MAGEA4 Coated Extracellular Vesicles Are Stable and Can Be Assembled In Vitro
Abstract
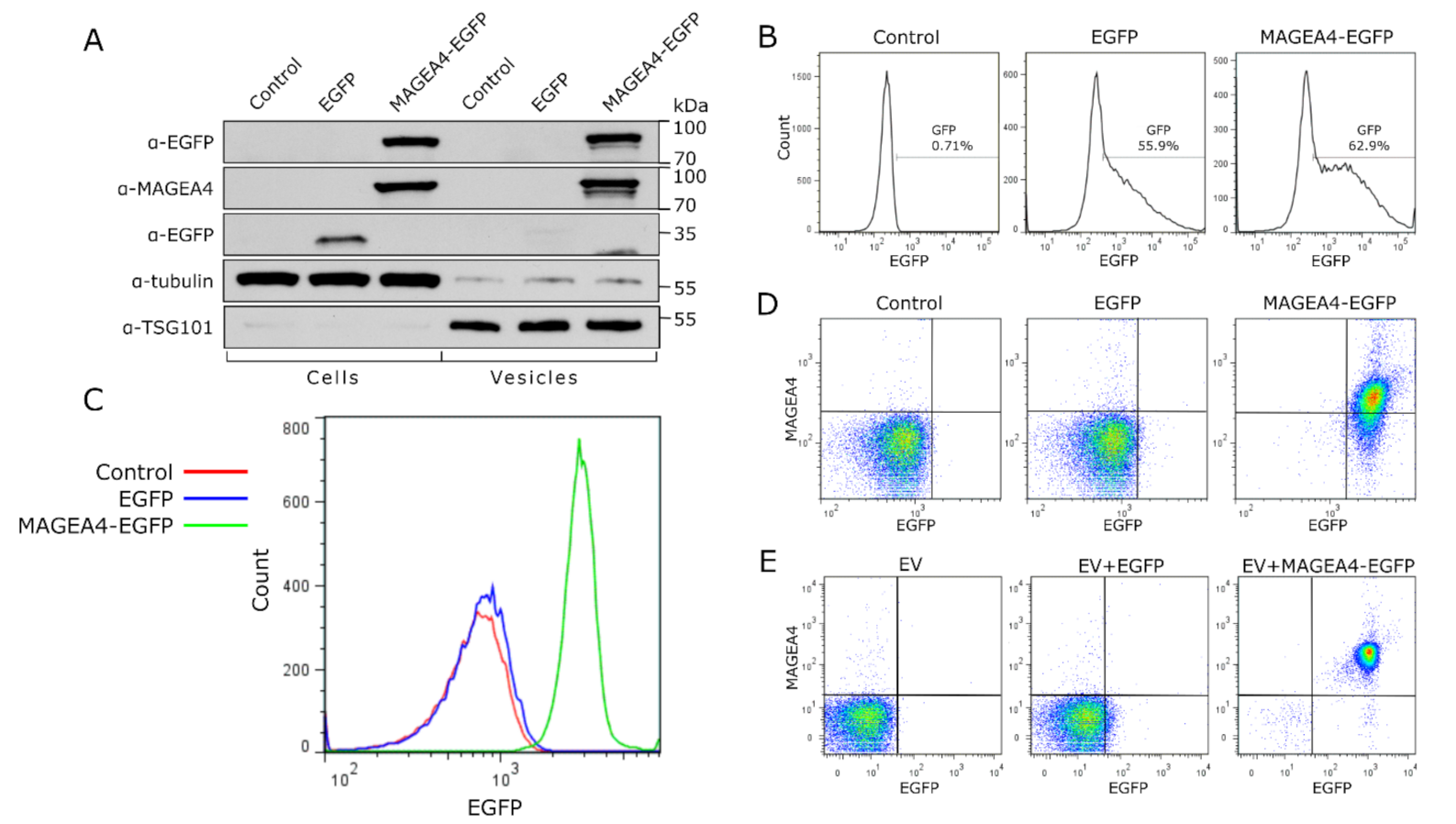
1. Introduction
2. Results
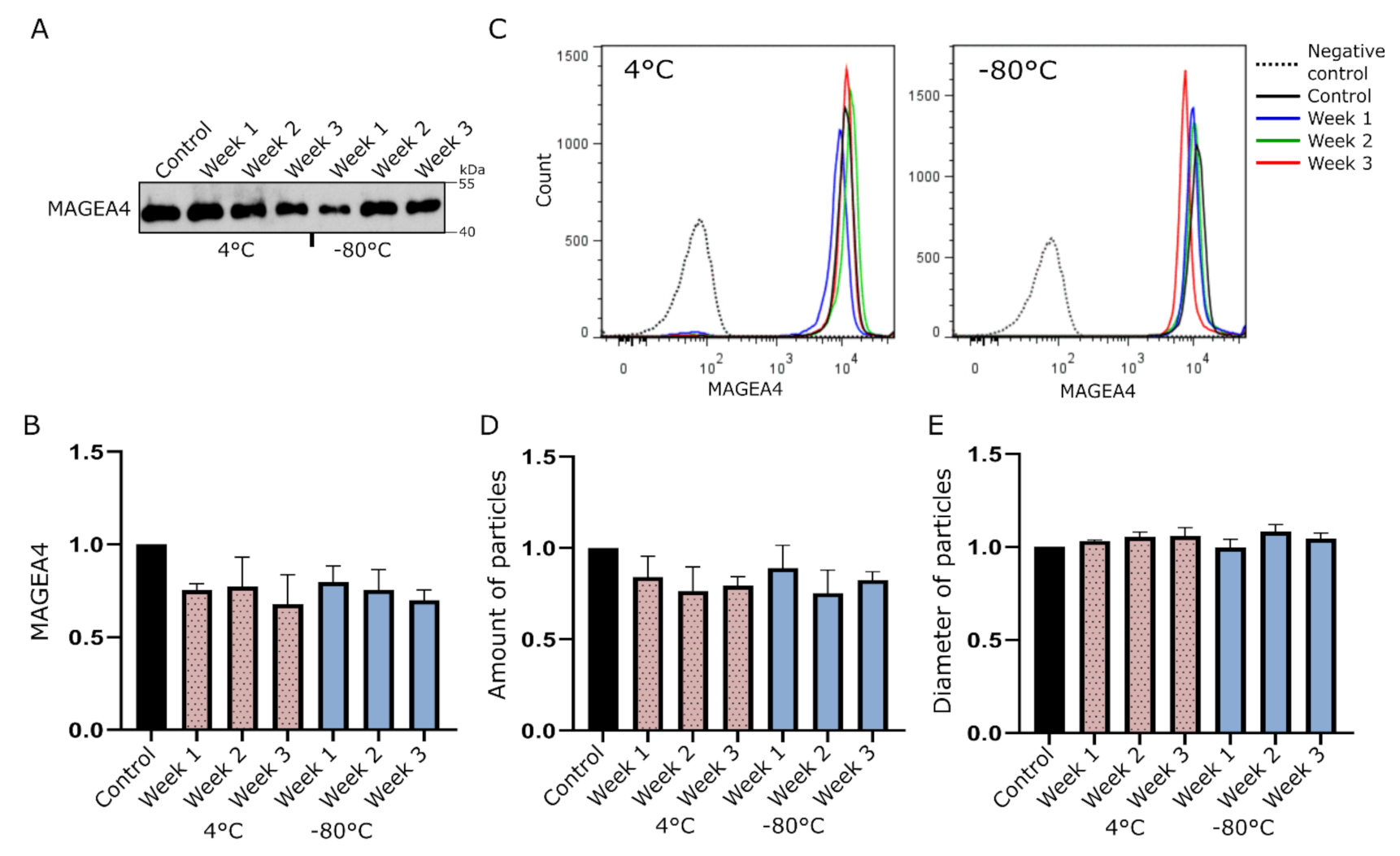
2.1. MAGEA4-EVs Are Stable under Common Storage Conditions
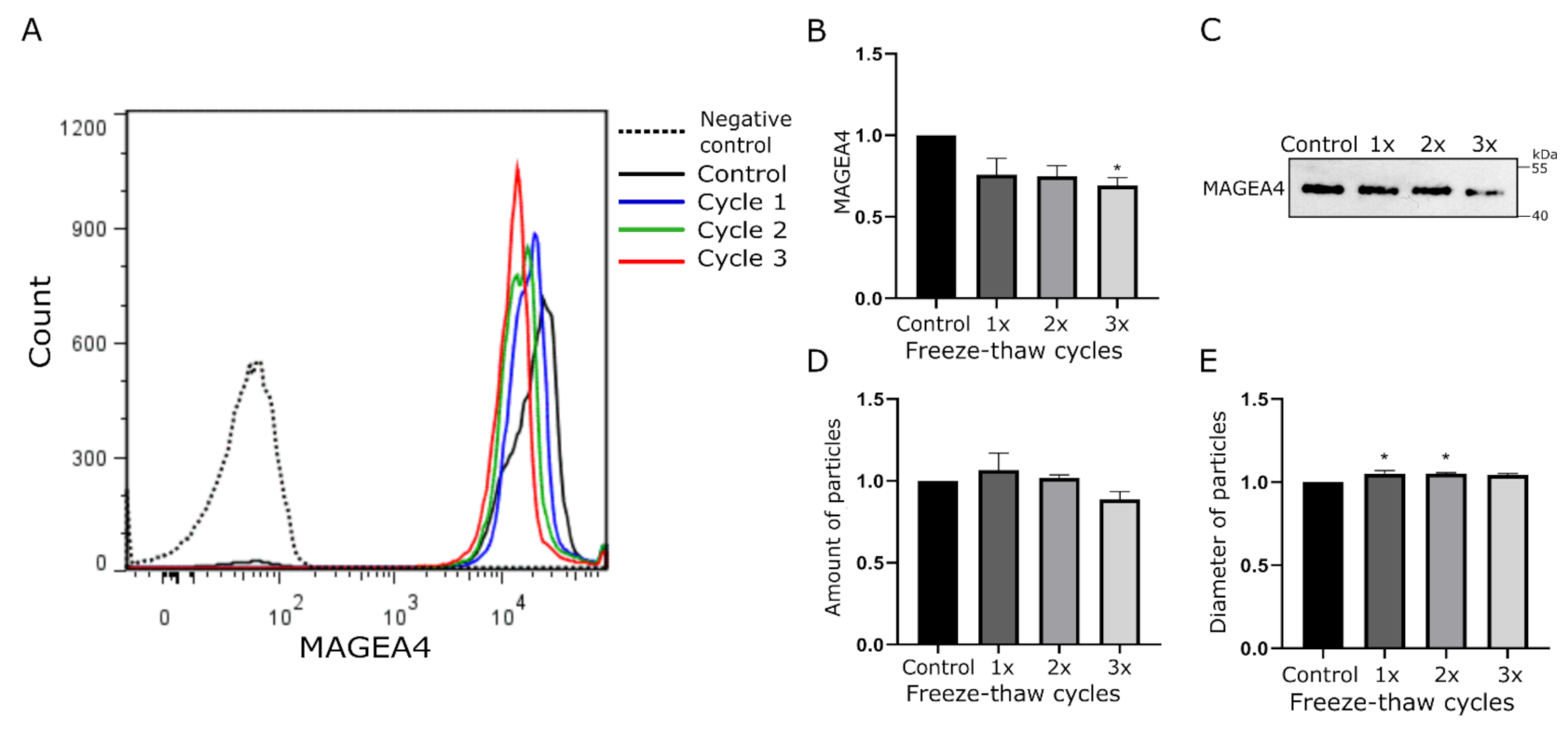
2.2. MAGEA4-EVs Are Stable to Freeze-Thaw Cycles
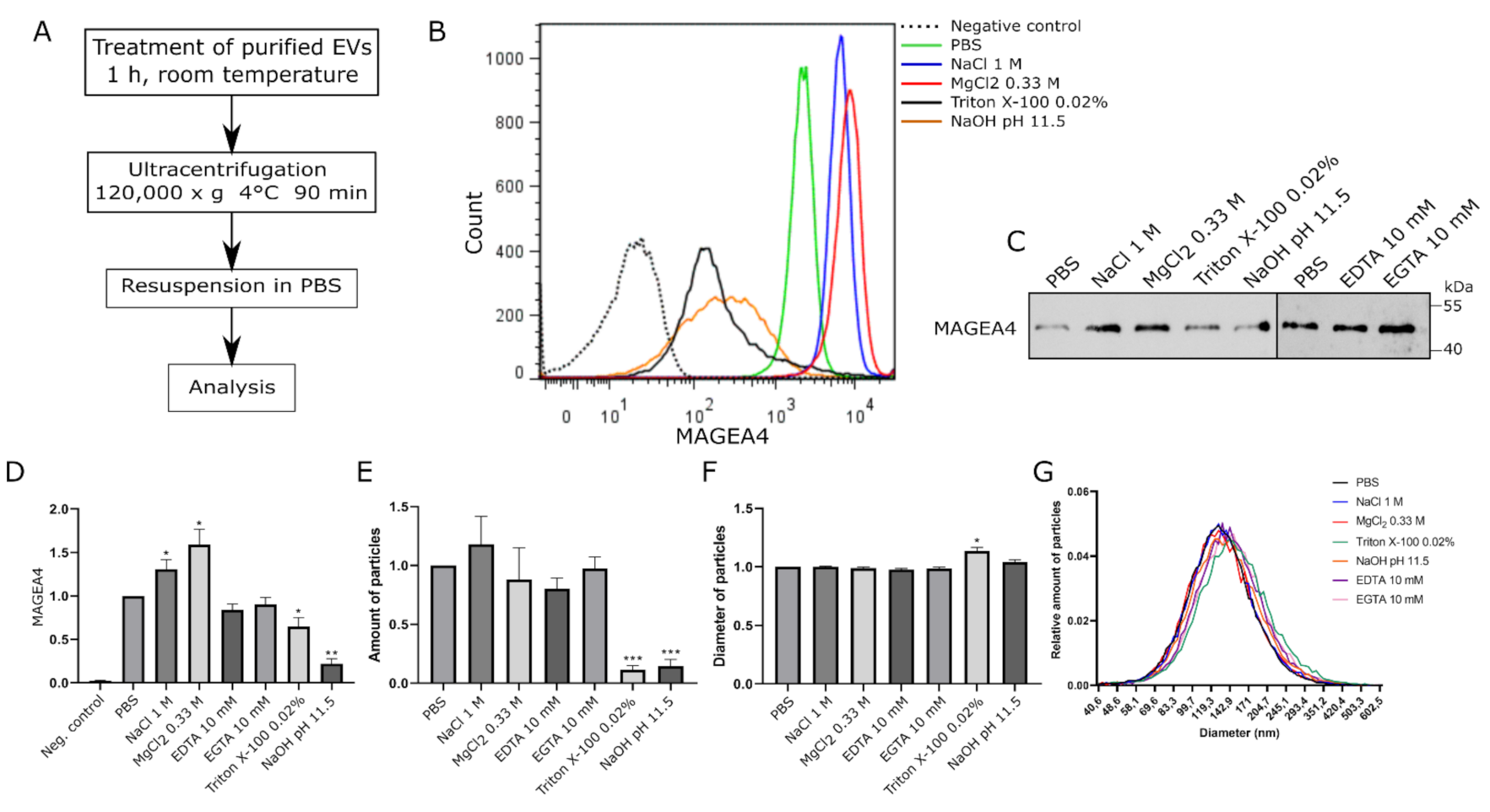
2.3. MAGEA4-EVs Are Resistant to the Treatment with High Salt
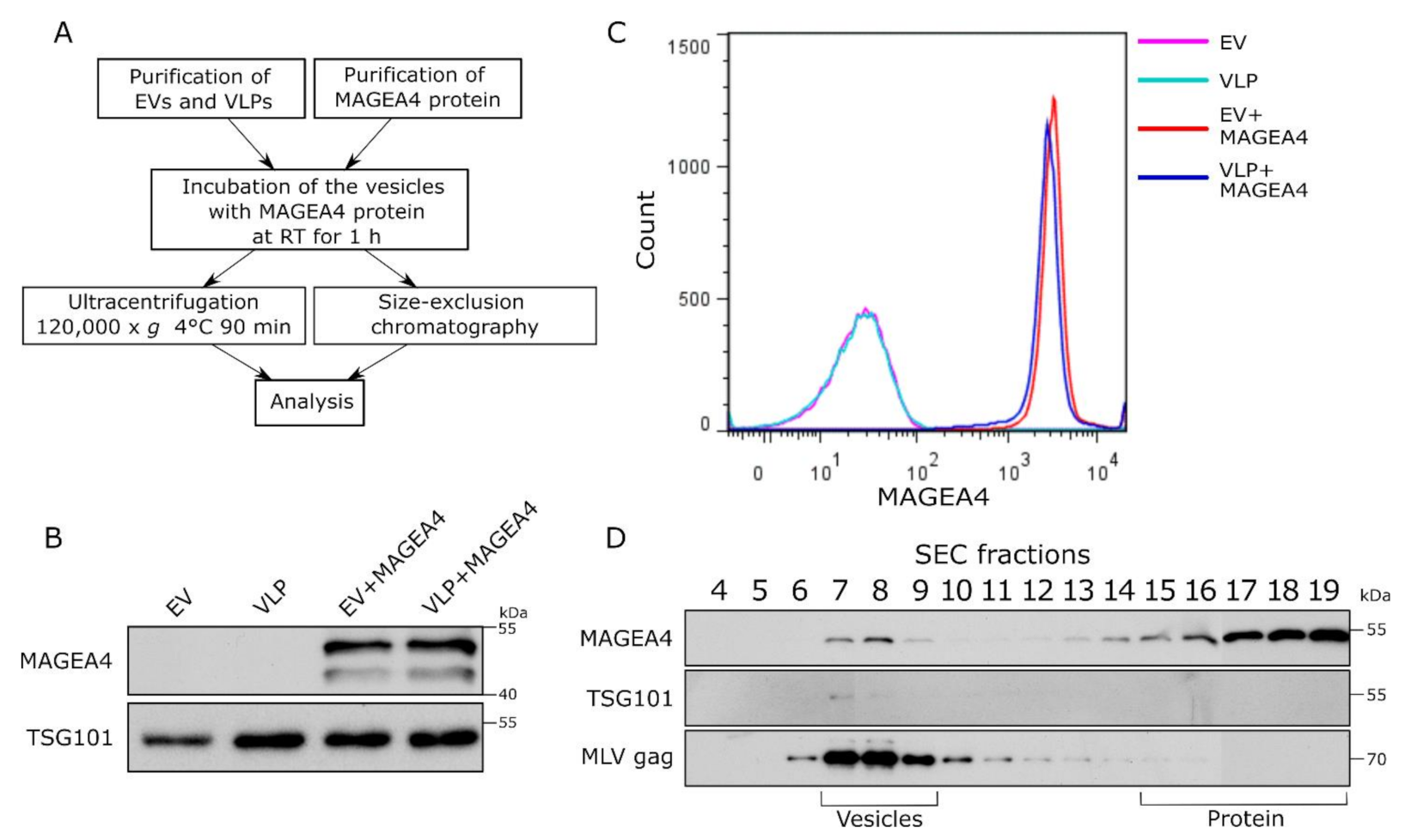
2.4. Purified MAGEA4 Protein Associates with Extracellular Vesicles In Vitro
2.5. MAGEA4 Can Be Used for Decorating EVs with Recombinant Proteins
3. Discussion
4. Materials and Methods
4.1. Cells and Plasmids
4.2. Proteins
4.3. Isolation and Purification of EVs and VLPs
4.4. Physico-Chemical Treatment of MAGEA4-EVs
4.5. In Vitro Binding Experiment
4.6. Western Blot Analysis
4.7. Flow Cytometry
4.8. Analysis of EVs by NTA
4.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Caby, M.-P.; Lankar, D.; Vincendeau-Scherrer, C.; Raposo, G.; Bonnerot, C. Exosomal-like Vesicles Are Present in Human Blood Plasma. Int. Immunol. 2005, 17, 879–887. [Google Scholar] [CrossRef] [PubMed]
- Pisitkun, T.; Shen, R.-F.; Knepper, M.A. Identification and Proteomic Profiling of Exosomes in Human Urine. Proc. Natl. Acad. Sci. USA 2004, 101, 13368–13373. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, Y.; Miura, Y.; Harazono, A.; Kanai-Azuma, M.; Akimoto, Y.; Kawakami, H.; Yamaguchi, T.; Toda, T.; Endo, T.; Tsubuki, M.; et al. Proteomic Analysis of Two Types of Exosomes in Human Whole Saliva. Biol. Pharm. Bull. 2011, 34, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, G.; Brody, I. The Prostasome: Its Secretion and Function in Man. Biochim. Biophys. Acta BBA-Rev. Biomembr. 1985, 822, 203–218. [Google Scholar] [CrossRef]
- Boilard, E.; Nigrovic, P.A.; Larabee, K.; Watts, G.F.M.; Coblyn, J.S.; Weinblatt, M.E.; Massarotti, E.M.; Remold-O’Donnell, E.; Farndale, R.W.; Ware, J.; et al. Platelets Amplify Inflammation in Arthritis via Collagen-Dependent Microparticle Production. Science 2010, 327, 580–583. [Google Scholar] [CrossRef]
