The Emerging Role of microRNA in Periodontitis: Pathophysiology, Clinical Potential and Future Molecular Perspectives
Abstract
1. Introduction
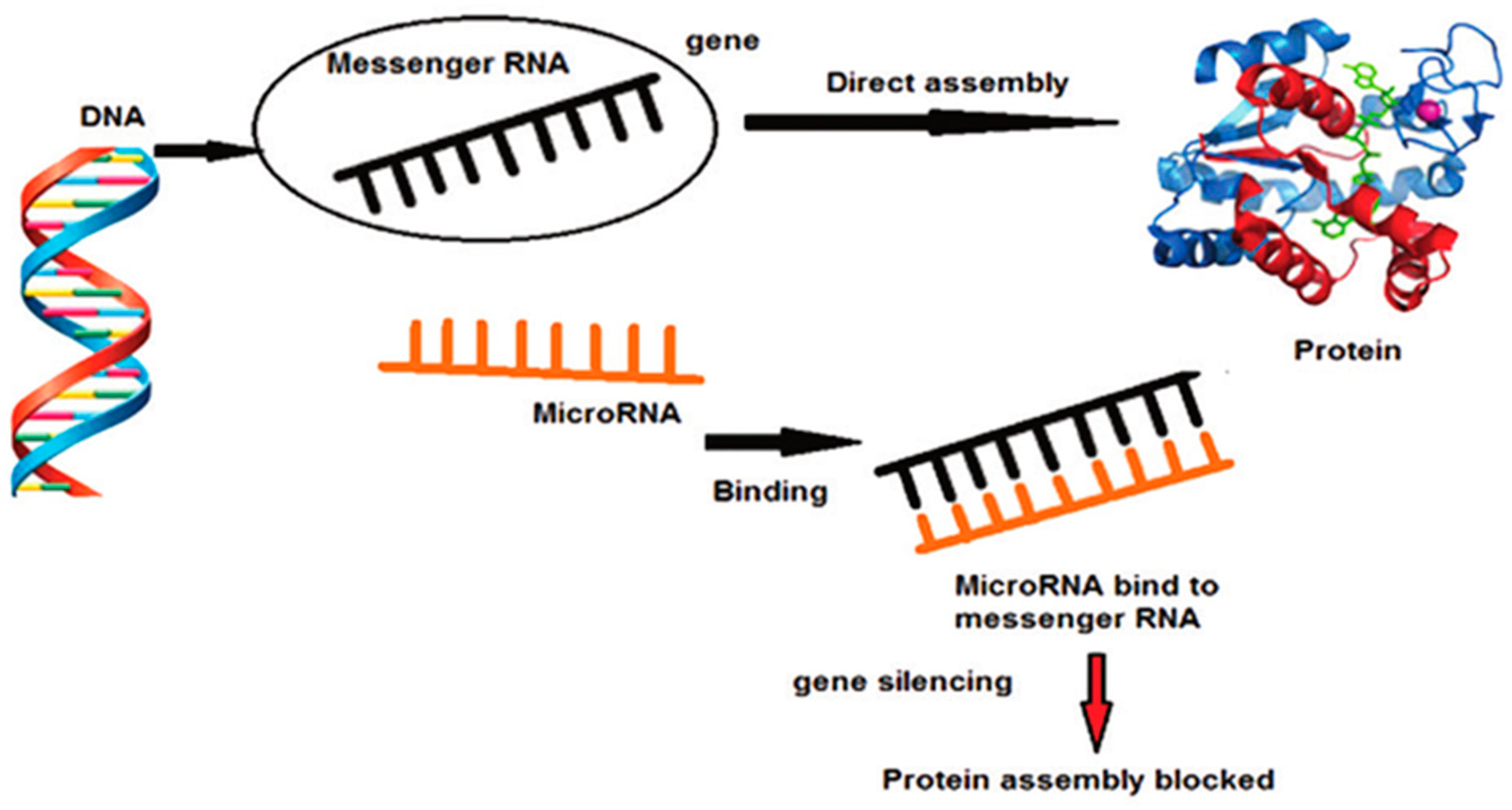
2. MiRNA, Molecular Signalling and Periodontal Disease
3. MiRNA as a Possible Link between Periodontal Disease and Various Systemic Disorders
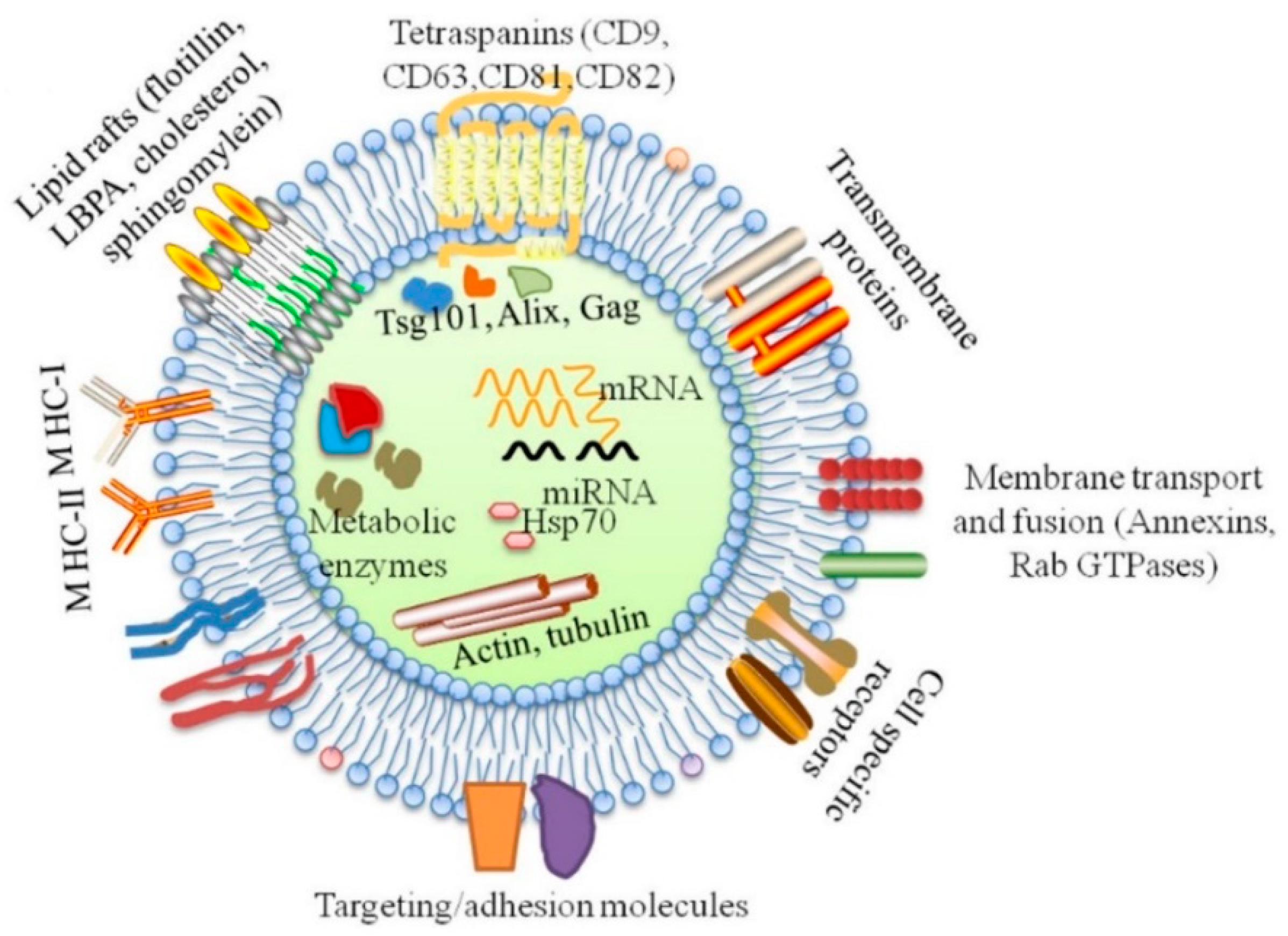
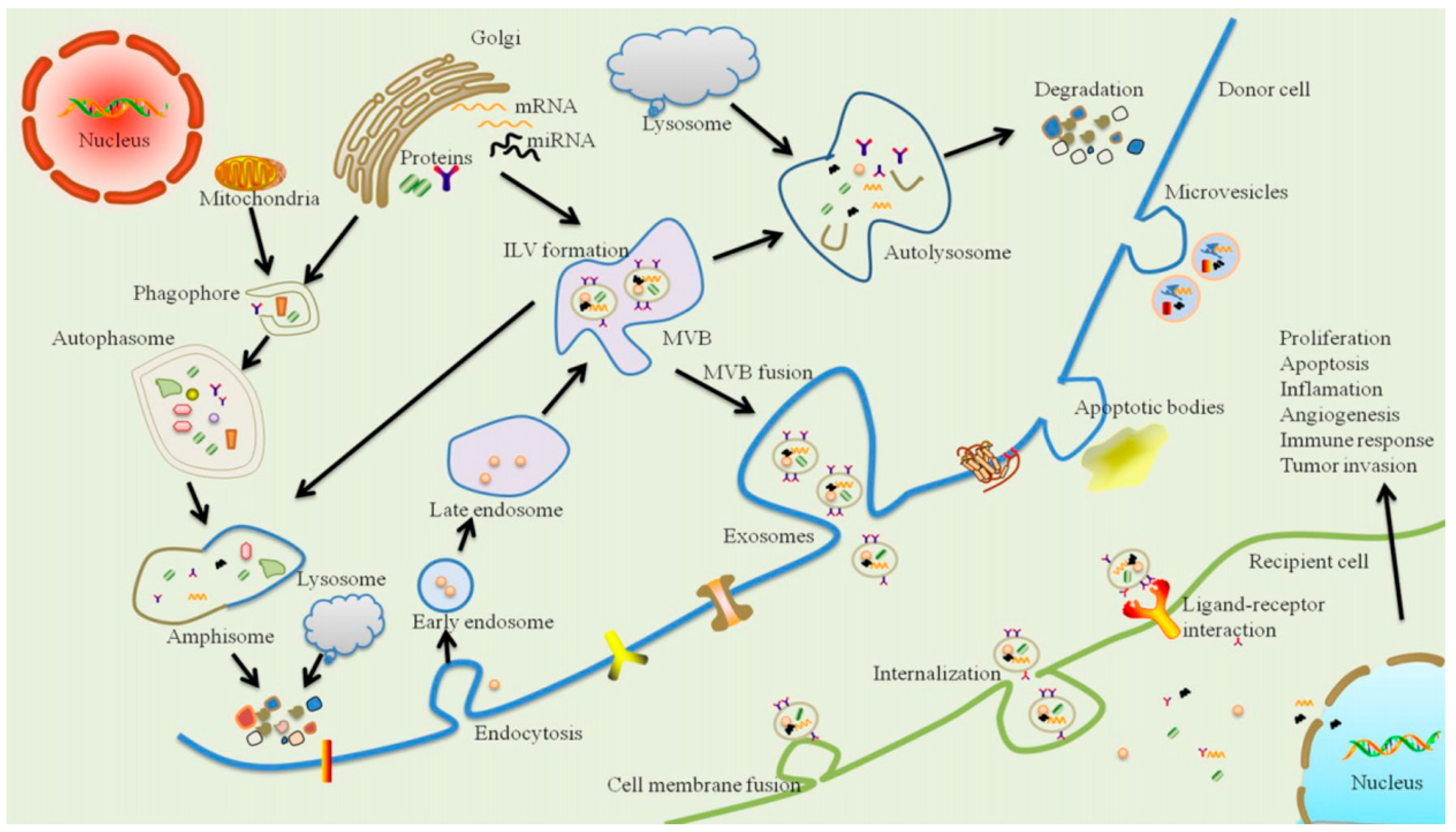
4. Exosomes and Periodontal Disease
5. Clinical Potential and Future Perspective in Periodontal Disease
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Crick, F.H. On protein synthesis. Symp. Soc. Exp. Biol. 1958, 12, 138–163. [Google Scholar] [PubMed]
- Crick, F. Central Dogma of Molecular Biology. Nat. Cell Biol. 1970, 227, 561–563. [Google Scholar] [CrossRef] [PubMed]
- Adcock, I.M.; Tsaprouni, L.; Bhavsar, P.; Ito, K. Epigenetic regulation of airway inflammation. Curr. Opin. Immunol. 2007, 19, 694–700. [Google Scholar] [CrossRef] [PubMed]
- Azevedo, A.M.; Rocha, L.P.C.; Amormino, S.A.D.F.; Gomes, C.C.; Dutra, W.O.; Gomez, R.S.; da Costa, J.E.; Moreira, P.R. DNA methylation profile of genes related to immune response in generalized periodontitis. J. Periodontal Res. 2020, 55, 426–431. [Google Scholar] [CrossRef] [PubMed]
- Barros, S.P.; Hefni, E.; Nepomuceno, R.; Offenbacher, S.; North, K. Targeting epigenetic mechanisms in periodontal diseases. Periodontology 2000 2018, 78, 174–184. [Google Scholar] [CrossRef] [PubMed]
- Isola, G.; Lo Giudice, A.; Polizzi, A.; Alibrandi, A.; Murabito, P.; Indelicato, F. Identification of the different salivary Interleukin-6 profiles in patients with periodontitis: A cross-sectional study. Arch Oral Biol. 2021, 122, 104997. [Google Scholar] [CrossRef] [PubMed]
- Kagiya, T. MicroRNAs: Potential Biomarkers and Therapeutic Targets for Alveolar Bone Loss in Periodontal Disease. Int. J. Mol. Sci. 2016, 17, 1317. [Google Scholar] [CrossRef]
- Friedman, J.M.; Jones, P.A. MicroRNAs: Critical mediators of differentiation, development and disease. Swiss Med. Wkly. 2009, 139, 466–472. [Google Scholar]
- Kebschull, M.; Papapanou, P.N. Mini but mighty: Micro RNA s in the pathobiology of periodontal disease. Periodontology 2000 2015, 69, 201–220. [Google Scholar] [CrossRef] [PubMed]
- Luan, X.; Zhou, X.; Naqvi, A.; Francis, M.; Foyle, D.; Nares, S.; Diekwisch, T.G.H. MicroRNAs and immunity in periodontal health and disease. Int. J. Oral Sci. 2018, 10, 1–14. [Google Scholar] [CrossRef]
- Kornman, K.S. Mapping the Pathogenesis of Periodontitis: A New Look. J. Periodontol. 2008, 79, 1560–1568. [Google Scholar] [CrossRef] [PubMed]
- Cekici, A.; Kantarci, A.; Hasturk, H.; van Dyke, T.E. Inflammatory and immune pathways in the pathogenesis of periodontal disease. Periodontology 2000 2014, 64, 57–80. [Google Scholar] [CrossRef] [PubMed]
- Olsen, I.; Singhrao, S.K.; Osmundsen, H. Periodontitis, pathogenesis and progression: miRNA-mediated cellular responses to Porphyromonas gingivalis. J. Oral Microbiol. 2017, 9, 1333396. [Google Scholar] [CrossRef]
- Maciotta, S.; Meregalli, M.; Torrente, Y. The involvement of microRNAs in neurodegenerative diseases. Front. Cell. Neurosci. 2013, 7, 265. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Shu, R.; Jiang, S.; Liu, D.L.; Zhang, X. Comparison of microRNA profiles of human periodontal diseased and healthy gingival tissues. Int. J. Oral Sci. 2011, 3, 125–134. [Google Scholar] [CrossRef]
- Ikeda, S.; Kong, S.W.; Lu, J.; Bisping, E.; Zhang, H.; Allen, P.D.; Golub, T.R.; Pieske, B.; Pu, W.T. Altered microRNA expression in human heart disease. Physiol. Genom. 2007, 31, 367–373. [Google Scholar] [CrossRef]
- Montecalvo, A.; Larregina, A.T.; Shufesky, W.J.; Stolz, D.B.; Sullivan, M.L.G.; Karlsson, J.M.; Baty, C.J.; Gibson, G.A.; Erdos, G.; Wang, Z.; et al. Mechanism of transfer of functional microRNAs between mouse dendritic cells via exosomes. Blood 2012, 119, 756–766. [Google Scholar] [CrossRef] [PubMed]
- Crescitelli, R.; Lässer, C.; Szabó, T.G.; Kittel, A.; Eldh, M.; Dianzani, I.; Buzás, E.I.; Lotvall, J. Distinct RNA profiles in subpopulations of extracellular vesicles: Apoptotic bodies, microvesicles and exosomes. J. Extracell. Vesicles 2013, 2. [Google Scholar] [CrossRef] [PubMed]
