Understanding LAG-3 Signaling
Abstract
1. Molecular Characterization of LAG-3
1.1. Function
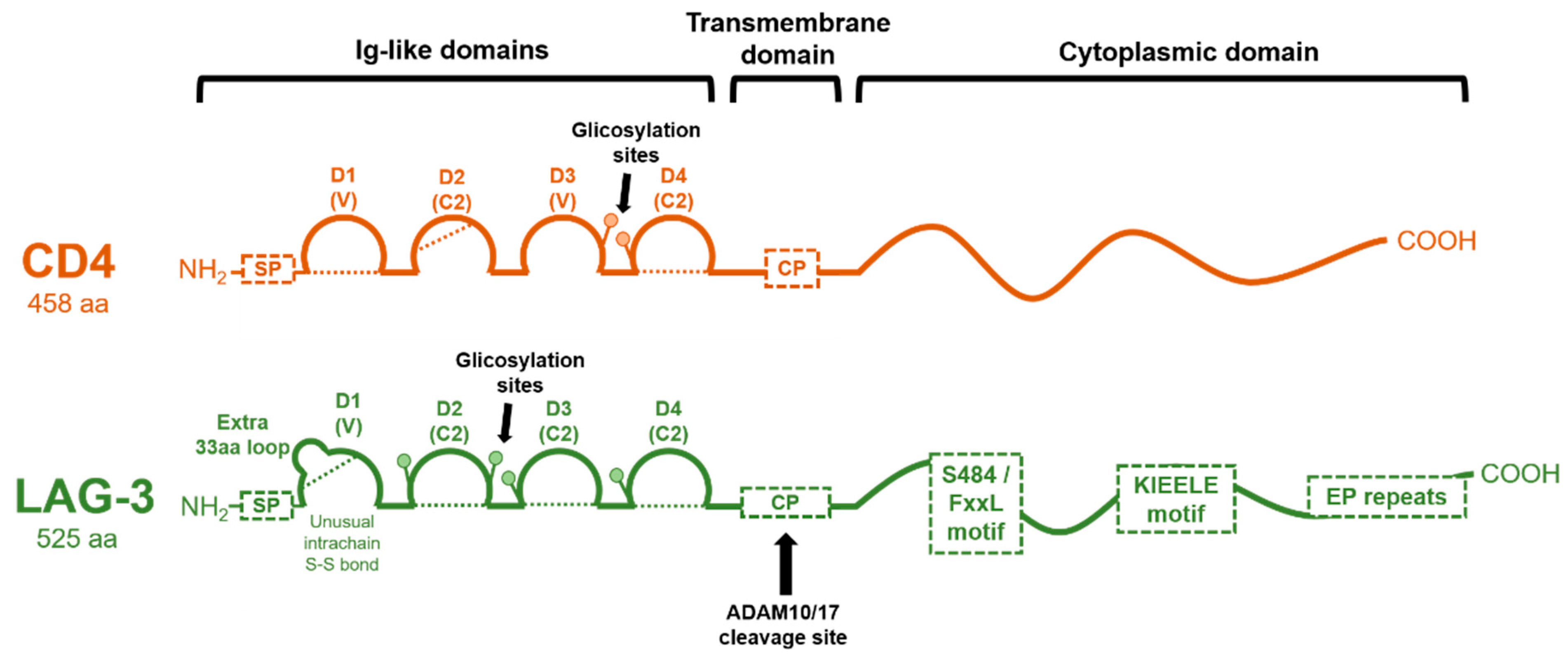
1.2. Genetic Structure of the LAG-3 Gene and Domain Organization
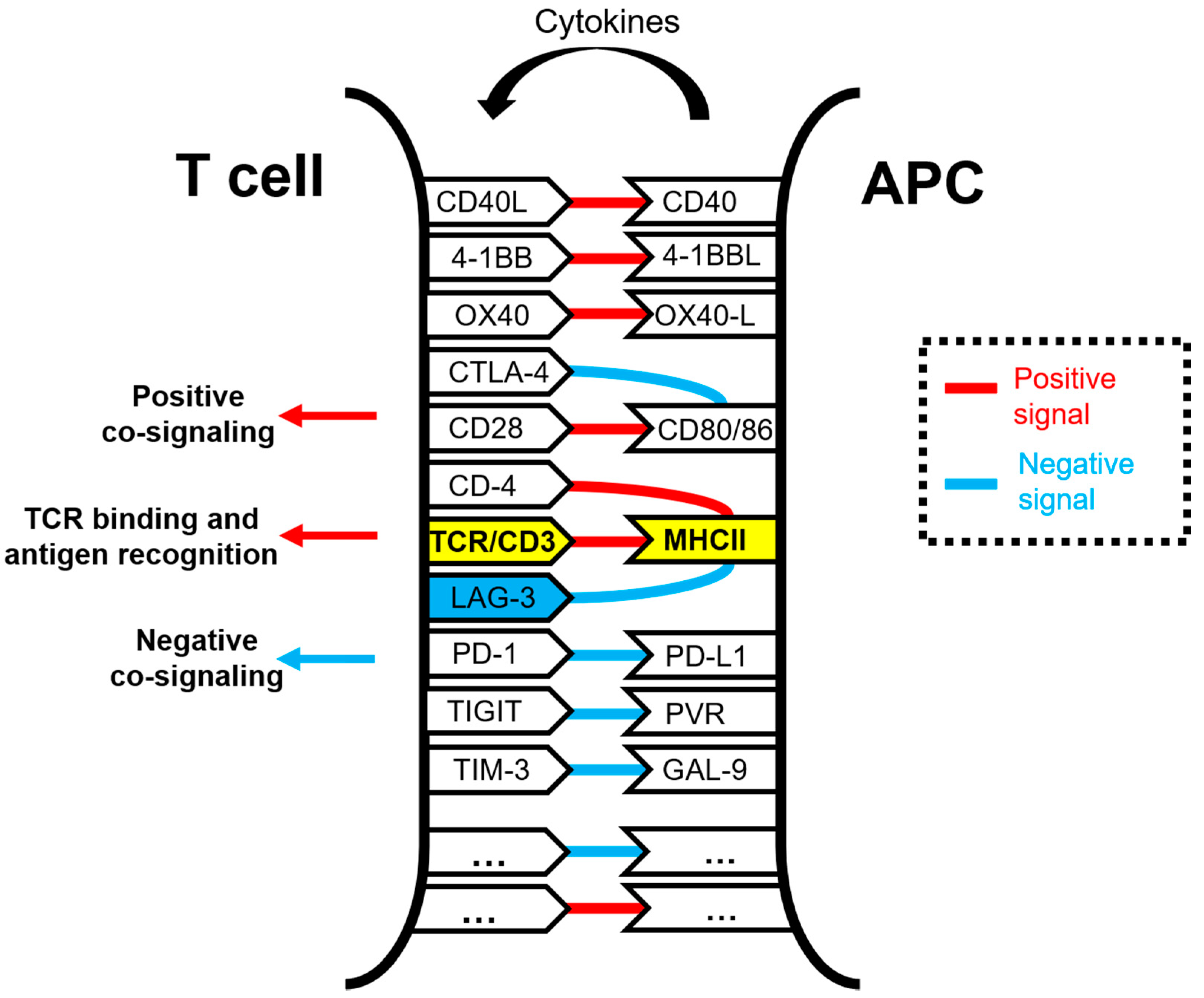
1.3. LAG-3 Ligands
2. Regulation of LAG-3 Expression
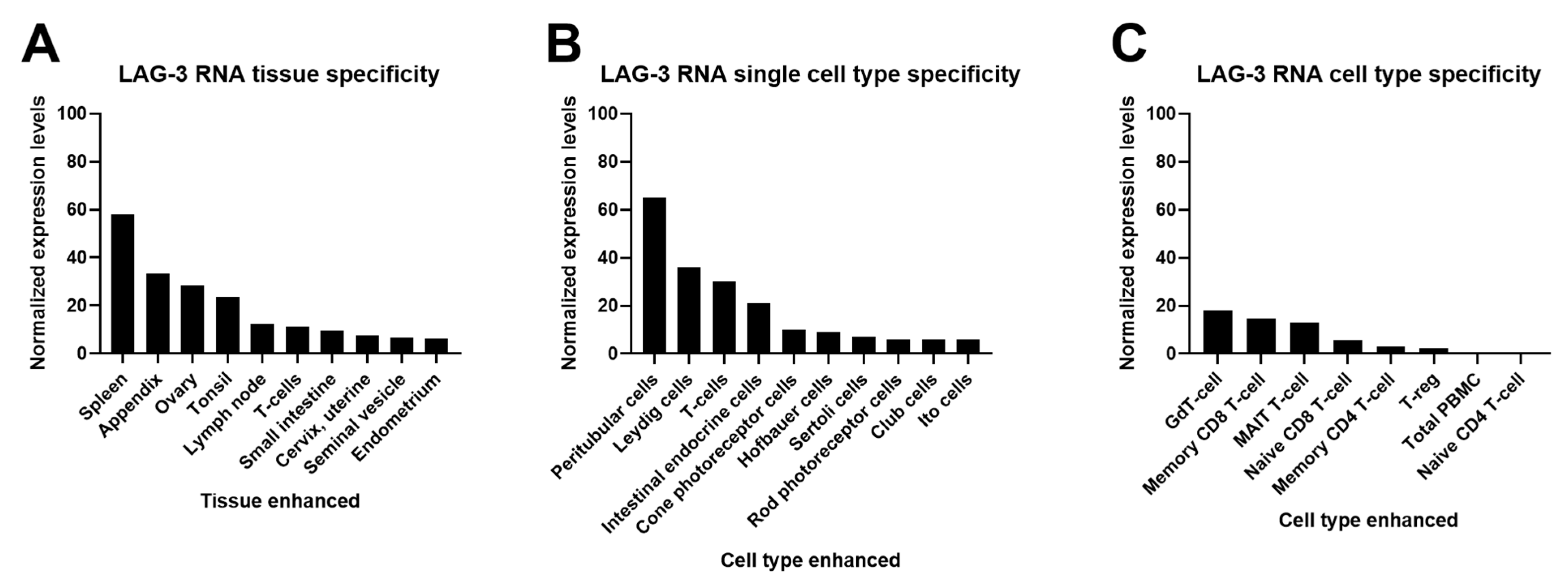
2.1. Cell and Tissue Expression
2.2. Genetic and Epigenetic Regulation of LAG-3 Expression
3. LAG-3 in Disease
3.1. Cancer
3.2. Parkinson’s Disease
3.3. Cardiovascular Diseases
3.4. HDL Hypercholesterolemia
3.5. Inflammatory Bowel Disease
3.6. Multiple Sclerosis
3.7. Diabetes Mellitus
3.8. LAG-3 in Infection
4. Clinical Landscape of LAG-3-Targeted Therapy
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Huard, B.; Tournier, M.; Hercend, T.; Triebel, F.; Faure, F. Lymphocyte-Activation Gene 3/Major Histocompatibility Complex Class II Interaction Modulates the Antigenic Response of CD4+ T Lymphocytes. Eur. J. Immunol. 1994, 24, 3216–3221. [Google Scholar] [CrossRef] [PubMed]
- Huard, B.; Prigent, P.; Pagès, F.; Bruniquel, D.; Triebel, F. T Cell Major Histocompatibility Complex Class II Molecules Down-Regulate CD4+ T Cell Clone Responses Following LAG-3 Binding. Eur. J. Immunol. 1996, 26, 1180–1186. [Google Scholar] [CrossRef] [PubMed]
- Workman, C.J.; Dugger, K.J.; Vignali, D.A.A. Cutting Edge: Molecular Analysis of the Negative Regulatory Function of Lymphocyte Activation Gene-3. J. Immunol. 2002, 169, 5392–5395. [Google Scholar] [CrossRef]
- Workman, C.J.; Rice, D.S.; Dugger, K.J.; Kurschner, C.; Vignali, D.A.A. Phenotypic Analysis of the Murine CD4-Related Glycoprotein, CD223 (LAG-3). Eur. J. Immunol. 2002, 32, 2255–2263. [Google Scholar] [CrossRef]
- Workman, C.J.; Cauley, L.S.; Kim, I.-J.; Blackman, M.A.; Woodland, D.L.; Vignali, D.A.A. Lymphocyte Activation Gene-3 (CD223) Regulates the Size of the Expanding T Cell Population Following Antigen Activation in vivo. J. Immunol. 2004, 172, 5450–5455. [Google Scholar] [CrossRef]
- Maçon-Lemaître, L.; Triebel, F. The Negative Regulatory Function of the Lymphocyte-Activation Gene-3 Co-Receptor (CD223) on Human T Cells. Immunology 2005, 115, 170–178. [Google Scholar] [CrossRef]
- Andrews, L.P.; Marciscano, A.E.; Drake, C.G.; Vignali, D.A.A. LAG3 (CD223) as a Cancer Immunotherapy Target. Immunol. Rev. 2017, 276, 80–96. [Google Scholar] [CrossRef]
- Triebel, F.; Jitsukawa, S.; Baixeras, E.; Roman-Roman, S.; Genevee, C.; Viegas-Pequignot, E.; Hercend, T. LAG-3, a Novel Lymphocyte Activation Gene Closely Related to CD4. J. Exp. Med. 1990, 171, 1393–1405. [Google Scholar] [CrossRef]
- Baixeras, B.E.; Huard, B.; Miossec, C.; Jitsukawa, S.; Martin, M.; Hercend, T.; Auffray, C.; Triebel, F.; Tonneau-Piatier, D. Characterization of the Lymphocyte Activation Gene 3-Encoded Protein. A New Ligand for Human Leukoc3~ Antigen Class H Antigens. Pharm. Res. 1992, 176, 327–337. [Google Scholar]
- Annunziato, F.; Manetti, R.; Tomasévic, I.; Giudizi, M.; Biagiotti, R.; Giannò, V.; Germano, P.; Mavilia, C.; Maggi, E.; Romagnani, S. Expression and Release of LAG-3-Encoded Protein by Human CD4 + T Cells Are Associated with IFN-Γ Production. FASEB J. 1996, 10, 769–776. [Google Scholar] [CrossRef]
- Avice, M.N.; Sarfati, M.; Triebel, F.; Delespesse, G.; Demeure, C.E. Lymphocyte Activation Gene-3, a MHC Class II Ligand Expressed on Activated T Cells, Stimulates TNF-Alpha and IL-12 Production by Monocytes and Dendritic Cells. J. Immunol. 1999, 162, 2748–2753. [Google Scholar]
- Slevin, S.M.; Garner, L.C.; Lahiff, C.; Tan, M.; Wang, L.M.; Ferry, H.; Greenaway, B.; Lynch, K.; Geremia, A.; Hughes, S.; et al. Lymphocyte Activation Gene (LAG)-3 Is Associated with Mucosal Inflammation and Disease Activity in Ulcerative Colitis. J. Crohn’s Colitis. 2020, 14, 1446–1461. [Google Scholar] [CrossRef] [PubMed]
- Hannier, S.; Tournier, M.; Bismuth, G.; Triebel, F. CD3/TCR Complex-Associated Lymphocyte Activation Gene-3 Molecules Inhibit CD3/TCR Signaling. J. Immunol. 1998, 161, 4058–4065. [Google Scholar] [PubMed]
- Blackburn, S.D.; Shin, H.; Haining, W.N.; Zou, T.; Workman, C.J.; Polley, A.; Betts, M.R.; Freeman, G.J.; Vignali, D.A.A.; Wherry, E.J. Coregulation of CD8+ T Cell Exhaustion by Multiple Inhibitory Receptors During Chronic Viral Infection. Nat. Immunol. 2009, 10, 29–37. [Google Scholar] [CrossRef]
- Chihara, N.; Madi, A.; Kondo, T.; Zhang, H.; Acharya, N.; Singer, M.; Nyman, J.; Marjanovic, N.D.; Kowalczyk, M.S.; Wang, C.; et al. Induction and Transcriptional Regulation of the Co-Inhibitory Gene Module in T Cells. Nature 2018, 558, 454–459. [Google Scholar] [CrossRef]
- Grosso, J.F.; Kelleher, C.C.; Harris, T.J.; Maris, C.H.; Hipkiss, E.L.; De Marzo, A.; Anders, R.; Netto, G.; Derese, G.; Bruno, T.C.; et al. LAG-3 Regulates CD8 + T Cell Accumulation and Effector Function in Murine Self—and Tumor-Tolerance Systems. J. Clin. Invest. 2007, 117, 3383–3392. [Google Scholar] [CrossRef] [PubMed]
- Grosso, J.F.; Goldberg, M.V.; Getnet, D.; Bruno, T.C.; Yen, H.-R.; Pyle, K.J.; Hipkiss, E.; Vignali, D.A.A.; Pardoll, D.M.; Drake, C.G. Functionally Distinct LAG-3 and PD-1 Subsets on Activated and Chronically Stimulated CD8 T Cells. J. Immunol. 2009, 182, 6659–6669. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.-Y.; Eppolito, C.; Lele, S.; Shrikant, P.; Matsuzaki, J.; Odunsi, K. LAG3 And PD1 Co-Inhibitory Molecules Collaborate to Limit CD8+ T Cell Signaling and Dampen Antitumor Immunity in a Murine Ovarian Cancer Model. Oncotarget 2015, 6, 27359. [Google Scholar] [CrossRef] [PubMed]