- Bobrie, A.; Colombo, M.; Raposo, G.; Théry, C. Exosome Secretion: Molecular Mechanisms and Roles in Immune Responses. Traffic 2011, 12, 1659–1668. [Google Scholar] [CrossRef]
- Chargaff, E.; West, R. The biological significance of the thromboplastic protein of blood. J. Biol. Chem. 1946, 166, 189–197. [Google Scholar] [CrossRef]
- Wolf, P. The Nature and Significance of Platelet Products in Human Plasma. Br. J. Haematol. 1967, 13, 269–288. [Google Scholar] [CrossRef]
- Bastida, E.; Ordinas, A.; Escolar, G.; Jamieson, G.A. Tissue Factor in Microvesicles Shed From U87MG Human Glioblastoma Cells Induces Coagulation, Platelet Aggregation, and Thrombogenesis. Blood 1984, 64, 177–184. [Google Scholar] [CrossRef]
- Kosaka, N.; Yoshioka, Y.; Hagiwara, K.; Tominaga, N.; Katsuda, T.; Ochiya, T. Trash or Treasure: Extracellular MicroRNAs and Cell-to-Cell Communication. Front. Genet. 2013, 4. [Google Scholar] [CrossRef]
- Mulcahy, L.A.; Pink, R.C.; Carter, D.R.F. Routes and Mechanisms of Extracellular Vesicle Uptake. J. Extracell. Vesicles 2014, 3, 24641. [Google Scholar] [CrossRef]
- Zitvogel, L.; Regnault, A.; Lozier, A.; Wolfers, J.; Flament, C.; Tenza, D.; Ricciardi-Castagnoli, P.; Raposo, G.; Amigorena, S. Eradication of Established Murine Tumors Using a Novel Cell-Free Vaccine: Dendritic Cell Derived Exosomes. Nat. Med. 1998, 4, 594–600. [Google Scholar] [CrossRef]
- Mathivanan, S.; Ji, H.; Simpson, R.J. Exosomes: Extracellular Organelles Important in Intercellular Communication. J. Proteom. 2010, 73, 1907–1920. [Google Scholar] [CrossRef]
- Record, M.; Carayon, K.; Poirot, M.; Silvente-Poirot, S. Exosomes as New Vesicular Lipid Transporters Involved in Cell–Cell Communication and Various Pathophysiologies. Biochim. Biophys. Acta BBA-Mol. Cell Biol. Lipids 2014, 1841, 108–120. [Google Scholar] [CrossRef]
- Yuana, Y.; Sturk, A.; Nieuwland, R. Extracellular Vesicles in Physiological and Pathological Conditions. Blood Rev. 2013, 27, 31–39. [Google Scholar] [CrossRef]
- van der Pol, E.; Böing, A.N.; Harrison, P.; Sturk, A.; Nieuwland, R. Classification, Functions, and Clinical Relevance of Extracellular Vesicles. Pharmacol. Rev. 2012, 64, 676–705. [Google Scholar] [CrossRef]