- Peng, Q.; Yang, J.-Y.; Zhou, G. Emerging functions and clinical applications of exosomes in human oral diseases. Cell Biosci. 2020, 10, 1–13. [Google Scholar] [CrossRef]
- Cheng, Q.; Shi, X.; Han, M.; Smbatyan, G.; Lenz, H.-J.; Zhang, Y. Reprogramming Exosomes as Nanoscale Controllers of Cellular Immunity. J. Am. Chem. Soc. 2018, 140, 16413–16417. [Google Scholar] [CrossRef]
- Barile, L.; Vassalli, G. Exosomes: Therapy delivery tools and biomarkers of diseases. Pharmacol. Ther. 2017, 174, 63–78. [Google Scholar] [CrossRef]
- Saeidimehr, S.; Ebrahimi, A.; Saki, N.; Goodarzi, P.; Rahim, F. MicroRNA-based linkage between aging and cancer: From epigenetics view point. Cell J. (Yakhteh) 2016, 18, 117. [Google Scholar]
- Nevins, M.; Chen, C.Y.; Kerr, E.; Mendoza-Azpur, G.; Isola, G.; Soto, C.P.; Stacchi, C.; Lombardi, T.; Kim, D.; Rocchietta, I. Comparison of a Novel Sonic Toothbrush to Manual Brushing on Plaque Control and Gingival Inflammation: A Multicenter, Randomized, Controlled Clinical Trial. Int. J. Periodontics Restor. Dent. 2021, 41, 99–104. [Google Scholar] [CrossRef] [PubMed]
- Grossi, S.G.; Zambon, J.J.; Ho, A.W.; Koch, G.; Dunford, R.G.; Machtei, E.E.; Norderyd, O.M.; Genco, R.J. Assessment of Risk for Periodontal Disease. I. Risk Indicators for Attachment Loss. J. Periodontol. 1994, 65, 260–267. [Google Scholar] [CrossRef] [PubMed]
- Ari, G.; Cherukuri, S.; Namasivayam, A. Epigenetics and Periodontitis: A Contemporary Review. J. Clin. Diagn. Res. 2016, 10, ZE07–ZE09. [Google Scholar] [CrossRef] [PubMed]
- Olsen, I. Organization of supragingival plaque at the micron scale. J. Oral Microbiol. 2018, 10, 1438722. [Google Scholar] [CrossRef] [PubMed]
- Iorio-Siciliano, V.; Ramaglia, L.; Isola, G.; Blasi, A.; Salvi, G.E.; Sculean, A. Changes in clinical parameters following adjunctive local sodium hypochlorite gel in minimally invasive nonsurgical therapy (MINST) of periodontal pockets: A 6-month randomized controlled clinical trial. Clin. Oral Investig. 2021. [Google Scholar] [CrossRef] [PubMed]
- Stoecklin-Wasmer, C.; Guarnieri, P.; Celenti, R.; Demmer, R.; Kebschull, M.; Papapanou, P. MicroRNAs and Their Target Genes in Gingival Tissues. J. Dent. Res. 2012, 91, 934–940. [Google Scholar] [CrossRef]
- Suárez, Y.; Wang, C.; Manes, T.D.; Pober, J.S. Cutting Edge: TNF-Induced MicroRNAs Regulate TNF-Induced Expression of E-Selectin and Intercellular Adhesion Molecule-1 on Human Endothelial Cells: Feedback Control of Inflammation. J. Immunol. 2009, 184, 21–25. [Google Scholar] [CrossRef]
- Gita, J.B.; George, A.V.; Pavithra, N.; Chandrasekaran, S.C.; Latchumanadhas, K.; Gnanamani, A. Dysregulation of miR-146a by periodontal pathogens: A risk for acute coronary syndrome. J. Periodontol. 2019, 90, 756–765. [Google Scholar] [CrossRef]
- Ghotloo, S.; Motedayyen, H.; Amani, D.; Saffari, M.; Sattari, M. Assessment of microRNA-146a in generalized aggressive periodontitis and its association with disease severity. J. Periodontal Res. 2019, 54, 27–32. [Google Scholar] [CrossRef] [PubMed]
- Kadkhodazadeh, M.; Jafari, A.R.; Amid, R.; Ebadian, A.R.; Alipour, M.M.; Mollaverdi, F.; Lafzi, A. MiR146a and MiR499 gene polymorphisms in Iranian periodontitis and peri-implantitis patients. J. Autom. Inf. Sci. 2013, 23, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Yoneda, T.; Tomofuji, T.; Ekuni, D.; Azuma, T.; Maruyama, T.; Fujimori, K.; Sugiura, Y.; Morita, M.; Yonedaa, T. Serum microRNAs and chronic periodontitis: A case-control study. Arch. Oral Biol. 2019, 101, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Motedayyen, H.; Ghotloo, S.; Saffari, M.; Sattari, M.; Amid, R. Evaluation of MicroRNA-146a and Its Targets in Gingival Tissues of Patients With Chronic Periodontitis. J. Periodontol. 2015, 86, 1380–1385. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, R.M.; Taganov, K.D.; Boldin, M.P.; Cheng, G.; Baltimore, D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc. Natl. Acad. Sci. USA 2007, 104, 1604–1609. [Google Scholar] [CrossRef] [PubMed]