- Williams, J.B.; Horton, B.L.; Zheng, Y.; Duan, Y.; Powell, J.D.; Gajewski, T.F. The EGR2 Targets LAG-3 and 4-1BB Describe and Regulate Dysfunctional Antigen-Specific CD8 + T Cells in the Tumor Microenvironment. J. Exp. Med. 2017, 214, 381–400. [Google Scholar] [CrossRef]
- Huard, B.; Mastrangeli, R.; Prigent, P.; Bruniquel, D.; Donini, S.; El-Tayar, N.; Maigret, B.; Dreano, M.; Triebel, F. Characterization of the Major Histocompatibility Complex Class II Binding Site on LAG-3 Protein. Proc. Natl. Acad. Sci. USA 1997, 94, 5744–5749. [Google Scholar] [CrossRef]
- Li, N.; Workman, C.J.; Martin, S.M.; Vignali, D.A.A. Biochemical Analysis of the Regulatory T Cell Protein Lymphocyte Activation Gene-3 (LAG-3; CD223). J. Immunol. 2004, 173, 6806–6812. [Google Scholar] [CrossRef]
- Li, N.; Wang, Y.; Forbes, K.; Vignali, K.M.; Heale, B.S.; Saftig, P.; Hartmann, D.; Black, R.A.; Rossi, J.J.; Blobel, C.P.; et al. Metalloproteases Regulate T-Cell Proliferation and Effector Function Via LAG-3. EMBO J. 2007, 26, 494–504. [Google Scholar] [CrossRef] [PubMed]
- Mastrangeli, R.; Micangeli, E.; Donini, S. Cloning of Murine LAG-3 by Magnetic Bead Bound Homologous Probes and PCR (GENE-CAPTURE PCR). Anal. Biochem. 1996, 241, 93–102. [Google Scholar] [CrossRef] [PubMed]
- Workman, C.J.; Vignali, D.A.A. The CD4-Related Molecule, LAG-3 (CD223), Regulates the Expansion of Activated T Cells. Eur. J. Immunol. 2003, 3, 970–979. [Google Scholar] [CrossRef]
- Maeda, T.K.; Sugiura, D.; Okazaki, M.T.; Okazaki, T. Atypical Motifs in the Cytoplasmic Region of the Inhibitory Immune Co-Receptor LAG-3 Inhibit T Cell Activation. J. Biol. Chem. 2019, 294, 6017–6026. [Google Scholar] [CrossRef] [PubMed]
- Iouzalen, N.; Andreae, S.; Hannier, S.; Triebel, F. LAP, A Lymphocyte Activation Gene-3 (LAG-3)-Associated Protein that Binds to a Repeated EP Motif in the Intracellular Region of LAG-3, May Participate in the Down-Regulation of the CD3/TCR Activation Pathway. Eur. J. Immunol. 2001, 31, 2885–2891. [Google Scholar] [CrossRef]
- Bae, J.; Lee, S.J.; Park, C.-G.; Lee, Y.S.; Chun, T. Trafficking of LAG-3 to the Surface on Activated T Cells via Its Cytoplasmic Domain and Protein Kinase C Signaling. J. Immunol. 2014, 193, 3101–3112. [Google Scholar] [CrossRef]
- Long, L.; Zhang, X.; Chen, F.; Pan, Q.; Phiphatwatchara, P. The Promising Immune Checkpoint LAG-3: From Tumor Microenvironment to Cancer Immunotherapy. Genes Cancer 2018, 9, 176. [Google Scholar] [CrossRef] [PubMed]
- Huard, B.; Prigent, P.; Tournier, M.; Bruniquel, D.; Triebel, F. CD4/Major Histocompatibility Complex Class II Interaction Analyzed with CD4—and Lymphocyte Activation Gene-3 (LAG-3)-Ig Fusion Proteins. Eur. J. Immunol. 1995, 25, 2718–2721. [Google Scholar] [CrossRef] [PubMed]
- Hemon, P.; Jean-Louis, F.; Ramgolam, K.; Brignone, C.; Viguier, M.; Bachelez, H.; Triebel, F.; Charron, D.; Aoudjit, F.; Al-Daccak, R.; et al. MHC Class II Engagement by Its Ligand LAG-3 (CD223) Contributes to Melanoma Resistance to Apoptosis. J. Immunol. 2011, 186, 5173–5183. [Google Scholar] [CrossRef]
- Donia, M.; Andersen, R.; Kjeldsen, J.W.; Fagone, P.; Munir, S.; Nicoletti, F.; Andersen, M.H.; Straten, P.T.; Svane, I.M. Aberrant Expression of MHC Class II in Melanoma Attracts Inflammatory Tumor-Specific CD4+T-Cells, which Dampen CD8+T-Cell Antitumor Reactivity. Cancer Res. 2015, 75, 3747–3759. [Google Scholar] [CrossRef] [PubMed]
- Chung, L.Y.; Tang, S.J.; Wu, Y.C.; Sun, G.H.; Liu, H.Y.; Sun, K.H. Galectin-3 Augments Tumor Initiating Property and Tumorigenicity pf Lung Cancer through Interaction with Β-Catenin. Oncotarget 2015, 6, 4936–4952. [Google Scholar] [CrossRef]
- Lu, W.; Wang, J.; Yang, G.; Yu, N.; Huang, Z.; Xu, H.; Li, J.; Qiu, J.; Zeng, X.; Chen, S.; et al. Posttranscriptional Regulation of Galectin-3 by miR—128 Contributes to Colorectal Cancer Progression. Oncotarget 2017, 8, 15242–15251. [Google Scholar] [CrossRef]
- Li, M.; Feng, Y.M.; Fang, S.Q. Overexpression of Ezrin and Galectin-3 as Predictors of Poor Prognosis of Cervical Cancer. Braz. J. Med. Biol. Res. 2017, 50. [Google Scholar] [CrossRef] [PubMed]
- Kouo, T.; Huang, L.; Pucsek, A.B.; Cao, M.; Solt, S.; Armstrong, T.; Jaffee, E. Galectin-3 Shapes Antitumor Immune Responses by Suppressing CD8+ T Cells via LAG-3 and Inhibiting Expansion of Plasmacytoid Dendritic Cells. Cancer Immunol. Res. 2015, 3, 412–423. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Sanmamed, M.F.; Datar, I.; Su, T.T.; Ji, L.; Sun, J.; Chen, L.; Chen, Y.; Zhu, G.; Yin, W.; et al. Fibrinogen-like Protein 1 Is a Major Immune Inhibitory Ligand of LAG-3. Cell 2019, 176, 334–347.e12. [Google Scholar] [CrossRef] [PubMed]
- Huard, B.; Gaulard, P.; Faure, F.; Hercend, T.; Triebel, F. Cellular Expression and Tissue Distribution of the Human LAG-3-Encoded Protein, An MHC Class II Ligand. Immunogenetics 1994, 39, 213–217. [Google Scholar] [CrossRef]
- Annunziato, F.; Manetti, R.; Cosmi, L.; Galli, G.; Heusser, C.H.; Romagnani, S.; Maggi, E. Opposite Role for Interleukin-4 and Interferon-Γ on CD30 and Lymphocyte Activation Gene-3 (LAG-3) Expression by Activated Naive T Cells. Eur. J. Immunol. 1997, 27, 2239–2244. [Google Scholar] [CrossRef] [PubMed]
- Bruniquel, D.; Borie, N.; Hannier, S.; Triebel, F. Regulation of Expression of the Human Lymphocyte Activation Gene-3 (LAG-3) Molecule, a Ligand for MHC Class II. Immunogenetics 1998, 48, 116–124. [Google Scholar] [CrossRef]
- Burton, B.R.; Britton, G.J.; Fang, H.; Verhagen, J.; Smithers, B.; Sabatos-Peyton, C.A.; Carney, L.J.; Gough, J.; Strobel, S.; Wraith, D.C. Sequential Transcriptional Changes Dictate Safe and Effective Antigen-Specific Immunotherapy. Nat. Commun. 2014. [Google Scholar] [CrossRef]