- van Balkom, B.W.M.; de Jong, O.G.; Smits, M.; Brummelman, J.; den Ouden, K.; de Bree, P.M.; van Eijndhoven, M.A.J.; Pegtel, D.M.; Stoorvogel, W.; Würdinger, T.; et al. Endothelial Cells Require MiR-214 to Secrete Exosomes That Suppress Senescence and Induce Angiogenesis in Human and Mouse Endothelial Cells. Blood 2013, 121, 3997–4006. [Google Scholar] [CrossRef]
- Barile, L.; Lionetti, V.; Cervio, E.; Matteucci, M.; Gherghiceanu, M.; Popescu, L.M.; Torre, T.; Siclari, F.; Moccetti, T.; Vassalli, G. Extracellular Vesicles from Human Cardiac Progenitor Cells Inhibit Cardiomyocyte Apoptosis and Improve Cardiac Function after Myocardial Infarction. Cardiovasc. Res. 2014, 103, 530–541. [Google Scholar] [CrossRef]
- Lopez-Verrilli, M.A.; Picou, F.; Court, F.A. Schwann Cell-Derived Exosomes Enhance Axonal Regeneration in the Peripheral Nervous System. Glia 2013, 61, 1795–1806. [Google Scholar] [CrossRef]
- Guescini, M.; Guidolin, D.; Vallorani, L.; Casadei, L.; Gioacchini, A.M.; Tibollo, P.; Battistelli, M.; Falcieri, E.; Battistin, L.; Agnati, L.F.; et al. C2C12 Myoblasts Release Micro-Vesicles Containing MtDNA and Proteins Involved in Signal Transduction. Exp. Cell Res. 2010, 316, 1977–1984. [Google Scholar] [CrossRef]
- Qiao, L.; Hu, S.; Liu, S.; Zhang, H.; Ma, H.; Huang, K.; Li, Z.; Su, T.; Vandergriff, A.; Tang, J.; et al. MicroRNA-21-5p Dysregulation in Exosomes Derived from Heart Failure Patients Impairs Regenerative Potential. J. Clin. Investig. 2019, 129, 2237–2250. [Google Scholar] [CrossRef] [PubMed]
- An, M.; Kwon, K.; Park, J.; Ryu, D.-R.; Shin, J.-A.; Lee Kang, J.; Choi, J.H.; Park, E.-M.; Lee, K.E.; Woo, M.; et al. Extracellular Matrix-Derived Extracellular Vesicles Promote Cardiomyocyte Growth and Electrical Activity in Engineered Cardiac Atria. Biomaterials 2017, 146, 49–59. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xu, C.; Nan, Y.; Nan, S. Microglia-Derived Extracellular Vesicles Carrying MiR-711 Alleviate Neurodegeneration in a Murine Alzheimer’s Disease Model by Binding to Itpkb. Front. Cell Dev. Biol. 2020, 8. [Google Scholar] [CrossRef] [PubMed]
- Dolcetti, E.; Bruno, A.; Guadalupi, L.; Rizzo, F.R.; Musella, A.; Gentile, A.; De Vito, F.; Caioli, S.; Bullitta, S.; Fresegna, D.; et al. Emerging Role of Extracellular Vesicles in the Pathophysiology of Multiple Sclerosis. Int. J. Mol. Sci. 2020, 21, 7336. [Google Scholar] [CrossRef]
- Hoshino, D.; Kirkbride, K.C.; Costello, K.; Clark, E.S.; Sinha, S.; Grega-Larson, N.; Tyska, M.J.; Weaver, A.M. Exosome Secretion Is Enhanced by Invadopodia and Drives Invasive Behavior. Cell Rep. 2013, 5, 1159–1168. [Google Scholar] [CrossRef]
- Peinado, H.; Alečković, M.; Lavotshkin, S.; Matei, I.; Costa-Silva, B.; Moreno-Bueno, G.; Hergueta-Redondo, M.; Williams, C.; García-Santos, G.; Ghajar, C.M.; et al. Melanoma Exosomes Educate Bone Marrow Progenitor Cells toward a Pro-Metastatic Phenotype through MET. Nat. Med. 2012, 18, 883–891. [Google Scholar] [CrossRef]
- Boelens, M.C.; Wu, T.J.; Nabet, B.Y.; Xu, B.; Qiu, Y.; Yoon, T.; Azzam, D.J.; Twyman-Saint Victor, C.; Wiemann, B.Z.; Ishwaran, H.; et al. Exosome Transfer from Stromal to Breast Cancer Cells Regulates Therapy Resistance Pathways. Cell 2014, 159, 499–513. [Google Scholar] [CrossRef]
- Fong, M.Y.; Zhou, W.; Liu, L.; Alontaga, A.Y.; Chandra, M.; Ashby, J.; Chow, A.; O’Connor, S.T.F.; Li, S.; Chin, A.R.; et al. Breast-Cancer-Secreted MiR-122 Reprograms Glucose Metabolism in Premetastatic Niche to Promote Metastasis. Nat. Cell Biol. 2015, 17, 183–194. [Google Scholar] [CrossRef]
- Perut, F.; Roncuzzi, L.; Baldini, N. The Emerging Roles of Extracellular Vesicles in Osteosarcoma. Front. Oncol. 2019, 9. [Google Scholar] [CrossRef]
- Lu, Z.; Zuo, B.; Jing, R.; Gao, X.; Rao, Q.; Liu, Z.; Qi, H.; Guo, H.; Yin, H. Dendritic Cell-Derived Exosomes Elicit Tumor Regression in Autochthonous Hepatocellular Carcinoma Mouse Models. J. Hepatol. 2017, 67, 739–748. [Google Scholar] [CrossRef]
- Delabranche, X.; Berger, A.; Boisramé-Helms, J.; Meziani, F. Microparticles and Infectious Diseases. Médecine Mal. Infect. 2012, 42, 335–343. [Google Scholar] [CrossRef]
- Quinn, Z.; Mao, W.; Xia, Y.; John, R.; Wan, Y. Conferring Receptors on Recipient Cells with Extracellular Vesicles for Targeted Drug Delivery. Bioact. Mater. 2021, 6, 749–756. [Google Scholar] [CrossRef]
- Schwarzenbach, H.; Hoon, D.S.B.; Pantel, K. Cell-Free Nucleic Acids as Biomarkers in Cancer Patients. Nat. Rev. Cancer 2011, 11, 426–437. [Google Scholar] [CrossRef]
- Perakis, S.; Speicher, M.R. Emerging Concepts in Liquid Biopsies. BMC Med. 2017, 15, 75. [Google Scholar] [CrossRef]
- Skog, J.; Würdinger, T.; van Rijn, S.; Meijer, D.H.; Gainche, L.; Curry, W.T.; Carter, B.S.; Krichevsky, A.M.; Breakefield, X.O. Glioblastoma Microvesicles Transport RNA and Proteins That Promote Tumour Growth and Provide Diagnostic Biomarkers. Nat. Cell Biol. 2008, 10, 1470–1476. [Google Scholar] [CrossRef]
- Melo, S.A.; Luecke, L.B.; Kahlert, C.; Fernandez, A.F.; Gammon, S.T.; Kaye, J.; LeBleu, V.S.; Mittendorf, E.A.; Weitz, J.; Rahbari, N.; et al. Glypican-1 Identifies Cancer Exosomes and Detects Early Pancreatic Cancer. Nature 2015, 523, 177–182. [Google Scholar] [CrossRef]
- Risha, Y.; Minic, Z.; Ghobadloo, S.M.; Berezovski, M.V. The Proteomic Analysis of Breast Cell Line Exosomes Reveals Disease Patterns and Potential Biomarkers. Sci. Rep. 2020, 10, 13572. [Google Scholar] [CrossRef]
- van der Bruggen, P.; Traversari, C.; Chomez, P.; Lurquin, C.; Plaen, E.D.; van den Eynde, B.; Knuth, A.; Boon, T. A Gene Encoding an Antigen Recognized by Cytolytic T Lymphocytes on a Human Melanoma. Science 1991, 254, 1643–1647. [Google Scholar] [CrossRef]
- Chen, Y.-T.; Scanlan, M.J.; Sahin, U.; Türeci, Ö.; Gure, A.O.; Tsang, S.; Williamson, B.; Stockert, E.; Pfreundschuh, M.; Old, L.J. A Testicular Antigen Aberrantly Expressed in Human Cancers Detected by Autologous Antibody Screening. Proc. Natl. Acad. Sci. USA 1997, 94, 1914–1918. [Google Scholar] [CrossRef]
- Brasseur, F.; Rimoldi, D.; Liénard, D.; Lethé, B.; Carrel, S.; Arienti, F.; Suter, L.; Vanwijck, R.; Bourlond, A.; Humblet, Y.; et al. Expression of MAGE Genes in Primary and Metastatic Cutaneous Melanoma. Int. J. Cancer 1995, 63, 375–380. [Google Scholar] [CrossRef]
- Barrow, C.; Browning, J.; MacGregor, D.; Davis, I.D.; Sturrock, S.; Jungbluth, A.A.; Cebon, J. Tumor Antigen Expression in Melanoma Varies According to Antigen and Stage. Clin. Cancer Res. 2006, 12, 764–771. [Google Scholar] [CrossRef]
- Zhang, L.; Li, B.; Peng, Y.; Wu, F.; Li, Q.; Lin, Z.; Xie, S.; Xiao, L.; Lin, X.; Ou, Z.; et al. The Prognostic Value of TMB and the Relationship between TMB and Immune Infiltration in Head and Neck Squamous Cell Carcinoma: A Gene Expression-Based Study. Oral Oncol. 2020, 110, 104943. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Shen, Y.; Wen, S.; Bajorin, D.F.; Reuter, V.E.; Old, L.J.; Jungbluth, A.A. Cancer-Testis Antigens: Expression and Correlation with Survival in Human Urothelial Carcinoma. Clin. Cancer Res. 2006, 12, 5442–5447. [Google Scholar] [CrossRef] [PubMed]
- Ries, J.; Schultze-Mosgau, S.; Neukam, F.; Diebel, E.; Wiltfang, J. Investigation of the Expression of Melanoma Antigen-Encoding Genes (MAGE-A1 to -A6) in Oral Squamous Cell Carcinomas to Determine Potential Targets for Gene-Based Cancer Immunotherapy. Int. J. Oncol. 2005, 26, 817–824. [Google Scholar] [CrossRef] [PubMed]
- Peikert, T.; Specks, U.; Farver, C.; Erzurum, S.C.; Comhair, S.A.A. Melanoma Antigen A4 Is Expressed in Non–Small Cell Lung Cancers and Promotes Apoptosis. Cancer Res. 2006, 66, 4693–4700. [Google Scholar] [CrossRef] [PubMed]
- Yakirevich, E.; Sabo, E.; Lavie, O.; Mazareb, S.; Spagnoli, G.C.; Resnick, M.B. Expression of the MAGE-A4 and NY-ESO-1 Cancer-Testis Antigens in Serous Ovarian Neoplasms. Clin. Cancer Res. 2003, 9, 6453–6460. [Google Scholar]
- Õunap, K.; Kurg, K.; Võsa, L.; Maiväli, Ü.; Teras, M.; Planken, A.; Ustav, M.; Kurg, R. Antibody Response against Cancer-testis Antigens MAGEA4 and MAGEA10 in Patients with Melanoma. Oncol. Lett. 2018, 16, 211–218. [Google Scholar] [CrossRef]
- Cruz, C.R.; Gerdemann, U.; Leen, A.M.; Shafer, J.A.; Ku, S.; Tzou, B.; Horton, T.M.; Sheehan, A.; Copeland, A.; Younes, A.; et al. Improving T-Cell Therapy for Relapsed EBV-Negative Hodgkin Lymphoma by Targeting Upregulated MAGE-A4. Clin. Cancer Res. 2011, 17, 7058–7066. [Google Scholar] [CrossRef]
- Jia, Z.-C.; Ni, B.; Huang, Z.-M.; Tian, Y.; Tang, J.; Wang, J.-X.; Fu, X.-L.; Wu, Y.-Z. Identification of Two Novel HLA-A*0201-Restricted CTL Epitopes Derived from MAGE-A4. Clin. Dev. Immunol. 2010, 2010, 567594. [Google Scholar] [CrossRef]
- Sanderson, J.P.; Crowley, D.J.; Wiedermann, G.E.; Quinn, L.L.; Crossland, K.L.; Tunbridge, H.M.; Cornforth, T.V.; Barnes, C.S.; Ahmed, T.; Howe, K.; et al. Preclinical Evaluation of an Affinity-Enhanced MAGE-A4-Specific T-Cell Receptor for Adoptive T-Cell Therapy. OncoImmunology 2020, 9, 1682381. [Google Scholar] [CrossRef]
- Meek, D.W.; Marcar, L. MAGE-A Antigens as Targets in Tumour Therapy. Cancer Lett. 2012, 324, 126–132. [Google Scholar] [CrossRef]
- Lulla, P.D.; Tzannou, I.; Vasileiou, S.; Carrum, G.; Ramos, C.A.; Kamble, R.; Wang, T.; Wu, M.; Bilgi, M.; Gee, A.P.; et al. The Safety and Clinical Effects of Administering a Multiantigen-Targeted T Cell Therapy to Patients with Multiple Myeloma. Sci. Transl. Med. 2020, 12. [Google Scholar] [CrossRef]
- Kurg, R.; Reinsalu, O.; Jagur, S.; Õunap, K.; Võsa, L.; Kasvandik, S.; Padari, K.; Gildemann, K.; Ustav, M. Biochemical and Proteomic Characterization of Retrovirus Gag Based Microparticles Carrying Melanoma Antigens. Sci. Rep. 2016, 6, 29425. [Google Scholar] [CrossRef]
- Kuldkepp, A.; Karakai, M.; Toomsoo, E.; Reinsalu, O.; Kurg, R. Cancer-Testis Antigens MAGEA Proteins Are Incorporated into Extracellular Vesicles Released by Cells. Oncotarget 2019, 10, 3694–3708. [Google Scholar] [CrossRef]
- Maroto, R.; Zhao, Y.; Jamaluddin, M.; Popov, V.L.; Wang, H.; Kalubowilage, M.; Zhang, Y.; Luisi, J.; Sun, H.; Culbertson, C.T.; et al. Effects of Storage Temperature on Airway Exosome Integrity for Diagnostic and Functional Analyses. J. Extracell. Vesicles 2017, 6, 1359478. [Google Scholar] [CrossRef]
- Smith, S.M. Strategies for the Purification of Membrane Proteins. In Protein Chromatography; Walls, D., Loughran, S.T., Eds.; Methods in Molecular Biology; Springer New York: New Yoker, NY, USA, 2017; Volume 1485, pp. 389–400. ISBN 978-1-4939-6410-9. [Google Scholar]
- Cray, J.A.; Russell, J.T.; Timson, D.J.; Singhal, R.S.; Hallsworth, J.E. A Universal Measure of Chaotropicity and Kosmotropicity: A Universal Measure of Chao- and Kosmotropicity. Environ. Microbiol. 2013, 15, 287–296. [Google Scholar] [CrossRef]
- Kräusslich, H.G.; Ochsenbauer, C.; Traenckner, A.-M.; Mergener, K.; Fäcke, M.; Gelderblom, H.R.; Bosch, V. Analysis of Protein Expression and Virus-like Particle Formation in Mammalian Cell Lines Stably Expressing HIV-1 Gag and Env Gene Products with or without Active HIV Proteinase. Virology 1993, 192, 605–617. [Google Scholar] [CrossRef]