- Taganov, K.D.; Boldin, M.P.; Chang, K.-J.; Baltimore, D. NF- B-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. USA 2006, 103, 12481–12486. [Google Scholar] [CrossRef] [PubMed]
- Tili, E.; Michaille, J.-J.; Cimino, A.; Costinean, S.; Dumitru, C.D.; Adair, B.; Fabbri, M.; Alder, H.; Liu, C.G.; Calin, G.A.; et al. Modulation of miR-155 and miR-125b Levels following Lipopolysaccharide/TNF-α Stimulation and Their Possible Roles in Regulating the Response to Endotoxin Shock. J. Immunol. 2007, 179, 5082–5089. [Google Scholar] [CrossRef] [PubMed]
- Akkouch, A.; Zhu, M.; Romero-Bustillos, M.; Eliason, S.; Qian, F.; Salem, A.K.; Amendt, B.A.; Hong, L. MicroRNA-200c Attenuates Periodontitis by Modulating Proinflammatory and Osteoclastogenic Mediators. Stem Cells Dev. 2019, 28, 1026–1036. [Google Scholar] [CrossRef]
- Ogata, Y.; Matsui, S.; Kato, A.; Zhou, L.; Nakayama, Y.; Takai, H. MicroRNA expression in inflamed and noninflamed gingival tissues from Japanese patients. J. Oral Sci. 2014, 56, 253–260. [Google Scholar] [CrossRef]
- Wendlandt, E.B.; Graff, J.W.; Gioannini, T.L.; McCaffrey, A.P.; Wilson, M.E. The role of microRNAs miR-200b and miR-200c in TLR4 signaling and NF-κB activation. Innate Immun. 2012, 18, 846–855. [Google Scholar] [CrossRef]
- Zhou, W.; Su, L.; Duan, X.; Chen, X.; Hays, A.; Upadhyayula, S.; Shivde, J.; Wang, H.; Li, Y.; Huang, D.; et al. MicroRNA-21 down-regulates inflammation and inhibits periodontitis. Mol. Immunol. 2018, 101, 608–614. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.-M.; Splinter, P.L.; O’Hara, S.P.; LaRusso, N.F. A Cellular Micro-RNA, let-7i, Regulates Toll-like Receptor 4 Expression and Contributes to Cholangiocyte Immune Responses against Cryptosporidium parvum Infection. J. Biol. Chem. 2007, 282, 28929–28938. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, A.; Mukherjee, B.; Dixit, M. MicroRNA Key to Angiogenesis Regulation: MiRNA Biology and Therapy. Curr. Cancer Drug Targets 2018, 18, 266–277. [Google Scholar] [CrossRef] [PubMed]
- Şaştım, Ç.Y.; Gürsoy, M.; Könönen, E.; Kasurinen, A.; Norvio, S.; Gürsoy, U.K.; Doğan, B. Salivary and serum markers of angiogenesis in periodontitis in relation to smoking. Clin. Oral Investig. 2021, 25, 1117–1126. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.H.; Na, H.S.; Jeong, S.Y.; Jeong, S.H.; Park, H.R.; Chung, J. Comparison of inflammatory microRNA expression in healthy and periodontitis tissues. Biocell 2011, 35, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Venugopal, P.; Koshy, T.; Lavu, V.; Rao, S.R.; Ramasamy, S.; Hariharan, S.; Venkatesan, V. Differential expression of microRNAs let-7a, miR-125b, miR-100, and miR-21 and interaction with NF-kB pathway genes in periodontitis pathogenesis. J. Cell. Physiol. 2018, 233, 5877–5884. [Google Scholar] [CrossRef] [PubMed]
- Perri, R.; Nares, S.; Zhang, S.; Barros, S.; Offenbacher, S. MicroRNA Modulation in Obesity and Periodontitis. J. Dent. Res. 2011, 91, 33–38. [Google Scholar] [CrossRef]
- Amaral, S.A.; Pereira, T.S.F.; Brito, J.A.R.; Cortelli, S.C.; Cortelli, J.R.; Gomez, R.S.; Costa, F.O.; Cota, L.O.M. Comparison of miRNA expression profiles in individuals with chronic or aggressive periodontitis. Oral Dis. 2018, 25, 561–568. [Google Scholar] [CrossRef] [PubMed]
- Tomofuji, T.; Yoneda, T.; Machida, T.; Ekuni, D.; Azuma, T.; Kataoka, K.; Maruyama, T.; Morita, M. Micro RNA s as serum biomarkers for periodontitis. J. Clin. Periodontol. 2016, 43, 418–425. [Google Scholar] [CrossRef]
- Aziz, F. The emerging role of miR-223 as novel potential diagnostic and therapeutic target for inflammatory disorders. Cell. Immunol. 2016, 303, 1–6. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, X. Downregulated miR-203 attenuates IL-β, IL-6, and TNF-α activation in TRAF6-treated human renal mesangial and tubular epithelial cells. Int. J. Clin. Exp. Pathol. 2020, 13, 324–331. [Google Scholar] [PubMed]
- Meyle, J.; Chapple, I. Molecular aspects of the pathogenesis of periodontitis. Periodontology 2000 2015, 69, 7–17. [Google Scholar] [CrossRef] [PubMed]
- Bellavia, D.; de Luca, A.; Carina, V.; Costa, V.; Raimondi, L.; Salamanna, F.; Alessandro, R.; Fini, M.; Giavaresi, G. Deregulated miRNAs in bone health: Epigenetic roles in osteoporosis. Bone 2019, 122, 52–75. [Google Scholar] [CrossRef]
- Loo, W.T.Y.; Jin, L.; Cheung, M.N.B.; Wang, M.; Chow, L.W.C. Epigenetic change in e-cardherin and COX-2 to predict chronic periodontitis. J. Transl. Med. 2010, 8, 110. [Google Scholar] [CrossRef] [PubMed]
- Choi, B.K.; Paster, B.J.; Dewhirst, F.E.; Göbel, U.B. Diversity of cultivable and uncultivable oral spirochetes from a patient with severe destructive periodontitis. Infect. Immun. 1994, 62, 1889–1895. [Google Scholar] [CrossRef]
- Kramer, C.D.; Genco, C.A. Microbiota, Immune Subversion, and Chronic Inflammation. Front. Immunol. 2017, 8, 255. [Google Scholar] [CrossRef] [PubMed]
- Tonetti, M.S.; Jepsen, S.; Jin, L.; Otomo-Corgel, J. Impact of the global burden of periodontal diseases on health, nutrition and wellbeing of mankind: A call for global action. J. Clin. Periodontol. 2017, 44, 456–462. [Google Scholar] [CrossRef]
- Isola, G.; Polizzi, A.; Santonocito, S.; Alibrandi, A.; Williams, R.C. Periodontitis activates the NLRP3 inflammasome in serum and saliva. J. Periodontol. 2021. [Google Scholar] [CrossRef]
- Isola, G.; Polizzi, A.; Alibrandi, A.; Williams, R.C.; Lo Giudice, A. Analysis of galectin-3 levels as a source of coronary heart disease risk during periodontitis. J. Periodontal Res. 2021, 56, 597–605. [Google Scholar] [CrossRef]
- Radović, N.; Jakoba, N.N.; Petrović, N.; Milosavljević, A.; Brković, B.; Roganović, J. MicroRNA-146a and microRNA-155 as novel crevicular fluid biomarkers for periodontitis in non-diabetic and type 2 diabetic patients. J. Clin. Periodontol. 2018, 45, 663–671. [Google Scholar] [CrossRef]
- Yagnik, K.; Mahendra, J.; Kurian, V.M. The Periodontal-Cardiovascular alliance: Evaluation of miRNA-146a in subgingival plaque samples of chronic periodontitis patients with and without coronary heart disease. J. Investig. Clin. Dent. 2019, 10, e12442. [Google Scholar] [CrossRef] [PubMed]
- Giannini, L.; Galbiati, G.; Rosso, G.L.; Maspero, C.; Farronato, G. Considerazioni sul trattamento ortodontico nei pazienti affetti da diabete mellito: Revisione della letteratura. Dent. Cadmos 2015, 83, 382–389. [Google Scholar] [CrossRef]
- Tsai, C.; Hayes, C.; Taylor, G.W. Glycemic control of type 2 diabetes and severe periodontal disease in the US adult population. Community Dent. Oral Epidemiol. 2002, 30, 182–192. [Google Scholar] [CrossRef] [PubMed]
- Polak, D.; Shapira, L. An update on the evidence for pathogenic mechanisms that may link periodontitis and diabetes. J. Clin. Periodontol. 2018, 45, 150–166. [Google Scholar] [CrossRef] [PubMed]
- Graziani, F.; Gennai, S.; Solini, A.; Petrini, M. A systematic review and meta-analysis of epidemiologic observational evidence on the effect of periodontitis on diabetes An update of the EFP-AAP review. J. Clin. Periodontol. 2018, 45, 167–187. [Google Scholar] [CrossRef]
- Monea, A.; Mezei, T.; Popsor, S.; Monea, M. Oxidative Stress: A Link between Diabetes Mellitus and Periodontal Disease. Int. J. Endocrinol. 2014, 2014, 917631. [Google Scholar] [CrossRef]
- Patil, V.P. Chronic Periodontitis in Type 2 Diabetes Mellitus: Oxidative Stress as a Common Factor in Periodontal Tissue Injury. J. Clin. Diagn. Res. 2016, 10, BC12. [Google Scholar] [CrossRef] [PubMed]
- Keating, S.T.; Plutzky, J.; El-Osta, A. Epigenetic Changes in Diabetes and Cardiovascular Risk. Circ. Res. 2016, 118, 1706–1722. [Google Scholar] [CrossRef]
- Ma, X.; Buscaglia, L.E.B.; Barker, J.R.; Li, Y. MicroRNAs in NF-κB signaling. J. Mol. Cell Biol. 2011, 3, 159–166. [Google Scholar] [CrossRef]
- Ji, G.; Lv, K.; Chen, H.; Wang, T.; Wang, Y.; Zhao, D.; Qu, L.; Li, Y. MiR-146a Regulates SOD2 Expression in H2O2 Stimulated PC12 Cells. PLoS ONE 2013, 8, e69351. [Google Scholar] [CrossRef]
- Elazazy, O.; Amr, K.; El Fattah, A.A.; Abouzaid, M. Evaluation of serum and gingival crevicular fluid microRNA-223, microRNA-203 and microRNA-200b expression in chronic periodontitis patients with and without diabetes type 2. Arch. Oral Biol. 2021, 121, 104949. [Google Scholar] [CrossRef]
- Chen, Z.; Yu, T.; Cabay, R.J.; Jin, Y.; Mahjabeen, I.; Luan, X.; Huang, L.; Dai, Y.; Zhou, X. miR-486-3p, miR-139-5p, and miR-21 as Biomarkers for the Detection of Oral Tongue Squamous Cell Carcinoma. Biomark. Cancer 2017, 9, 1179299X1700900001. [Google Scholar] [CrossRef]
- Kalea, A.; Hoteit, R.; Suvan, J.; Lovering, R.; Palmen, J.; Cooper, J.; Khodiyar, V.; Harrington, Z.; Humphries, S.; D’Aiuto, F. Upregulation of Gingival Tissue miR-200b in Obese Periodontitis Subjects. J. Dent. Res. 2015, 94, 59S–69S. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, N.; Tanaka, T. Role of miRNAs in the pathogenesis and susceptibility of diabetes mellitus. J. Hum. Genet. 2016, 62, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Loos, B.G.; Papantonopoulos, G.; Jepsen, S.; Laine, M.L. What is the Contribution of Genetics to Periodontal Risk? Dent. Clin. N. Am. 2015, 59, 761–780. [Google Scholar] [CrossRef] [PubMed]
- Newton, J.T.; Asimakopoulou, K. Managing oral hygiene as a risk factor for periodontal disease: A systematic review of psychological approaches to behaviour change for improved plaque control in periodontal management. J. Clin. Periodontol. 2015, 42, S36–S46. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Yu, T.; Ma, C.; Wang, Y.; Xie, B.; Xuan, D.; Zhang, J. Macrophages Play a Key Role in the Obesity-Induced Periodontal Innate Immune Dysfunction via Nucleotide-Binding Oligomerization Domain-Like Receptor Protein 3 Pathway. J. Periodontol. 2016, 87, 1195–1205. [Google Scholar] [CrossRef]
- Chu, X.; Newman, J.; Park, B.; Nares, S.; Ordonez, G.; Iacopino, A. In Vitro alteration of macrophage phenotype and function by serum lipids. Cell Tissue Res. 1999, 296, 331–337. [Google Scholar] [CrossRef]
- Tomikawa, K.; Yamamoto, T.; Shiomi, N.; Shimoe, M.; Hongo, S.; Yamashiro, K.; Yamaguchi, T.; Maeda, H.; Takashiba, S. Smad2 Decelerates Re-epithelialization during Gingival Wound Healing. J. Dent. Res. 2012, 91, 764–770. [Google Scholar] [CrossRef]
- Paraskevi, A.; Theodoropoulos, G.; Papaconstantinou, I.; Mantzaris, G.; Nikiteas, N.; Gazouli, M. Circulating MicroRNA in inflammatory bowel disease. J. Crohn’s Colitis 2012, 6, 900–904. [Google Scholar] [CrossRef]
- Tryndyak, V.P.; Latendresse, J.R.; Montgomery, B.; Ross, S.A.; Beland, F.A.; Rusyn, I.; Pogribny, I.P. Plasma microRNAs are sensitive indicators of inter-strain differences in the severity of liver injury induced in mice by a choline- and folate-deficient diet. Toxicol. Appl. Pharmacol. 2012, 262, 52–59. [Google Scholar] [CrossRef] [PubMed]
- Alisi, A.; da Sacco, L.; Bruscalupi, G.; Piemonte, F.; Panera, N.; de Vito, R.; Leoni, S.; Bottazzo, G.F.; Masotti, A.; Nobili, V. Mirnome analysis reveals novel molecular determinants in the pathogenesis of diet-induced nonalcoholic fatty liver disease. Lab. Investig. 2010, 91, 283–293. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Zhang, R.; Zhang, Y.; Li, J.; Grossmann, R.; Zhao, R. In Ovo leptin administration affects hepatic lipid metabolism and microRNA expression in newly hatched broiler chickens. J. Anim. Sci. Biotechnol. 2012, 3, 16. [Google Scholar] [CrossRef] [PubMed]
- Nokhbehsaim, M.; Keser, S.; Nogueira, A.V.B.; Jäger, A.; Jepsen, S.; Cirelli, J.A.; Bourauel, C.; Eick, S.; Deschner, J. Leptin Effects on the Regenerative Capacity of Human Periodontal Cells. Int. J. Endocrinol. 2014, 2014, 180304. [Google Scholar] [CrossRef] [PubMed]
- Artese, L.; Piattelli, A.; Cardoso, L.A.D.G.; Ferrari, D.S.; Onuma, T.; Piccirilli, M.; Faveri, M.; Perrotti, V.; Simon, M.; Shibli, J.A. Immunoexpression of Angiogenesis, Nitric Oxide Synthase, and Proliferation Markers in Gingival Samples of Patients with Aggressive and Chronic Periodontitis. J. Periodontol. 2010, 81, 718–726. [Google Scholar] [CrossRef] [PubMed]
- Chan, Y.C.; Roy, S.; Khanna, S.; Sen, C.K. Downregulation of Endothelial MicroRNA-200b Supports Cutaneous Wound Angiogenesis By Desilencing GATA Binding Protein 2 and Vascular Endothelial Growth Factor Receptor 2. Arter. Thromb. Vasc. Biol. 2012, 32, 1372–1382. [Google Scholar] [CrossRef] [PubMed]
- Pols, M.S.; Klumperman, J. Trafficking and function of the tetraspanin CD63. Exp. Cell Res. 2009, 315, 1584–1592. [Google Scholar] [CrossRef]
- Edgar, J.R. Q&A: What are exosomes, exactly? BMC Biol. 2016, 14, 46. [Google Scholar] [CrossRef]
- Wang, Z.; Chen, J.-Q.; Liu, J.-L.; Tian, L. Exosomes in tumor microenvironment: Novel transporters and biomarkers. J. Transl. Med. 2016, 14, 297. [Google Scholar] [CrossRef]
- Wan, M.; Ning, B.; Spiegel, S.; Lyon, C.J.; Hu, T.Y. Tumor-derived exosomes (TDEs): How to avoid the sting in the tail. Med. Res. Rev. 2020, 40, 385–412. [Google Scholar] [CrossRef]
- Tan, L.; Wu, H.; Liu, Y.; Zhao, M.; Li, D.; Lu, Q. Recent advances of exosomes in immune modulation and autoimmune diseases. Autoimmunity 2016, 49, 357–365. [Google Scholar] [CrossRef] [PubMed]
- Graves, D.; Cochran, D. The Contribution of Interleukin-1 and Tumor Necrosis Factor to Periodontal Tissue Destruction. J. Periodontol. 2003, 74, 391–401. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Dai, W.; Wang, H.; Xue, C.; Feng, J.; He, Y.; Wang, P.; Li, S.; Bai, D.; Shu, R. Periodontal ligament fibroblasts regulate osteoblasts by exosome secretion induced by inflammatory stimuli. Arch. Oral Biol. 2019, 105, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Dong, C.; Yang, J.; Jin, Y.; Zheng, W.; Zhou, Q.; Liang, Y.; Bao, L.; Feng, G.; Ji, J.; et al. Exosomal microRNA-155-5p from PDLSCs regulated Th17/Treg balance by targeting sirtuin-1 in chronic periodontitis. J. Cell. Physiol. 2019, 234, 20662–20674. [Google Scholar] [CrossRef] [PubMed]
- Fujimori, K.; Yoneda, T.; Tomofuji, T.; Ekuni, D.; Azuma, T.; Maruyama, T.; Mizuno, H.; Sugiura, Y.; Morita, M. Detection of Salivary miRNAs Reflecting Chronic Periodontitis: A Pilot Study. Molecules 2019, 24, 1034. [Google Scholar] [CrossRef] [PubMed]
- Saito, A.; Horie, M.; Ejiri, K.; Aoki, A.; Katagiri, S.; Maekawa, S.; Suzuki, S.; Kong, S.; Yamauchi, T.; Yamaguchi, Y.; et al. MicroRNA profiling in gingival crevicular fluid of periodontitis-a pilot study. FEBS Open Bio 2017, 7, 981–994. [Google Scholar] [CrossRef] [PubMed]
- Kamal, N.N.S.N.M.; Awang, R.A.R.; Mohamad, S.; Shahidan, W.N.S. Plasma- and Saliva Exosome Profile Reveals a Distinct MicroRNA Signature in Chronic Periodontitis. Front. Physiol. 2020, 11, 11. [Google Scholar] [CrossRef]
- Shin, H.; Oh, S.; Hong, S.; Kang, M.; Kang, D.; Ji, Y.-G.; Choi, B.H.; Kang, K.-W.; Jeong, H.; Park, Y.; et al. Early-Stage Lung Cancer Diagnosis by Deep Learning-Based Spectroscopic Analysis of Circulating Exosomes. ACS Nano 2020, 14, 5435–5444. [Google Scholar] [CrossRef]
- Palanisamy, V.; Sharma, S.; Deshpande, A.; Zhou, H.; Gimzewski, J.; Wong, D.T. Nanostructural and Transcriptomic Analyses of Human Saliva Derived Exosomes. PLoS ONE 2010, 5, e8577. [Google Scholar] [CrossRef]
- Han, Y.; Jia, L.; Zheng, Y.; Li, W. Salivary Exosomes: Emerging Roles in Systemic Disease. Int. J. Biol. Sci. 2018, 14, 633–643. [Google Scholar] [CrossRef]
- Huang, J.; Kang, B.; Qu, Y.; Mu, D. Protective effect of exosome on organs after ischemia-reperfusion injury. Zhongguo Xiu Fu Chong Jian Wai Ke Za Zhi 2017, 31, 751–754. [Google Scholar] [PubMed]
- Wu, P.; Zhang, B.; Shi, H.; Qian, H.; Xu, W. MSC-exosome: A novel cell-free therapy for cutaneous regeneration. Cytotherapy 2018, 20, 291–301. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Wu, X.; Zhang, X.; Sun, Y.; Yan, Y.; Shi, H.; Zhu, Y.; Wu, L.; Pan, Z.; Zhu, W.; et al. Human Umbilical Cord Mesenchymal Stem Cell Exosomes Enhance Angiogenesis Through the Wnt4/β-Catenin Pathway. Stem Cells Transl. Med. 2015, 4, 513–522. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Sun, R.; Wu, C.; Wang, L.; Zhang, C. Exosome: A Novel Approach to Stimulate Bone Regeneration through Regulation of Osteogenesis and Angiogenesis. Int. J. Mol. Sci. 2016, 17, 712. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Nasser, M.I.; Shen, J.; Qu, S.; He, Q.; Zhao, M. Functions of Exosomes in the Triangular Relationship between the Tumor, Inflammation, and Immunity in the Tumor Microenvironment. J. Immunol. Res. 2019, 2019, 4197829. [Google Scholar] [CrossRef]
- Gegout, P.-Y.; Stutz, C.; Olson, J.; Batool, F.; Petit, C.; Tenenbaum, H.; Benkirane-Jessel, N.; Huck, O. Interests of Exosomes in Bone and Periodontal Regeneration: A Systematic Review; Springer: New York, NY, USA, 2020; pp. 1–21. [Google Scholar]
- Polizzi, A.; Santonocito, S.; Vaccaro, M.; Fichera, G.; Torrisi, S.; Ronsivalle, V.; Palazzo, G.; Sicari, F.; Indelicato, F. Relationship between periodontitis and psychosocial impact in patients with systemic sclerosis: A clinical study. Mediterr. J. Clin. Psychol. 2020, 8. [Google Scholar] [CrossRef]
- Santonocito, S.; Ronsivalle, V.; Fichera, G.; Indelicato, F. Psychological impact and patient perception of occlusion and orthodontic treatment in periodontitis patients. Mediterr. J. Clin. Psychol. 2020, 8. [Google Scholar] [CrossRef]
- Kim, S.-H.; Lee, S.-Y.; Lee, Y.-M.; Lee, Y.-K. MicroRNAs as biomarkers for dental diseases. Singap. Dent. J. 2015, 36, 18–22. [Google Scholar] [CrossRef]
- García-Giménez, J.; Sanchis-Gomar, F.; Lippi, G.; Mena, S.; Ivars, D.; Gomez-Cabrera, M.; Viña, J.; Pallardó, F. Epigenetic biomarkers: A new perspective in laboratory diagnostics. Clin. Chim. Acta 2012, 413, 1576–1582. [Google Scholar] [CrossRef]
- Testa, U.; Pelosi, E.; Castelli, G.; Labbaye, C. miR-146 and miR-155: Two Key Modulators of Immune Response and Tumor Development. Non-Coding RNA 2017, 3, 22. [Google Scholar] [CrossRef]
- O’Brien, S.J.; Carter, J.V.; Burton, J.F.; Oxford, B.G.; Schmidt, M.N.; Hallion, J.C.; Galandiuk, S. The role of the miR-200 family in epithelial-mesenchymal transition in colorectal cancer: A systematic review. Int. J. Cancer 2018, 142, 2501–2511. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, Z.; Gemeinhart, R.A. Progress in microRNA delivery. J. Control. Release 2013, 172, 962–974. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Martin, P.; Fogarty, B.; Brown, A.; Schurman, K.; Phipps, R.; Yin, V.P.; Lockman, P.; Bai, S. Exosome Delivered Anticancer Drugs Across the Blood-Brain Barrier for Brain Cancer Therapy in Danio Rerio. Pharm. Res. 2015, 32, 2003–2014. [Google Scholar] [CrossRef] [PubMed]
- Mishra, S.; Yadav, T.; Rani, V. Exploring miRNA based approaches in cancer diagnostics and therapeutics. Crit. Rev. Oncol. 2016, 98, 12–23. [Google Scholar] [CrossRef] [PubMed]
- Xavier, J.R.; Thakur, T.; Desai, P.; Jaiswal, M.K.; Sears, N.; Cosgriff-Hernandez, E.; Kaunas, R.; Gaharwar, A.K. Bioactive Nanoengineered Hydrogels for Bone Tissue Engineering: A Growth-Factor-Free Approach. ACS Nano 2015, 9, 3109–3118. [Google Scholar] [CrossRef]
- Gaharwar, A.K.; Peppas, N.A.; Khademhosseini, A. Nanocomposite hydrogels for biomedical applications. Biotechnol. Bioeng. 2014, 111, 441–453. [Google Scholar] [CrossRef]
- Munagala, R.; Aqil, F.; Jeyabalan, J.; Gupta, R.C. Bovine milk-derived exosomes for drug delivery. Cancer Lett. 2016, 371, 48–61. [Google Scholar] [CrossRef]
- Tian, T.; Zhang, H.-X.; He, C.-P.; Fan, S.; Zhu, Y.-L.; Qi, C.; Huang, N.-P.; Xiao, Z.-D.; Lu, Z.-H.; Tannous, B.A.; et al. Surface functionalized exosomes as targeted drug delivery vehicles for cerebral ischemia therapy. Biomaterials 2018, 150, 137–149. [Google Scholar] [CrossRef]
- Chew, J.R.J.; Chuah, S.J.; Teo, K.Y.W.; Zhang, S.; Lai, R.C.; Fu, J.H.; Lim, L.P.; Lim, S.K.; Toh, W.S. Mesenchymal stem cell exosomes enhance periodontal ligament cell functions and promote periodontal regeneration. Acta Biomater. 2019, 89, 252–264. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, E.; Khalil, E.; Sabry, D. Effect of Adipose-Derived Stem Cells and Their Exo as Adjunctive Therapy to Nonsurgical Periodontal Treatment: A Histologic and Histomorphometric Study in Rats. Biomolecules 2018, 8, 167. [Google Scholar] [CrossRef]
| miRNAs | miRNA in Diseased Tissues | Functions | Reference |
|---|---|---|---|
| miRNA-548 | Upregulation | Upregulation of IL-8 within the periodontal pocket | [26,27] |
| miRNA-31 | Upregulation | Regulates the expression of ICAM-1, which controls the migration of leukocytes from the bloodstream to the tissues | [29] |
| miRNA-17 | Upregulation | Regulates the expression of E- Selectin, which controls the migration of leukocytes from the bloodstream to the tissues | [15] |
| miRNA146 | Upregulation | Negatively regulates the TLR signalling pathway | [30,31] |
| miRNA-146a | Upregulation | Negatively regulates TLR signalling; reduced expression of NF-κb, TNFα, IL-1β and IL-6, which induce osteoclastogenesis | [54] |
| miRNA-146b | Upregulation | Negatively regulates TLR signalling | [54] |
| miRNA-155 | Downregulation | Regulates TLR release in inflamed tissues | [54] |
| miRNA-200 | Upregulation | Reduces the release of IL-6, IL-8, IFRD1 and NF-κb | [38] |
| miRNA-200c | Upregulation | Regulatory effect on TLR4-mediated signalling in macrophages | [39] |
| miRNA-21 | Upregulation | Decreases NF-κb activation | [41] |
| miRNA-let-7 | Upregulation | Inhibits TLR4 | [42] |
| miRNA-203 | Downregulation | Promotes neo-angiogenesis and regulates innate immunity | [10,28] |
| miRNA-223 | Upregulation | Plays a role in alveolar bone loss | [48,50] |
| mRNAs | Correlation with Systemic Disease | Activity | Expression | References |
|---|---|---|---|---|
| mRNA-146a | Heart disease | Chronic inflammatory disorders, both cardiac and periodontal diseases. Acts against Th1 and Th2 cells, shifting the balance towards Th1. | Upregulation in tissues of patient with periodontal and heart diseases | [30] |
| mRNA-let7 | Heart disease | Inhibition of angiogenesis through up-regulation of TSP-1. | Upregulation in tissues of patient with periodontal and heart diseases. | [43,45] |
| mRNA-146 | Diabetes | Negative feedback control of NFκB target genes and are involved in oxidative stress by targeting SOD. | Overexpression in crevicular fluid of diabetic patients with periodontitis. | [60] |
| mRNA-155 | Diabetes | Negative feedback control of NFκB target genes and are involved in oxidative stress by targeting SOD. | Overexpression in crevicular fluid of diabetic patients with periodontitis. | [60] |
| mRNA-223 | Diabetes | Increased expression of TNF-α. | Upregulation in tissue, crevicular fluid and serum in patients with diabetes and periodontitis. | [71] |
| mRNA-203 | Diabetes | Reduced expression of TNF-α. | Downregulation in tissue, crevicular fluid and serum in patients with diabetes and periodontitis | [71] |
| mRNA-200-3p | Diabetes | Increased expression of TNF-α. | Upregulation in tissue, crevicular fluid and serum in patients with diabetes and periodontitis. | [71] |
| mRNA-200b-5p | Obesity | Reduced expression of its target genes ZEB1, ZEB2, GATA2 and KDR involved in re-epithelisation. | Upregulation in tissue of obese patients with periodontitis. | [73] |
| mRNA-200b/c | Obesity | Alters TLR4 signalling in macrophages. | Upregulation in tissue of obese patients with periodontitis. | [40] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santonocito, S.; Polizzi, A.; Palazzo, G.; Isola, G. The Emerging Role of microRNA in Periodontitis: Pathophysiology, Clinical Potential and Future Molecular Perspectives. Int. J. Mol. Sci. 2021, 22, 5456. https://doi.org/10.3390/ijms22115456
Santonocito S, Polizzi A, Palazzo G, Isola G. The Emerging Role of microRNA in Periodontitis: Pathophysiology, Clinical Potential and Future Molecular Perspectives. International Journal of Molecular Sciences. 2021; 22(11):5456. https://doi.org/10.3390/ijms22115456
Chicago/Turabian StyleSantonocito, Simona, Alessandro Polizzi, Giuseppe Palazzo, and Gaetano Isola. 2021. "The Emerging Role of microRNA in Periodontitis: Pathophysiology, Clinical Potential and Future Molecular Perspectives" International Journal of Molecular Sciences 22, no. 11: 5456. https://doi.org/10.3390/ijms22115456
APA StyleSantonocito, S., Polizzi, A., Palazzo, G., & Isola, G. (2021). The Emerging Role of microRNA in Periodontitis: Pathophysiology, Clinical Potential and Future Molecular Perspectives. International Journal of Molecular Sciences, 22(11), 5456. https://doi.org/10.3390/ijms22115456