- Matsuzaki, J.; Gnjatic, S.; Mhawech-Fauceglia, P.; Beck, A.; Miller, A.; Tsuji, T.; Eppolito, C.; Qian, F.; Lele, S.; Shrikant, P.; et al. Tumor-Infiltrating NY-ESO-1–Specific CD8 + T Cells are Negatively Regulated by LAG-3 and PD-1 in Human Ovarian Cancer. Proc. Natl. Acad. Sci USA 2010, 107, 7875–7880. [Google Scholar] [CrossRef]
- Zuazo, M.; Arasanz, H.; Fernández-Hinojal, G.; García-Granda, M.J.; Gato, M.; Bocanegra, A.; Martínez, M.; Hernández, B.; Teijeira, L.; Morilla, I.; et al. Functional Systemic CD4 Immunity Is Required for Clinical Responses to PD-L1/PD-1 Blockade Therapy. EMBO Mol. Med. 2019, 11, e10293. [Google Scholar] [CrossRef]
- Huard, B.; Tournier, M.; Triebel, F. LAG-3 Does Not Define a Specific Mode of Natural Killing in Human. Immunol. Lett. 1998, 61, 109–112. [Google Scholar] [CrossRef]
- Huang, C.T.; Workman, C.J.; Flies, D.; Pan, X.; Marson, A.L.; Zhou, G.; Hipkiss, E.L.; Ravi, S.; Kowalski, J.; Levitsky, H.I.; et al. Role of LAG-3 in Regulatory T cells. Immunity 2004, 21, 503–513. [Google Scholar] [CrossRef] [PubMed]
- Gagliani, N.; Magnani, C.F.; Huber, S.; Gianolini, M.E.; Pala, M.; Licona-Limon, P.; Guo, B.; Herbert, D.B.R.; Bulfone, A.; Trentini, F.; et al. Coexpression of CD49b and LAG-3 Identifies Human and Mouse T Regulatory Type 1 Cells. Nat. Med. 2013, 19, 739–746. [Google Scholar] [CrossRef] [PubMed]
- Alfen, J.S.; Larghi, P.; Facciotti, F.; Gagliani, N.; Bosotti, R.; Paroni, M.; Maglie, S.; Gruarin, P.; Vasco, C.M.; Ranzani, V.; et al. Intestinal IFN-Γ–Producing Type 1 Regulatory T Cells Coexpress CCR5 and Programmed Cell Death Protein 1 and Downregulate IL-10 in the Inflamed Guts of Patients with Inflammatory Bowel Disease. J. Allergy Clin. Immunol. 2018, 142, 1537–1547.e8. [Google Scholar] [CrossRef] [PubMed]
- White, A.M.; Wraith, D.C. Tr1-Like T Cells—An Enigmatic Regulatory T Cell Lineage. Front. Immunol. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Do, J.S.; Visperas, A.; Sanogo, Y.O.; Bechtel, J.J.; Dvorina, N.; Kim, S.; Jang, E.; Stohlman, S.A.; Shen, B.; Fairchild, R.L.; et al. An IL-27/Lag3 Axis Enhances Foxp3+ Regulatory T Cell-Suppressive Function and Therapeutic Efficacy. Mucosal. Immunol. 2016, 9, 137–145. [Google Scholar] [CrossRef]
- Workman, C.J.; Vignali, D.A.A. Negative Regulation of T Cell Homeostasis by Lymphocyte Activation Gene-3 (CD223). J. Immunol. 2005, 174, 688–695. [Google Scholar] [CrossRef]
- Camisaschi, C.; Casati, C.; Rini, F.; Perego, M.; De Filippo, A.; Triebel, F.; Parmiani, G.; Belli, F.; Rivoltini, L.; Castellli, C. LAG-3 Expression Defines a Subset of CD4 (+) CD25(High)Foxp3 (+) Regulatory T Cells that Are Expanded at Tumor Sites. J. Immunol. 2010, 184, 6545–6551. [Google Scholar] [CrossRef] [PubMed]
- Gandhi, M.K.; Lambley, E.; Duraiswamy, J.; Dua, U.; Smith, C.; Elliott, S.; Gill, D.; Marlton, P.; Seymour, J.; Khanna, R. Expression of LAG-3 by Tumor-Infiltrating Lymphocytes Is Coincident with the Suppression of Latent Membrane Antigen-Specific CD8+ T-Cell Function in Hodgkin Lymphoma Patients. Blood 2006, 108, 2280–2289. [Google Scholar] [CrossRef]
- Hald, S.M.; Rakaee, M.; Martinez, I.; Richardsen, E.; Al-Saad, S.; Paulsen, E.E.; Blix, E.S.; Kilvaer, T.; Andersen, S.; Busund, L.T.; et al. LAG-3 in Non–Small-cell Lung Cancer: Expression in Primary Tumors and Metastatic Lymph Nodes Is Associated with Improved Survival. Clin. Lung Cancer 2018, 19, 249–259.e2. [Google Scholar] [CrossRef]
- Lee, S.J.; Jun, S.Y.; Lee, I.H.; Kang, B.W.; Park, S.Y.; Kim, H.J.; Park, J.S.; Choi, G.-S.; Yoon, G.; Kim, J.G. CD274, LAG3, and IDO1 Expressions in Tumor-Infiltrating Immune Cells as Prognostic Biomarker for Patients with MSI-High Colon Cancer. J. Cancer Res. Clin. Oncol. 2018, 144, 1005–1014. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yongdong, L.; Yiling, L.; Binliu, L.; Qitao, H.; Wang, F.; Zhong, Q. Prognostic Value of Lymphocyte Activation Gene-3 (LAG-3) Expression in Esophageal Squamous Cell Carcinoma. J. Cancer 2018, 9, 4287–4293. [Google Scholar] [CrossRef] [PubMed]
- Kisielow, M.; Kisielow, J.; Capoferri-Sollami, G.; Karjalainen, K. Expression of Lymphocyte Activation Gene 3 (LAG-3) on B Cells Is Induced by T Cells. Eur. J. Immunol. 2005, 35, 2081–2088. [Google Scholar] [CrossRef] [PubMed]
- Lino, A.C.; Dang, V.D.; Lampropoulou, V.; Welle, A.; Joedicke, J.; Pohar, J.; Simon, Q.; Thalmensi, J.; Baures, A.; Fluhler, V.; et al. LAG-3 Inhibitory Receptor Expression Identifies Immunosuppressive Natural Regulatory Plasma Cells. Immunity 2018, 49, 120–133.e9. [Google Scholar] [CrossRef] [PubMed]
- Khsheibun, R.; Paperna, T.; Volkowich, A.; Lejbkowicz, I.; Avidan, N.; Miller, A. Gene Expression Profiling of the Response to Interferon Beta in Epstein-Barr-Transformed and Primary B Cells of Patients with Multiple Sclerosis. PLoS ONE 2014, 9, 102331. [Google Scholar] [CrossRef] [PubMed]
- Andreae, S.; Piras, F.; Burdin, N.; Triebel, F. Maturation and Activation of Dendritic Cells Induced by Lymphocyte Activation Gene-3 (CD223). J. Immunol. 2002, 168, 3874–3880. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Zhou, M.; Qiu, J.; Lin, Y.; Chen, X.; Huang, S.; Mo, M.; Liu, H.; Peng, G.; Zhu, X.; et al. Association of LAG3 Genetic Variation with an Increased Risk of PD in Chinese Female Population. J. Neuroinflammation 2019, 16. [Google Scholar] [CrossRef]
- Angelopoulou, E.; Paudel, Y.N.; Villa, C.; Shaikh, M.F.; Piperi, C. Lymphocyte-Activation Gene 3 (LAG3) Protein as a Possible Therapeutic Target for Parkinson’s Disease: Molecular Mechanisms Connecting Neuroinflammation to α-Synuclein Spreading Pathology. Biology 2020, 9, 86. [Google Scholar] [CrossRef]
- Mao, X.; Ou, M.T.; Karuppagounder, S.S.; Kam, T.I.; Yin, X.; Xiong, Y.; Ge, P.; Umanah, G.E.; Brahmachari, S.; Shin, J.; et al. Pathological α-Synuclein Transmission Initiated by Binding Lymphocyte-Activation Gene 3. Science. 2016, 353, 6307. [Google Scholar] [CrossRef]
- Cunningham, F.; Achuthan, P.; Akanni, W.; Allen, J.; Amode, M.R.; Armean, I.M.; Bennett, R.; Bhai, J.; Billis, K.; Boddu, S.; et al. Ensembl 2019. Nucleic Acids Res. 2019, 47. [Google Scholar] [CrossRef]
- Saleh, R.; Toor, S.M.; Nair, V.S.; Elkord, E. Role of Epigenetic Modifications in Inhibitory Immune Checkpoints in Cancer Development and Progression. Front. Immunol. 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Goltz, D.; Gevensleben, H.; Grünen, S.; Dietrich, J.; Kristiansen, G.; Landsberg, J.; Dietrich, D. PD-L1 (CD274) Promoter Methylation Predicts Survival in Patients with Acute Myeloid Leukemia. Leukemia 2017, 31, 738–743. [Google Scholar] [CrossRef]
- Goltz, D.; Gevensleben, H.; Dietrich, J.; Ellinger, J.; Landsberg, J.; Kristiansen, G.; Dietrich, D. Promoter Methylation of the Immune Checkpoint Receptor PD-1 (PDCD1) Is an Independent Prognostic Biomarker for Biochemical Recurrence-Free Survival in Prostate Cancer Patients Following Radical Prostatectomy. Oncoimmunology 2016, 5, e1221555. [Google Scholar] [CrossRef] [PubMed]
- Goltz, D.; Gevensleben, H.; Dietrich, J.; Schroeck, F.; de Vos, L.; Droege, F.; Kristiansen, G.; Schroeck, A.; Landsberg, J.; Bootz, F.; et al. PDCD1 (PD-1) Promoter Methylation Predicts Outcome in Head and Neck Squamous Cell Carcinoma Patients. Oncotarget 2017, 8, 41011–41020. [Google Scholar] [CrossRef] [PubMed]
- Goltz, D.; Gevensleben, H.; Vogt, T.J.; Dietrich, J.; Golletz, C.; Bootz, F.; Kristiansen, G.; Landsberg, J.; Dietrich, D. CTLA4 Methylation Predicts Response to Anti-PD-1 and Anti-CTLA-4 Immunotherapy in Melanoma Patients. JCI Insight. 2018, 3. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Lv, D.; Cai, C.; Zhao, Z.; Wang, M.; Chen, W.; Liu, Y. A TP53-Associated Immune Prognostic Signature for the Prediction of Overall Survival and Therapeutic Responses in Muscle-Invasive Bladder Cancer. Front. Immunol. 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Clifford, R.L.; Fishbane, N.; Patel, J.; MacIsaac, J.L.; McEwen, L.M.; Fisher, A.J.; Brandsma, C.; Nair, P.; Kobor, M.S.; Hackett, T.L.; et al. Altered DNA Methylation is Associated with Aberrant Gene Expression in Parenchymal but not Airway Fibroblasts Isolated from Individuals with COPD. Clin. Epigenetics 2018, 10. [Google Scholar] [CrossRef]
- Klümper, N.; Ralser, D.J.; Bawden, E.G.; Landsberg, J.; Zarbl, R.; Kristiansen, G.; Toma, M.; Ritter, M.; Hölzel, M.; Ellinger, J.; et al. LAG3 (LAG-3, CD223) DNA Methylation Correlates with LAG3 Expression by Tumor and Immune Cells, Immune Cell Infiltration, and Overall Survival in Clear Cell Renal Cell Carcinoma. J. Immunother. Cancer 2020, 8. [Google Scholar] [CrossRef]
- Querfeld, C.; Wu, X.; Sanchez, J.F.; Palmer, J.M.; Motevalli, A.; Zain, J.; Rosen, S.T. The miRNA Profile of Cutaneous T Cell Lymphoma Correlates with the Dysfunctional Immunophenotype of the Disease. Blood 2016, 128, 4132. [Google Scholar] [CrossRef]
- Laino, A.S.; Betts, B.C.; Veerapathran, A.; Dolgalev, I.; Sarnaik, A.; Quayle, S.N.; Jones, S.S.; Weber, J.S.; Woods, D.M. HDAC6 Selective Inhibition of Melanoma Patient T-Cells Augments Anti-Tumor Characteristics. J. Immunother. Cancer 2019, 7. [Google Scholar] [CrossRef] [PubMed]
- Okamura, T.; Fujio, K.; Shibuya, M.; Sumitomo, S.; Shoda, H.; Sakaguchi, S.; Yamamoto, K. CD4+CD25-LAG3+ Regulatory T Cells Controlled by the Transcription Factor Egr-2. Proc. Natl. Acad. Sci USA 2009, 106, 13974–13979. [Google Scholar] [CrossRef]
- Huang, K.; Pang, T.; Tong, C.; Chen, H.; Nie, Y.; Wu, J.; Zhang, Y.; Chen, G.; Zhou, W.; Yang, D.; et al. Integrative Expression and Prognosis Analysis of DHX37 in Human Cancers by Data Mining. Biomed. Res. Int. 2021, 2021. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Z.; Wei, F.; Ren, X. Exhausted T Cells and Epigenetic Status. Cancer Biology and Medicine. Cancer Biol. Med. 2020, 17, 923–936. [Google Scholar] [CrossRef] [PubMed]
- Deng, W.W.; Mao, L.; Yu, G.T.; Bu, L.L.; Ma, S.R.; Liu, B.; Gutkind, J.S.; Kulkarni, A.B.; Zhang, W.; Sun, Z.; et al. LAG-3 Confers Poor Prognosis and Its Blockade Reshapes Antitumor Response in Head And Neck Squamous Cell Carcinoma. Oncoimmunology 2016, 5, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Marcq, E.; De Waele, J.; Van Audenaerde, J.; Lion, E.; Santermans, E.; Hens, N.; Pauwels, P.; Meerbeeck, J.P.V.; Smits, E.L.J. Abundant Expression Of TIM-3, LAG-3, PD-1 and PD-L1 As Immunotherapy Checkpoint Targets in Effusions of Mesothelioma Patients. Oncotarget 2017, 8, 89722–89735. [Google Scholar] [CrossRef]
- Burugu, S.; Gao, D.; Leung, S.; Chia, S.K.; Nielsen, T.O. LAG-3+ Tumor Infiltrating Lymphocytes in Breast Cancer: Clinical Correlates and Association with PD-1/PD-L1 + Tumors. Ann. Oncol. 2017, 28, 2977–2984. [Google Scholar] [CrossRef]
- Yanik, E.L.; Kaunitz, G.J.; Cottrell, T.R.; Succaria, F.; McMiller, T.L.; Ascierto, M.L.; Esandrio, J.; Xu, H.; Ogurtsova, A.; Cornish, T.; et al. Association of HIV Status with Local Immune Response to Anal Squamous Cell Carcinoma: Implications for Immunotherapy. JAMA Oncol. 2017, 3, 974–978. [Google Scholar] [CrossRef]
- Saka, D.; Gökalp, M.; Piyade, B.; Cevik, N.C.; Sever, E.; Unutmaz, D.; Ceyhan, G.O.; Demir, I.E.; Asimgil, H. Mechanisms of T-Cell Exhaustion in Pancreatic Cancer. Cancers 2020, 12, 2274. [Google Scholar] [CrossRef]
- Wuerdemann, N.; Pütz, K.; Eckel, H.; Jain, R.; Wittekindt, C.; Huebbers, C.U.; Sharma, S.J.; Langer, C.; Gattenlöhner, S.; Büttner, R.; et al. LAG-3, TIM-3 and Vista Expression on Tumor-Infiltrating Lymphocytes in Oropharyngeal Squamous Cell Carcinoma-Potential Biomarkers for Targeted Therapy Concepts. Int. J. Mol. Sci. 2021, 22, 1–17. [Google Scholar]
- Shapiro, M.; Herishanu, Y.; Katz, B.Z.; Dezorella, N.; Sun, C.; Kay, S.; Polliack, A.; Avivi, I.; Wiestner, A.; Perry, C. Lymphocyte Activation Gene 3: A Novel Therapeutic Target in Chronic Lymphocytic Leukemia. Haematol. 2017, 102, 874–882. [Google Scholar] [CrossRef]
- Chen, J.; Chen, Z. The Effect of Immune Microenvironment on the Progression and Prognosis of Colorectal Cancer. Med. Oncol. 2014, 31. [Google Scholar] [CrossRef] [PubMed]
- Li, F.J.; Zhang, Y.; Jin, G.X.; Yao, L.; Wu, D.Q. Expression Of LAG-3 Is Coincident with the Impaired Effector Function of HBV-Specific CD8 + T Cell in HCC Patients. Immunol. Lett. 2013, 150, 116–122. [Google Scholar] [CrossRef] [PubMed]
- Giraldo, N.A.; Becht, E.; Pagès, F.; Skliris, G.; Verkarre, V.; Vano, Y.; Mejean, A.; Saint-Aubert, N.; Lacroix, L.; Natario, I.; et al. Orchestration and Prognostic Significance of Immune Checkpoints in the Microenvironment of Primary and Metastatic Renal Cell Cancer. Clin. Cancer Res. 2015, 21, 3031–3040. [Google Scholar] [CrossRef] [PubMed]
- Takaya, S.; Saito, H.; Ikeguchi, M. Upregulation of Immune Checkpoint Molecules, PD-1 and LAG-3, On CD4 + and CD8+ T Cells After Gastric Cancer Surgery. Yonago Acta. Med. 2015, 58, 39–44. [Google Scholar] [PubMed]
- Yang, Z.Z.; Kim, H.J.; Villasboas, J.C.; Chen, Y.P.; Price-Troska, T.P.; Jalali, S.; Wilson, M.; Novak, A.J.; Ansell, S.M. Expression of LAG-3 Defines Exhaustion of Intratumoral PD-1+ T Cells and Correlates with Poor Outcome in Follicular Lymphoma. Oncotarget 2017, 8, 61425–61439. [Google Scholar] [CrossRef]
- Saleh, R.R.; Peinado, P.; Fuentes-Antrás, J.; Pérez-Segura, P.; Pandiella, A.; Amir, E.; Ocaña, A. Prognostic Value of Lymphocyte-Activation Gene 3 (LAG3) in Cancer: A Meta-Analysis. Front. Oncol. 2019, 9, 1040. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Dong, T.; Xuan, Q.; Zhao, H.; Qin, L.; Zhang, Q. Lymphocyte-Activation Gene-3 Expression and Prognostic Value in Neoadjuvant-Treated Triple-Negative Breast Cancer. J. Breast Cancer 2018, 21, 124–133. [Google Scholar] [CrossRef]
- Datar, I.; Sanmamed, M.F.; Wang, J.; Henick, B.S.; Choi, J.; Badri, T.; Dong, W.; Mani, N.; Toki, M.; Mejías, L.D.; et al. Expression Analysis and Significance of PD-1, LAG-3, and TIM-3 in Human Non–Small Cell Lung Cancer Using Spatially Resolved and Multiparametric Single-Cell Analysis. Clin. Cancer Res. 2019, 25, 4663–4673. [Google Scholar] [CrossRef]
- Sobottka, B.; Moch, H.; Varga, Z. Differential PD-1/LAG-3 Expression and Immune Phenotypes in Metastatic Sites of Breast Cancer. Breast Cancer Res. 2021, 23. [Google Scholar] [CrossRef]
- Zhang, X.; Zhao, H.; Shi, X.; Jia, X.; Yang, Y. Identification and Validation of an Immune-Related Gene Signature Predictive of Overall Survival in Colon Cancer. Aging 2020, 12, 26095–26120. [Google Scholar] [CrossRef] [PubMed]
- Lichtenegger, F.S.; Rothe, M.; Schnorfeil, F.M.; Deiser, K.; Krupka, C.; Augsberger, C.; Schlüter, M.; Neitz, J.; Subklewe, M. Targeting LAG-3 and PD-1 to Enhance T Cell Activation by Antigen-Presenting Cells. Front. Immunol. 2018, 9, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Jing, W.; Gershan, J.A.; Weber, J.; Tlomak, D.; McOlash, L.; Sabatos-Peyton, C.; Johnson, B.D. Combined Immune Checkpoint Protein Blockade and Low Dose Whole Body Irradiation as Immunotherapy for Myeloma. J. Immunother. Cancer 2015, 3. [Google Scholar] [CrossRef] [PubMed]
- Haudebourg, T.; Dugast, A.S.; Coulon, F.; Usual, C.; Triebel, F.; Vanhove, B. Depletion of LAG-3 Positive Cells in Cardiac Allograft Reveals Their Role in Rejection and Tolerance. Transplant 2007, 84, 1500–1506. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Ye, J.; Ma, Y.; Hua, P.; Huang, Y.; Fu, X.; Lie, D.; Yuan, M.; Xiag, Z. Function of T Regulatory Type 1 Cells Is Down-Regulated and Is Associated with the Clinical Presentation of Coronary Artery Disease. Hum. Immunol. 2018, 79, 564–570. [Google Scholar] [CrossRef]
- Golden, D.; Kolmakova, A.; Sura, S.; Vella, A.T.; Manichaikul, A.; Wang, X.-Q.; Bielinski, S.J.; Taylor, K.D.; Chen, Y.I.; Rich, S.S.; et al. Lymphocyte Activation Gene 3 and Coronary Artery Disease. JCI Insight 2016, 1, 88628. [Google Scholar] [CrossRef]
- Rodriguez, A. High HDL-Cholesterol Paradox: SCARB1-LAG3-HDL Axis. Curr. Atheroscler. Rep. 2021, 23. [Google Scholar] [CrossRef]
- Bauché, D.; Joyce-Shaikh, B.; Jain, R.; Grein, J.; Ku, K.S.; Blumenschein, W.M.; Ganal-Vonarburg, S.C.; Wilson, D.C.; McClanahan, T.K.; Malefyt, R.D.W.; et al. LAG3 + Regulatory T Cells Restrain Interleukin-23-Producing CX3CR1 + Gut-Resident Macrophages during Group 3 Innate Lymphoid Cell-Driven Colitis. Immunity 2018, 49, 342–352.e5. [Google Scholar] [CrossRef]
- Zhang, Z.; Duvefelt, K.; Svensson, F.; Masterman, T.; Jonasdottir, G.; Salter, H.; Emahazion, T.; Hellgren, D.; Falk, G.; Olsson, T.; et al. Two Genes Encoding Immune-Regulatory Molecules (LAG3 And IL7R) Confer Susceptibility to Multiple Sclerosis. Genes Immun. 2005, 6, 145–152. [Google Scholar] [CrossRef]
- Bettini, M.; Szymczak-Workman, A.L.; Forbes, K.; Castellaw, A.H.; Selby, M.; Pan, X.; Drake, C.G.; Korman, A.J.; Dario, A.; Vignali, A. Cutting Edge: Accelerated Autoimmune Diabetes in the Absence of LAG-3. J. Immunol. 2011, 187, 3493–3498. [Google Scholar] [CrossRef]
- Delmastro, M.M.; Styche, A.J.; Trucco, M.M.; Workman, C.J.; Vignali, D.A.A.; Piganelli, J.D. Modulation of Redox Balance Leaves Murine Diabetogenic TH1 T Cells “LAG-3-Ing” Behind. Diabetes 2012, 61, 1760–1768. [Google Scholar] [CrossRef]
- Doe, H.T.; Kimura, D.; Miyakoda, M.; Kimura, K.; Akbari, M.; Yui, K. Expression Of PD-1/LAG-3 and Cytokine Production by CD4 + T Cells During Infection with Plasmodium Parasites. Microbiol. Immunol. 2016, 60, 21–31. [Google Scholar] [CrossRef] [PubMed]
- Phillips, B.L.; Mehra, S.; Ahsan, M.H.; Selman, M.; Khader, S.A.; Kaushal, D. LAG3 Expression in Active Mycobacterium Tuberculosis Infections. Am. J. Pathol. 2015, 185, 820–833. [Google Scholar] [CrossRef] [PubMed]
- Graydon, C.G.; Balasko, A.L.; Fowke, K.R. Roles, Function and Relevance of LAG3 in HIV Infection. PLoS Pathog. 2019, 15, e1007429. [Google Scholar] [CrossRef] [PubMed]
- Jochems, S.P.; Jacquelin, B.; Tchitchek, N.; Busato, F.; Pichon, F.; Huot, N.; Liu, Y.; Ploquin, M.J.; Roché, E.; Cheynier, R.; et al. DNA Methylation Changes in Metabolic and Immune-Regulatory Pathways in Blood and Lymph Node CD4 + T Cells in Response to SIV Infections. Clin. Epigenetics. 2020, 12. [Google Scholar] [CrossRef]
- McLane, L.M.; Abdel-Hakeem, M.S.; Wherry, E.J. CD8 T Cell Exhaustion During Chronic Viral Infection and Cancer; Annual Review of Immunology; Annual Reviews Inc.: Palo Alto, CA, USA, 2019; Volume 37, pp. 457–495. [Google Scholar]
- Anderson, A.C.; Joller, N.; Kuchroo, V.K. Lag-3, Tim-3, and TIGIT: Co-inhibitory Receptors with Specialized Functions in Immune Regulation. Immun. Cell Press 2016, 44, 989–1004. [Google Scholar] [CrossRef]
- Richter, K.; Agnellini, P.; Oxenius, A. On the Role of the Inhibitory Receptor LAG-3 in Acute and Chronic LCMV Infection. Int. Immunol. 2009, 22, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Coulon, P.-G.; Prakash, S.; Srivastava, R.; Geertsema, R.; Dhanushkodi, N.; Lam, C.; Nguyen, V.; Gorospe, E.; Nguyen, A.M.; et al. Blockade of PD-1 and LAG-3 Immune Checkpoints Combined with Vaccination Restores the Function of Antiviral Tissue-Resident CD8 + T RM Cells and Reduces Ocular Herpes Simplex Infection and Disease in HLA Transgenic Rabbits. J. Virol. 2009, 93. [Google Scholar] [CrossRef]
- Roy, S.; Coulon, P.G.; Srivastava, R.; Vahed, H.; Kim, G.J.; Walia, S.S.; Yamada, T.; Fouladi, M.A.; Ly, V.T.; BenMohamed, L. Blockade of LAG-3 Immune Checkpoint Combined With Therapeutic Vaccination Restore the Function of Tissue-Resident Anti-Viral CD8 + T Cells and Protect Against Recurrent Ocular Herpes Simplex Infection and Disease. Front. Immunol. 2018, 9, 2922. [Google Scholar] [CrossRef]
- Liu, Y.; Source, S.; Nuvolone, M.; Domange, J.; Aguzzi, A. Lymphocyte Activation Gene 3 (Lag3) Expression Is Increased in Prion Infections but Does Not Modify Disease Progression. Sci. Rep. 2018, 8. [Google Scholar] [CrossRef]
- Martins, F.; Sofiya, L.; Sykiotis, G.P.; Lamine, F.; Maillard, M.; Shabafrouz, K.; Shabafrouz, K.; Ribi, C.; Cairoli, A.; Guex-Crosier, Y.; et al. Adverse effects of immune—checkpoint inhibitors: Epidemiology, management and surveillance. Nat. Rev. Clin. Oncol. 2019, 16, 563–580. [Google Scholar] [CrossRef] [PubMed]
- Nishino, M.; Ramaiya, N.H.; Hatabu, H.; Hodi, F.S. Monitoring Immune-Checkpoint Blockade: Response Evaluation and Biomarker Development. Nat. Rev. Clin. Oncol. 2018, 14, 655–668. [Google Scholar] [CrossRef] [PubMed]
- Prasad, V.; De Jesús, K.; Mailankody, S. The High Price of Anticancer Drugs: Origins, Implications, Barriers, Solutions. Nat. Rev. Clin. Oncol. 2017, 14, 381–390. [Google Scholar] [CrossRef]
- Topalian, S.L.; Weiner, G.J.; Pardoll, D.M. Cancer Immunotherapy Comes of Age. J. Clin. Oncol. 2011, 29, 4828. [Google Scholar] [CrossRef] [PubMed]
- Topalian, S.L.; Drake, C.G.; Pardoll, D.M. Perspective Immune Checkpoint Blockade: A Common Denominator Approach to Cancer Therapy. Cancer Cell. 2015, 27, 450–461. [Google Scholar] [CrossRef] [PubMed]
- Chocarro de Erauso, L.; Zuazo, M.; Arasanz, H.; Bocanegra, A.; Hernandez, C.; Fernandez, G.; Garcia-Granda, M.J.; Blanco, E.; Vera, R.; Kochan, G.; et al. Resistance to PD-L1/PD-1 Blockade Immunotherapy. A Tumor-Intrinsic or Tumor-Extrinsic Phenomenon? Front. Pharmacol. 2020, 11. [Google Scholar] [CrossRef]
- Zuazo, M.; Arasanz, H.; Bocanegra, A.; Fernandez, G.; Chocarro, L.; Vera, R.; Kochan, G.; Escors, D. Systemic CD4 Immunity as a Key Contributor to PD-L1/PD-1 Blockade Immunotherapy Efficacy. Front. Immunol. 2020, 11. [Google Scholar] [CrossRef]
- Zuazo, M.; Arasanz, H.; Bocanegra, A.; Chocarro, L.; Vera, R.; Escors, D.; Kagamu, H.; Kochan, G. Systemic CD4 Immunity: A Powerful Clinical Biomarker for PD-L1/PD-1 Immunotherapy. EMBO Mol. Med. 2020, 12, e12706. [Google Scholar] [CrossRef]
- Bocanegra, A.; Fernandez-Hinojal, G.; Zuazo-Ibarra, M.; Arasanz, H.; Garcia-Granda, M.J.; Hernandez, C.; Ibañez, M.; Hernandez-Marin, B.; Martinez-Aguillo, M.; Lecumberri, M.J.; et al. PD-L1 Expression in Systemic Immune Cell Populations as a Potential Predictive Biomarker of Responses to PD-L1/PD-1 Blockade Therapy in Lung Cancer. Int. J. Mol. Sci. 2019, 20, 1631. [Google Scholar] [CrossRef]
- Rotte, A.; Jin, J.Y.; Lemaire, V. Mechanistic overview of immune checkpoints to support the rational design of their combinations in cancer immunotherapy. Ann. Oncol. 2018, 29, 71–83. [Google Scholar] [CrossRef] [PubMed]
- Legg, J.W.; McGuinness, B.; Arasanz, H.; Bocanegra, A.; Bartlett, P.; Benedetti, G.; Birkett, N.; Cox, C.; De Juan, E.; Enever, C.; et al. Abstract 930: CB213: A Half-Life Extended Bispecific Humabody V H Delivering Dual Checkpoint Blockade to Reverse the Dysfunction of LAG3 + PD-1 + Double-Positive T Cells. Am. Assoc. Cancer Res. 2020. [Google Scholar] [CrossRef]
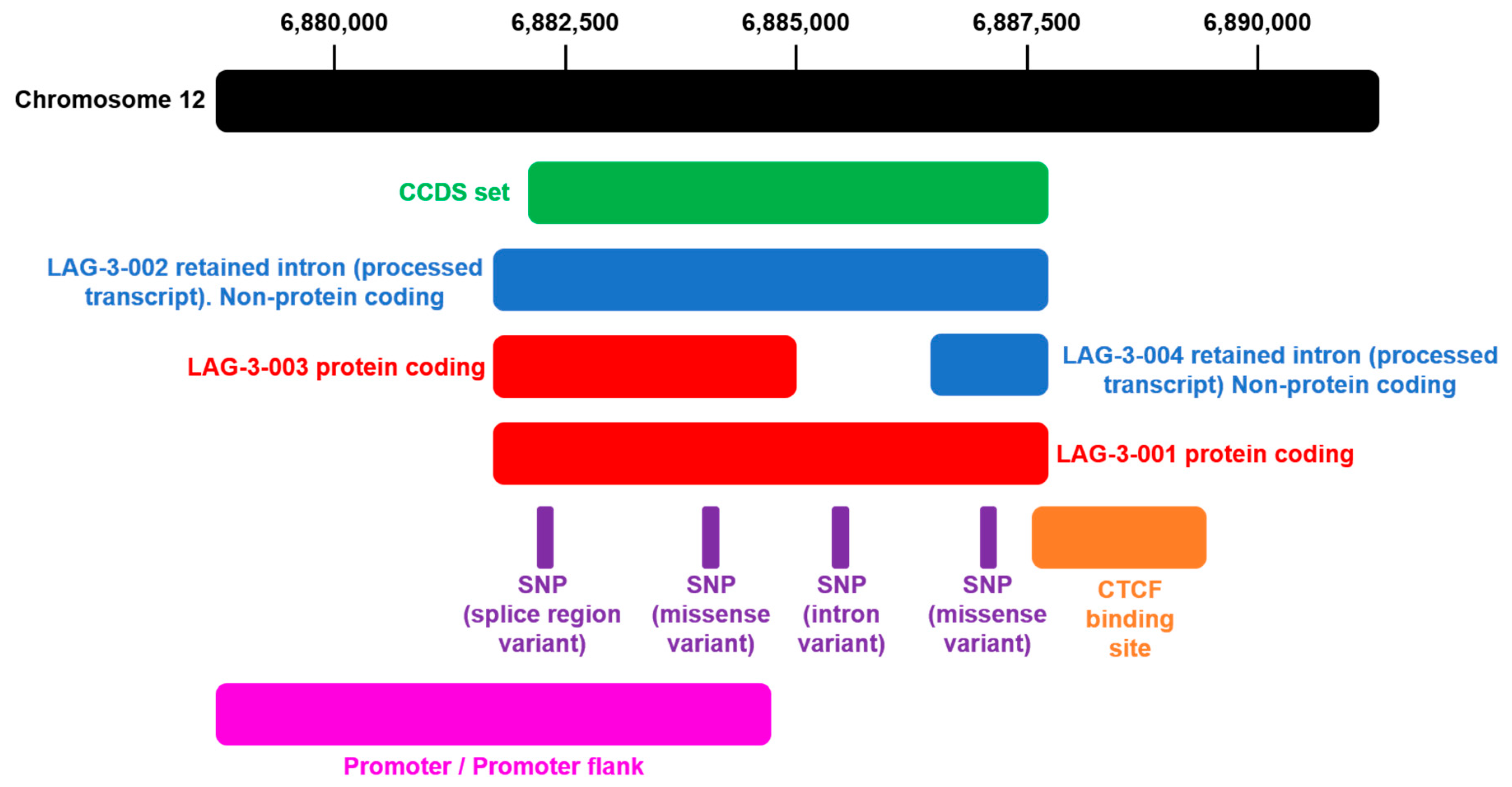
| Transcript (ID) | Length | Biotype | Location | Exons | Annotation |
|---|---|---|---|---|---|
| Lag3-201 (ENST00000203629.3) | Transcript length: 1976 bps Translation length: 525 residues | Protein coding | Chromosome 12: 6,772,520-6,778,455 | 8 (all coding) | 24 domains and features 2174 variant alleles |
| Lag3-202 (ENST00000441671.6) | Transcript length: 1576 bps Translation length: 360 residues | Protein coding | Chromosome 12: 6,772,519-6,775,733 | 5 (all coding) | 19 domains and features 1365 variant alleles |
| Lag3-203 (ENST00000538079.1) | Transcript length: 2587 bps | Retained intron | Chromosome 12: 6,772,512-6,778,455 | 6 (non-coding) | 2174 variant alleles |
| Lag3-204 (ENST00000541049.1) | Transcript length: 684 bps | Retained intron | Chromosome 12: 6,777,450-6,778,455 | 2 (non-coding) | 404 variant alleles |
| Phase | Therapy | NCT Identifier | Intervention/Treatment Tested | Condition or Disease |
|---|---|---|---|---|
| Early I | Monotherapy | NCT04566978 | Anti-LAG-3 (89Zr-DFO-REGN3767) | Large B-cell Lymphoma, DLBCL |
| I | Monotherapy | NCT03489369 | Anti-LAG-3 (Sym022) | Metastatic Cancer, Solid Tumor, Lymphoma |
| NCT03965533 | Anti-LAG-3 (GSK2831781) | Healthy Volunteers | ||
| NCT02195349 | Anti-LAG-3 (GSK2831781) | Psoriasis | ||
| NCT03538028 | Anti-LAG-3 (INCAGN02385) | Select Advanced Malignancies | ||
| NCT00351949 | LAG-3-Ig (Eftilagimod Alpha, IMP321) | Metastatic Renal Cell Carcinoma (MRCC) | ||
| Monotherapy and Combination | NCT03252938 | LAG-3-Ig (Eftilagimod alpha, IMP321), anti-PD-L1 (Avelumab), standard-of-care chemotherapy | Solid Tumors, Peritoneal Carcinomatosis | |
| NCT02658981 | Anti-LAG-3 (BMS-986016), Anti-PD-1 (Nivolumab, BMS-936558), Anti-CD137 (Urelumab, BMS-663513) | Glioblastoma, Gliosarcoma, Recurrent Brain Neoplasm | ||
| NCT03250832 | Anti-LAG-3 (TSR-033), Anti-PD-1 | Advanced Solid tumors, Colorectal Cancer | ||
| NCT03005782 | Anti-LAG-3 (REGN3767), Anti-PD-1 (Cemiplimab, REGN2810) | Advanced Cancers | ||
| NCT02966548 | Anti-LAG-3 (Retalimab, BMS-986016), Anti-PD-1 (Nivolumab, BMS-936558) | Advanced Solid Tumors | ||
| NCT00354263 | LAG-3-Ig (Eftilagimod Alpha, IMP321), Agrippal Reference Flu Antigen (commercially available flu vaccine) | Healthy Volunteers | ||
| Combination | NCT04658147 | Anti-LAG-3 (Relatlimab), Anti-PD-1 (Nivolumab) | Hepatocellular Carcinoma | |
| NCT03219268 | Anti-PD-1/Anti-LAG-3 DART protein MGD013, Anti-HER2 (Margetuximab, MGAH22) | Advanced Solid Tumors, Hematologic Neoplasms, Gastric Cancer, Ovarian Cancer, Gastroesophageal Cancer, HER2-positive Breast Cancer, HER2-positive Gastric Cancer | ||
| NCT03440437 | Anti-LAG-3/PD-L1 Bispecific Antibody (FS118) | Advanced Cancer, Metastatic Cancer | ||
| NCT00732082 | LAG-3-Ig (Eftilagimod Alpha, IMP321), Gemcitabine (Gemzar) | Pancreatic Neoplasms | ||
| NCT03742349 | Anti-LAG-3 (LAG525, IMP701), Anti-PD-L1 (Spartalizumab, PDR001), Anti-A2AR (NIR178), MET inhibitor (capmatinib, INC280), Anti-M-CSF (MCS110), Anti-IL-1-beta (canakinumab, ACZ885) | Triple Negative Breast Cancer (TNBC) | ||
| NCT04140500 | Anti-PD1-LAG-3 Bispecific Antibody (RO7247669) | Solid Tumors | ||
| NCT03849469 | Anti-CTLA4-LAG-3 Bispecific Antibody (XmAb®22841), Anti-PD-1 (Pembrolizumab (Keytruda®)) | Selected Advanced Solid Tumors | ||
| NCT03311412 | Anti-LAG-3 (Sym022), Anti-TIM-3 (Sym023), Anti-PD-1 (Sym021) | Metastatic Cancer, Solid Tumors, Lymphoma | ||
| NCT02817633 | Anti-LAG-3 (TSR-033), Anti-PD-1 (TSR-042), Anti-TIM-3 (TSR-022) | Advanced or Metastatic Solid Tumors | ||
| NCT04252768 | LAG-3-Ig (Eftilagimod Alpha, IMP321), Paclitaxel | Metastatic Breast Cancer | ||
| NCT02676869 | LAG-3-Ig (Eftilagimod Alpha, IMP321), Anti-PD-1 (Pembrolizumab) | Unresectable or Metastatic Melanoma | ||
| NCT00349934 | LAG-3-Ig (Eftilagimod Alpha, IMP321), Paclitaxel | Metastatic Breast Carcinoma | ||
| NCT00354861 | LAG-3-Ig (Eftilagimod Alpha, IMP321), hepatitis B antigen (without alum), Engerix B (hepatitis B antigen absorbed on alum) | Healthy Volunteers | ||
| NCT03493932 | Anti-LAG-3 (Relatimab, BMS-986016), Anti-PD-1 (Nivolumab) | Recurrent Glioblastoma Patients | ||
| NCT03044613 | Anti-LAG-3 (Relatlimab, BMS-986016) Anti-PD-1 (Nivolumab, Opdivo), Carboplatin, Paclitaxel, Radiation | Gastric Cancer, Esophageal Cancer, Gastroesophageal Cancer | ||
| NCT04641871 | Anti-LAG-3 (Sym022), Anti-TIM-3 (Sym023), Anti-PD-1 (Sym01) | Advanced Solid Tumor Malignancies | ||
| I/II | Monotherapy | NCT04618393 | Anti-PD-1-LAG-3 Bi-specific Antibody (EMB-02) | Advanced Solid Tumors |
| Monotherapy and Combination | NCT04706715 | Anti-LAG-3 (89Zr-DFO-REGN3767), Cemiplimab | Metastatic Solid Tumor | |
| NCT02460224 | Anti-LAG-3 (LAG525), Anti-PD1 (PDR001) | Advanced Solid Tumors | ||
| NCT01968109 | Anti-LAG-3 (Relatlimab, BMS-986016), Anti-PD-1 (Nivolumab, BMS-936558), (BMS-986213) | Neoplasms by Site, Solid Tumors | ||
| Combination | NCT04611126 | Anti-LAG-3 (Relatlimab), Anti-PD-L1 (Ipilimumab), Anti-PD-1 (Nivolumab), Cyclophosphamid, Fludarabine Phosphate, Tumor Infiltrating Lymphocytes infusion | Metastatic Ovarian Cancer, Metastatic Fallopian Tube Cancer, Peritoneal Cancer | |
| NCT04150965 | Anti-LAG-3 (Relatlimab, BMS-986016), Elotuzumab (Empliciti), Pomalidomide, Dexamethasone, Anti-TIGIT (BMS-986207) | Multiple Myeloma, Relapsed Refractory Multiple Myeloma | ||
| NCT01308294 | LAG-3-Ig (ImmuFact-IMP321), Tumor Antigenic Peptides (NA-17, MAGE-3.A2, NY-ESO-1, Melan-A, MAGE-A3, MAGE-A3-DP4), Montanide ISA-51 | Melanoma | ||
| NCT00365937 | LAG-3-Ig (Eftilagimod Alpha, IMP321), Immunological peptides and adjuvants, HLA-A2 peptides (Tyrosinase.A2, MAGE-C2.A2, MAGE-3.A2, MAGE-1.A2, NA17.A2 (GnTV), MAGE-10.A2), Montanide ISA51 | Melanoma | ||
| NCT03459222 | Anti-LAG-3 (Relatlimab, BMS-986016), Anti-PD-1 (Nivolumab, Opdivo, BMS-936558), IDO1 Inhibitor (BMS-986205), Anti-CTLA-4 (Ipilimumab, Yervoy, BMS-734016) | Advanced Solid Cancers | ||
| NCT02488759 | Anti-LAG-3 (Relatlimab, BMS-986016), Anti-PD-1 (Nivolumab), Anti-CTLA4 (Ipilimumab), Anti-CD38 (Daratumumab, Darzalex) | Various Advanced Cancers | ||
| NCT02061761 | Anti-LAG-3 (BMS-986016), Anti-PD-1 (Nivolumab, BMS-936558) | Hematologic Neoplasm (Refractory B-Cell Malignancies) | ||
| NCT03610711 | Anti-LAG-3 (Relatlimab), Anti-PD-1 (Nivolumab, Optivo) | Gastroesophageal Cancer | ||
| NCT04370704 | Anti-PD-1 (NCMGA00012), Anti-LAG-3 (INCAGN02385), Anti-TIM-3 (INCAGN02390) | Melanoma | ||
| II | Monotherapy | NCT03893565 | Anti-LAG-3 (GSK2831781) | Ulcerative Colitis |
| Monotherapy and Combination | NCT04080804 | Anti-LAG-3 (Relatlimab, BMS-986016), Anti-PD1 (Nivolumab, OPDIVO), Anti-CTLA4 (Ipilimumab, Yervoy) | Head and Neck Squamous Cell Carcinoma (HNSCC) | |
| NCT03743766 | Anti-LAG-3 (Relatlimab, BMS-986016), Anti-PD1 (Nivolumab, BMS-936558) | Melanoma | ||
| Combination | NCT04567615 | Anti-LAG-3 (Anti-LAG-3 (Relatlimab, BMS-986016), Anti-PD1 (Nivolumab, BMS-936558) | Advanced Hepatocellular Carcinoma | |
| NCT03484923 | Anti-LAG-3 IgG4 (LAG525), Anti-PD-1 (Spartalizumab, PDR001), Capmatinib (INC280), Canakinumab (ACZ885), Ribociclib (LEE011) | Melanoma | ||
| NCT04634825 | PD-1XLAG-3 bispecific DARTmolecule (Tebotelimab, MGD013), Anti-B7-H3 (Enoblituzumab, MGA271), Anti-PD-1 (Retifanlimab, INCMGA00012, MGA012) | Head and Neck Neoplasms | ||
| NCT04326257 | Anti-LAG-3 (Relatlimab), Anti-PD-1 (Nivolumab); Anti-PD-L1 (Ipilimumab) | Squamous Cell Carcinoma of the Head and Neck | ||
| NCT03625323 | LAG-3-Ig (Eftilagimod Alpha, IMP321), Anti-PD-1 (Pembrolizumab, Keytruda, MK-3475) | Non-Small Cell Lung Carcinoma (NSCLC) and Head and Neck Carcinoma (HNSCC) | ||
| NCT03623854 | Anti-LAG-3 (Relatlimab, BMS-986016), Anti-PD-1 (Nivolumab, BMS-936558) | Chordoma | ||
| NCT03662659 | Anti-LAG-3 (Relatlimab, BMS-986016), Anti-PD-1 (Nivolumab, Opdivo, BMS-936558), (BMS-986213), XELOX (Oxaliplatin + capecitabine), FOLFOX (Oxaliplatin + leucovorin + fluorouracil), SOX (Oxaliplatin + tegafur/gimeracil/oteracil potassium) | Gastric or Gastroesophageal Junction (GEJ) Cancers | ||
| NCT02614833 | LAG-3-Ig (Eftilagimod Alpha, IMP321), Paclitaxel | Adenocarcinoma Breast Stage IV | ||
| NCT03365791 | Anti-LAG-3 (LAG525), Anti-PD-1 (PDR001) | Advanced Solid and Hematologic Malignancies | ||
| NCT02060188 | Anti-LAG-3 (BMS-986016), Anti-PD-1 (Nivolumab, Opdivo, BMS-936558) | Colorectal Cancer | ||
| NCT02519322 | Anti-LAG-3 (Relatlimab, BMS-986016), Anti-PD-1 (Nivolumab, BMS-936558), surgery | Melanoma | ||
| NCT03642067 | Anti-LAG-3 (Relatlimab, BMS-986016), Anti-PD-1 (Nivolumab, OPDIVO) | Advanced Colorectal Cancer | ||
| NCT03607890 | Anti-LAG-3 (Relatlimab, BMS-986016), Anti-PD-1 (Nivolumab, OPDIVO) | Advanced Mismatch Repair-Deficient Cancers | ||
| II/III | Combination | NCT04129320 | PD-1XLAG-3 bispecific DART protein (MGD013) Anti-B7-H3 (Enoblituzumab, MGA271), anti-PD-1 (MGA012, INCMGA00012) | Head and Neck Cancer |
| NCT04082364 | Anti-HER2 (margetuximab, MGAH22), Anti-PD-1/anti-LAG-3 dual checkpoint inhibitor DART molecule (MGD013), chemotherapy (XELOX (Capecitabine + Oxaliplatin), mFOLFOX-6 (Leucovorin + 5-FU + Oxaliplatin) | Gastric Cancer, Gastroesophageal Junction Cancer, HER2-positive Gastric Cancer |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chocarro, L.; Blanco, E.; Zuazo, M.; Arasanz, H.; Bocanegra, A.; Fernández-Rubio, L.; Morente, P.; Fernández-Hinojal, G.; Echaide, M.; Garnica, M.; et al. Understanding LAG-3 Signaling. Int. J. Mol. Sci. 2021, 22, 5282. https://doi.org/10.3390/ijms22105282
Chocarro L, Blanco E, Zuazo M, Arasanz H, Bocanegra A, Fernández-Rubio L, Morente P, Fernández-Hinojal G, Echaide M, Garnica M, et al. Understanding LAG-3 Signaling. International Journal of Molecular Sciences. 2021; 22(10):5282. https://doi.org/10.3390/ijms22105282
Chicago/Turabian StyleChocarro, Luisa, Ester Blanco, Miren Zuazo, Hugo Arasanz, Ana Bocanegra, Leticia Fernández-Rubio, Pilar Morente, Gonzalo Fernández-Hinojal, Miriam Echaide, Maider Garnica, and et al. 2021. "Understanding LAG-3 Signaling" International Journal of Molecular Sciences 22, no. 10: 5282. https://doi.org/10.3390/ijms22105282
APA StyleChocarro, L., Blanco, E., Zuazo, M., Arasanz, H., Bocanegra, A., Fernández-Rubio, L., Morente, P., Fernández-Hinojal, G., Echaide, M., Garnica, M., Ramos, P., Vera, R., Kochan, G., & Escors, D. (2021). Understanding LAG-3 Signaling. International Journal of Molecular Sciences, 22(10), 5282. https://doi.org/10.3390/ijms22105282