- Pusic, A.D.; Pusic, K.M.; Clayton, B.L.L.; Kraig, R.P. IFNγ-Stimulated Dendritic Cell Exosomes as a Potential Therapeutic for Remyelination. J. Neuroimmunol. 2014, 266, 12–23. [Google Scholar] [CrossRef]
- Bu, N.; Wu, H.-Q.; Zhang, G.-L.; Zhan, S.-Q.; Zhang, R.; Fan, Q.-Y.; Li, Y.-L.; Zhai, Y.-F.; Ren, H.-W. Immature Dendritic Cell Exosomes Suppress Experimental Autoimmune Myasthenia Gravis. J. Neuroimmunol. 2015, 285, 71–75. [Google Scholar] [CrossRef]
- Wang, B.; Yao, K.; Huuskes, B.M.; Shen, H.-H.; Zhuang, J.; Godson, C.; Brennan, E.P.; Wilkinson-Berka, J.L.; Wise, A.F.; Ricardo, S.D. Mesenchymal Stem Cells Deliver Exogenous MicroRNA-Let7c via Exosomes to Attenuate Renal Fibrosis. Mol. Ther. 2016, 24, 1290–1301. [Google Scholar] [CrossRef]
- Tan, J.L.; Lau, S.N.; Leaw, B.; Nguyen, H.P.T.; Salamonsen, L.A.; Saad, M.I.; Chan, S.T.; Zhu, D.; Krause, M.; Kim, C.; et al. Amnion Epithelial Cell-Derived Exosomes Restrict Lung Injury and Enhance Endogenous Lung Repair. STEM CELLS Transl. Med. 2018, 7, 180–196. [Google Scholar] [CrossRef] [PubMed]
- Khatua, A.K.; Taylor, H.E.; Hildreth, J.E.K.; Popik, W. Exosomes Packaging APOBEC3G Confer Human Immunodeficiency Virus Resistance to Recipient Cells. J. Virol. 2009, 83, 512–521. [Google Scholar] [CrossRef] [PubMed]
- Kuate, S.; Cinatl, J.; Doerr, H.W.; Überla, K. Exosomal Vaccines Containing the S Protein of the SARS Coronavirus Induce High Levels of Neutralizing Antibodies. Virology 2007, 362, 26–37. [Google Scholar] [CrossRef] [PubMed]
- Dai, S.; Wei, D.; Wu, Z.; Zhou, X.; Wei, X.; Huang, H.; Li, G. Phase I Clinical Trial of Autologous Ascites-Derived Exosomes Combined With GM-CSF for Colorectal Cancer. Mol. Ther. 2008, 16, 782–790. [Google Scholar] [CrossRef]
- Yang, T.; Martin, P.; Fogarty, B.; Brown, A.; Schurman, K.; Phipps, R.; Yin, V.P.; Lockman, P.; Bai, S. Exosome Delivered Anticancer Drugs across the Blood-Brain Barrier for Brain Cancer Therapy in Danio Rerio. Pharm. Res. 2015, 32, 2003–2014. [Google Scholar] [CrossRef]
- Wang, G.; Hu, W.; Chen, H.; Shou, X.; Ye, T.; Xu, Y. Cocktail Strategy Based on NK Cell-Derived Exosomes and Their Biomimetic Nanoparticles for Dual Tumor Therapy. Cancers 2019, 11, 1560. [Google Scholar] [CrossRef]
- Kusuma, G.D.; