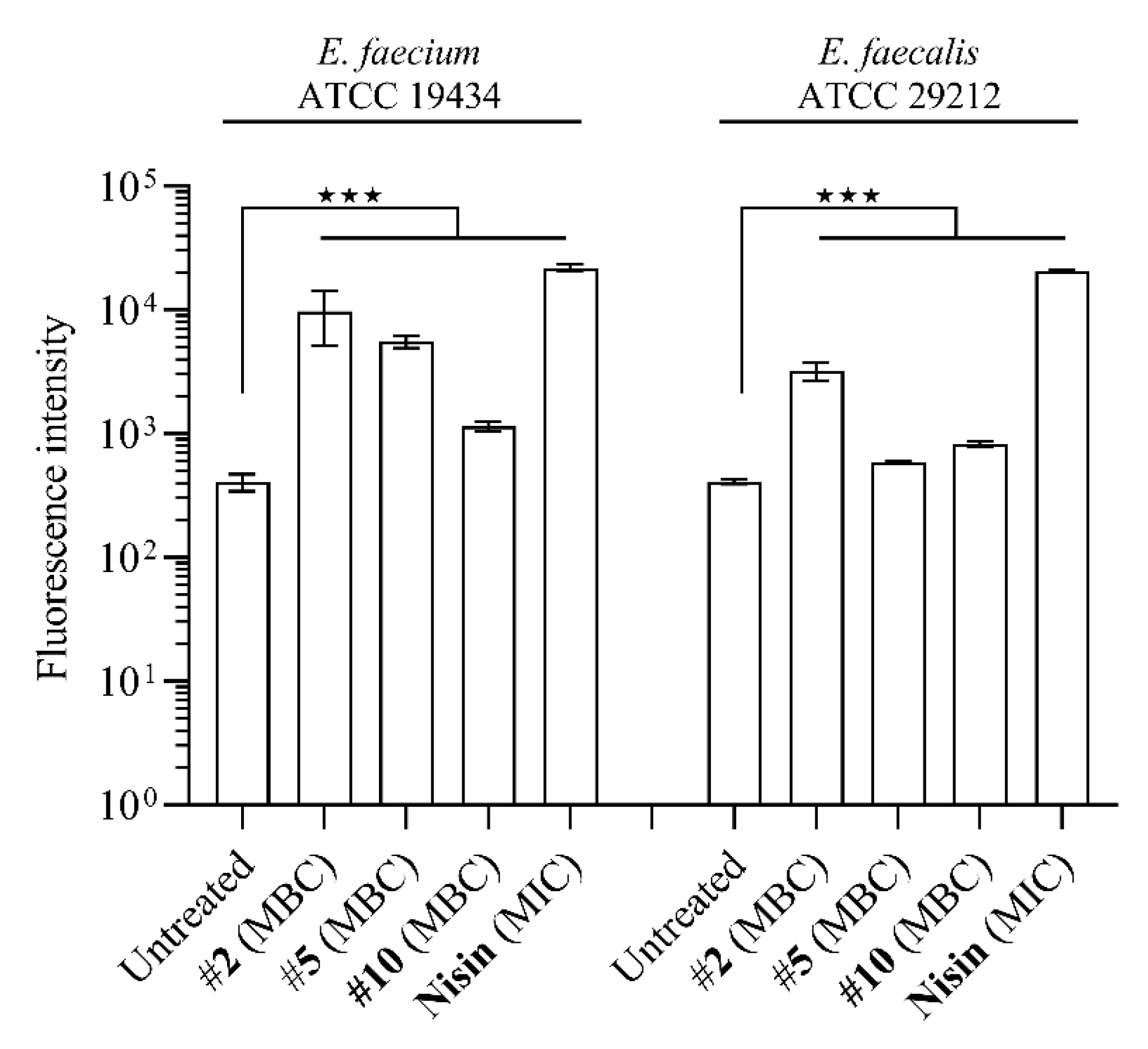
2.5. Compounds 2, 5 and 10 Compromise Membrane Integrity to a Different Degree
3. Conclusions
4. Materials and Methods
4.1. Analytical HPLC
4.2. Bacterial Strains, Growth Conditions and Chemicals
4.3. Determination of Minimum Inhibitory Concentration (MIC) and Minimum Bactericidal Concentration (MBC)
4.4. Combination with Conventional Antibiotics
4.5. Time–Kill Kinetics
4.6. Assessment of Membrane Integrity Using Flow Cytometry
4.7. Statistics
4.8. Determination of Hemolytic Activity
4.9. Determination of Antiproliferative Activity on HepG2 Cell Line
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Paphitou, N.I. Antimicrobial resistance: Action to combat the rising microbial challenges. Int. J. Antimicrob. Agents 2013, 42, S25–S28. [Google Scholar] [CrossRef]
- Lewis, K. Platforms for antibiotic discovery. Nat. Rev. Drug Discov. 2013, 12, 371–387. [Google Scholar] [CrossRef] [PubMed]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Centers for Disease Control. Antibiotic Resistance Threats in the United States. Centers for Disease Control and Prevention; U.S. Department of Health and Human Services: Atlanta, GA, USA, 2019. [CrossRef]
- García-Solache, M.; Rice, L.B. The Enterococcus: A model of adaptability to its environment. Clin. Microbiol. Rev. 2019, 32, e00058-18. [Google Scholar] [CrossRef] [PubMed]
- Fiore, E.; Van Tyne, D.; Gilmore, M.S. Pathogenicity of Enterococci. Gram-Positive Pathog. 2019, 378–397. [Google Scholar] [CrossRef]
- Weiner, L.M.; Webb, A.K.; Limbago, B.; Dudeck, M.A.; Patel, J.; Kallen, A.J.; Edwards, J.R.; Sievert, D.M. Antimicrobial-resistant pathogens associated with healthcare-associated infections: Summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2011–2014. Infect. Control. Hosp. Epidemiol. 2016, 37, 1288–1301. [Google Scholar] [CrossRef] [PubMed]
- Miller, W.R.; Munita, J.M.; Arias, C.A. Mechanisms of antibiotic resistance in enterococci. Expert Rev. Anti-Infect. Ther. 2014, 12, 1221–1236. [Google Scholar] [CrossRef] [PubMed]
- Arias, C.A.; Contreras, G.A.; Murray, B.E. Management of multidrug-resistant enterococcal infections. Clin. Microbiol. Infect. 2010, 16, 555–562. [Google Scholar] [CrossRef]
- Cassini, A.; Högberg, L.D.; Plachouras, D.; Quattrocchi, A.; Hoxha, A.; Simonsen, G.S.; Colomb-Cotinat, M.; Kretzschmar, M.E.; Devleesschauwer, B.; Cecchini, M.; et al. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: A population-level modelling analysis. Lancet Infect. Dis. 2019, 19, 56–66. [Google Scholar] [CrossRef]
- Ayobami, O.; Willrich, N.; Reuss, A.; Eckmanns, T.; Markwart, R. The ongoing challenge of vancomycin-resistant Enterococcus faecium and Enterococcus faecalis in Europe: An epidemiological analysis of bloodstream infections. Emerg. Microbes Infect. 2020, 9, 1180–1193. [Google Scholar] [CrossRef]
- Magana, M.; Pushpanathan, M.; Santos, A.L.; Leanse, L.; Fernandez, M.; Ioannidis, A.; Giulianotti, M.A.; Apidianakis, Y.; Bradfute, S.; Ferguson, A.L.; et al. The value of antimicrobial peptides in the age of resistance. Lancet Infect. Dis. 2020, 20, e216–e230. [Google Scholar] [CrossRef]
- Vlieghe, P.; Lisowski, V.; Martinez, J.; Khrestchatisky, M. Synthetic therapeutic peptides: Science and market. Drug Discov. Today 2010, 15, 40–56. [Google Scholar] [CrossRef] [PubMed]
- Molchanova, N.; Hansen, P.R.; Franzyk, H. Advances in development of antimicrobial peptidomimetics as potential drugs. Molecules 2017, 22, 1430. [Google Scholar] [CrossRef] [PubMed]
- Chongsiriwatana, N.P.; Patch, J.A.; Czyzewski, A.M.; Dohm, M.T.; Ivankin, A.; Gidalevitz, D.; Zuckermann, R.N.; Barron, A.E. Peptoids that mimic the structure, function, and mechanism of helical antimicrobial peptides. Proc. Natl. Acad. Sci. USA 2008, 105, 2794–2799. [Google Scholar] [CrossRef] [PubMed]
- Shuey, S.W.; Delaney, W.J.; Shah, M.C.; Scialdone, M.A. Antimicrobial β-peptoids by a block synthesis approach. Bioorg. Med. Chem. Lett. 2006, 16, 1245–1248. [Google Scholar] [CrossRef]
- Liu, D.; DeGrado, W.F. De novo design, synthesis, and characterization of antimicrobial β-peptides. J. Am. Chem. Soc. 2001, 123, 7553–7559. [Google Scholar] [CrossRef]
- Schmitt, M.A.; Weisblum, B.; Gellman, S.H. Interplay among folding, sequence, and lipophilicity in the antibacterial and hemolytic activities of α/β-peptides. J. Am. Chem. Soc. 2007, 129, 417–428. [Google Scholar] [CrossRef]
- Niu, Y.; Wu, H.; Li, Y.; Hu, Y.; Padhee, S.; Li, Q.; Cao, C.; Cai, J. AApeptides as a new class of antimicrobial agents. Org. Biomol. Chem. 2013, 11, 4283–4290. [Google Scholar] [CrossRef]
- Olsen, C.A.; Ziegler, H.L.; Nielsen, H.M.; Frimodt-Møller, N.; Jaroszewski, J.W.; Franzyk, H. Antimicrobial, hemolytic, and cytotoxic activities of β-peptoid–peptide hybrid oligomers: Improved properties compared to natural AMPs. ChemBioChem 2010, 11, 1356–1360. [Google Scholar] [CrossRef]
- Jahnsen, R.D.; Frimodt-Møller, N.; Franzyk, H. Antimicrobial activity of peptidomimetics against multidrug-resistant Escherichia coli: A comparative study of different backbones. J. Med. Chem. 2012, 55, 7253–7261. [Google Scholar] [CrossRef]
- Frederiksen, N.; Hansen, P.R.; Björkling, F.; Franzyk, H. Peptide/peptoid hybrid oligomers: The influence of hydrophobicity and relative side-chain length on antibacterial activity and cell selectivity. Molecules 2019, 24, 4429. [Google Scholar] [CrossRef]
- Hein-Kristensen, L.; Knapp, K.M.; Franzyk, H.; Gram, L. Bacterial membrane activity of α-peptide/β-peptoid chimeras: Influence of amino acid composition and chain length on the activity against different bacterial strains. BMC Microbiol. 2011, 11, 144. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Knapp, K.M.; Yang, L.; Molin, S.; Franzyk, H.; Folkesson, A. High in vitro antimicrobial activity of β-peptoid–peptide hybrid oligomers against planktonic and biofilm cultures of Staphylococcus epidermidis. Int. J. Antimicrob. Agents 2013, 41, 20–27. [Google Scholar] [CrossRef]
- Citterio, L.; Franzyk, H.; Palarasah, Y.; Andersen, T.E.; Mateiu, R.V.; Gram, L. Improved in vitro evaluation of novel antimicrobials: Potential synergy between human plasma and antibacterial peptidomimetics, AMPs and antibiotics against human pathogenic bacteria. Res. Microbiol. 2016, 167, 72–82. [Google Scholar] [CrossRef] [PubMed]
- Rice, L.B. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: No ESKAPE. J. Infect. Dis. 2008, 197, 1079–1081. [Google Scholar] [CrossRef]
- Jahnsen, R.O.; Sandberg-Schaal, A.; Frimodt-Møller, N.; Nielsen, H.M.; Franzyk, H. End group modification: Efficient tool for improving activity of antimicrobial peptide analogues towards Gram-positive bacteria. Eur. J. Pharm. Biopharm. 2015, 95, 40–46. [Google Scholar] [CrossRef] [PubMed]
- Molchanova, N.; Hansen, P.R.; Damborg, P.; Nielsen, H.M.; Franzyk, H. Lysine-based α-peptide/β-peptoid peptidomimetics: Influence of hydrophobicity, fluorination, and distribution of cationic charge on antimicrobial activity and cytotoxicity. ChemMedChem 2017, 12, 312–318. [Google Scholar] [CrossRef] [PubMed]
- Skovbakke, S.L.; Larsen, C.J.; Heegaard, P.M.; Moesby, L.; Franzyk, H. Lipidated α-Peptide/β-peptoid hybrids with potent anti-inflammatory activity. J. Med. Chem. 2015, 58, 801–813. [Google Scholar] [CrossRef]
- Holdfeldt, A.; Skovbakke, S.L.; Winther, M.; Gabl, M.; Nielsen, C.; Perez-Gassol, I.; Larsen, C.J.; Wang, J.M.; Karlsson, A.; Dahlgren, C.; et al. The lipidated peptidomimetic Lau-((S)-Aoc)-(Lys-βNphe)6-NH2 is a novel formyl peptide receptor 2 agonist that activates both human and mouse neutrophil NADPH oxidase. J. Biol. Chem. 2016, 291, 19888–19899. [Google Scholar] [CrossRef]
- Chen, C.; Hu, J.; Yang, C.; Zhang, Y.; Wang, F.; Mu, Q.; Pan, F.; Xu, H.; Lu, J.R. Amino acid side chains affect the bioactivity of designed short peptide amphiphiles. J. Mater. Chem. B 2016, 4, 2359–2368. [Google Scholar] [CrossRef]
- Frederiksen, N.; Hansen, P.R.; Zabicka, D.; Tomczak, M.; Urbas, M.; Domraceva, I.; Björkling, F.; Franzyk, H. Alternating cationic-hydrophobic peptide/peptoid hybrids: Influence of hydrophobicity on antibacterial activity and cell selectivity. ChemMedChem 2020, 15, 2544–2561. [Google Scholar] [CrossRef]
- Bychowska, A.; Theilacker, C.; Czerwicka, M.; Marszewska, K.; Huebner, J.; Holst, O.; Stepnowski, P.; Kaczyński, Z. Chemical structure of wall teichoic acid isolated from Enterococcus faecium strain U0317. Carbohydr. Res. 2011, 346, 2816–2819. [Google Scholar] [CrossRef]
- Theilacker, C.; Holst, O.; Lindner, B.; Huebner, J.; Kaczyński, Z. The structure of the wall teichoic acid isolated from Enterococcus faecalis strain 12030. Carbohydr. Res. 2012, 354, 106–109. [Google Scholar] [CrossRef] [PubMed]
- Baker, K.R.; Jana, B.; Hansen, A.M.; Nielsen, H.M.; Franzyk, H.; Guardabassi, L. Repurposing azithromycin and rifampicin against Gram-negative pathogens by combination with peptidomimetics. Front. Cell. Infect. Microbiol. 2019, 9, 236. [Google Scholar] [CrossRef] [PubMed]
- Vehreschild, M.J.; Haverkamp, M.; Biehl, L.M.; Lemmen, S.; Fätkenheuer, G. Vancomycin-resistant enterococci (VRE): A reason to isolate? Infection 2019, 47, 7–11. [Google Scholar] [CrossRef] [PubMed]
- Chow, J.W. Aminoglycoside resistance in enterococci. Clin. Infect. Dis. 2000, 31, 586–589. [Google Scholar] [CrossRef]
- Jahnsen, R.D.; Haney, E.F.; Franzyk, H.; Hancock, R.E. Characterization of a proteolytically stable multifunctional host defense peptidomimetic. Chem. Biol. 2013, 20, 1286–1295. [Google Scholar] [CrossRef]
- Stiefel, P.; Schmidt-Emrich, S.; Maniura-Weber, K.; Ren, Q. Critical aspects of using bacterial cell viability assays with the fluorophores SYTO9 and propidium iodide. BMC Microbiol. 2015, 15, 36. [Google Scholar] [CrossRef]
- Odds, F.C. Synergy, antagonism, and what the chequerboard puts between them. J. Antimicrob. Chemother. 2003, 52, 1. [Google Scholar] [CrossRef]
- Jensen, C.; Li, H.; Vestergaard, M.; Dalsgaard, A.; Frees, D.; Leisner, J.J. Nisin damages the septal membrane and triggers DNA condensation in methicillin-resistant Staphylococcus aureus. Front. Microbiol. 2020, 11, 1007. [Google Scholar] [CrossRef]
- Williamson, R.; Le Bouguénec, C.; Gutmann, L.; Horaud, T. One or two low affinity penicillin-binding proteins may be responsible for the range of susceptibility of Enterococcus faecium to benzylpenicillin. Microbiology 1985, 131, 1933–1940. [Google Scholar] [CrossRef] [PubMed]
| No. | E. faecium | E. faecalis | ||||||
| D344R 1 | ATCC 19434 | 3978 1 | 2961 1 | 1798 1 | ATCC 29212 | 38262 2 | 39002 2 | |
| Subclass I | ||||||||
| 1 | 4 | 8 | 4 | 2 | 2 | 32- > 32 | 16 | 32 |
| 2 | 2 | 2 | 2 | 2 | 2 | 2–4 | 4 | 2 |
| Subclass II | ||||||||
| 3 | 4–8 | 16–32 | 8 | 8 | 4–8 | >32 | 32- > 32 | 32- > 32 |
| 4 | 4 | 8 | 4–8 | 4 | 4 | >32 | 16 | 32 |
| 5 | 2 | 4 | 2 | 2 | 2 | 4–8 | 8–16 | 8–16 |
| 6 | 4 | 8 | 4 | 4 | 4 | 16 | 16 | 16 |
| Subclass III | ||||||||
| 7 | 4–8 | 8–16 | 4–8 | 2–4 | 2–4 | 8–16 | >32 | 8–16 |
| 8 | 2 | 2–4 | 2 | 2 | 2–4 | 2–4 | 8–16 | 2 |
| 9 | 2 | 2–4 | 2–4 | 2–4 | 2–4 | 2–4 | 8 | 4 |
| 10 | 4 | 8 | 2–4 | 4 | 2–4 | 8–16 | 16 | 8 |
| 11 | 2–4 | 4 | 2–4 | 2–4 | 2–4 | 4 | 4–8 | 4–8 |
| Vancomycin | 2 | 1–2 | 16 | >256 | >256 | 2–4 | 1 | 1 |
| No. | Luna C18(2) HST 1 (0–60% B; 10 min) | Hemolysis (at 400 µg/mL) | HepG2 IC50 (µg/mL) |
|---|---|---|---|
| Subclass I | |||
| 1 | 41.8 | 6.1% | 9 |
| 2 | 51.6 | 98.4% | 15 |
| Subclass II | |||
| 3 | 40.2 | 0.6% | 41 |
| 4 | 43.6 | 2.6% | 42 |
| 5 | 46.6 | 9.4% | 73 |
| 6 | 45.9 | 15.7% | 42 |
| Subclass III | |||
| 7 | 41.3 | 1.5% | 50 |
| 8 | 45.9 | 28.8% | 27 |
| 9 | 46.8 | 23.0% | 15 |
| 10 | 47.2 | 6.9% | 41 |
| 11 | 50.5 | 95.6% | 22 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vestergaard, M.; Skive, B.; Domraceva, I.; Ingmer, H.; Franzyk, H. Peptide/β-Peptoid Hybrids with Activity against Vancomycin-Resistant Enterococci: Influence of Hydrophobicity and Structural Features on Antibacterial and Hemolytic Properties. Int. J. Mol. Sci. 2021, 22, 5617. https://doi.org/10.3390/ijms22115617
Vestergaard M, Skive B, Domraceva I, Ingmer H, Franzyk H. Peptide/β-Peptoid Hybrids with Activity against Vancomycin-Resistant Enterococci: Influence of Hydrophobicity and Structural Features on Antibacterial and Hemolytic Properties. International Journal of Molecular Sciences. 2021; 22(11):5617. https://doi.org/10.3390/ijms22115617
Chicago/Turabian StyleVestergaard, Martin, Bolette Skive, Ilona Domraceva, Hanne Ingmer, and Henrik Franzyk. 2021. "Peptide/β-Peptoid Hybrids with Activity against Vancomycin-Resistant Enterococci: Influence of Hydrophobicity and Structural Features on Antibacterial and Hemolytic Properties" International Journal of Molecular Sciences 22, no. 11: 5617. https://doi.org/10.3390/ijms22115617
APA StyleVestergaard, M., Skive, B., Domraceva, I., Ingmer, H., & Franzyk, H. (2021). Peptide/β-Peptoid Hybrids with Activity against Vancomycin-Resistant Enterococci: Influence of Hydrophobicity and Structural Features on Antibacterial and Hemolytic Properties. International Journal of Molecular Sciences, 22(11), 5617. https://doi.org/10.3390/ijms22115617






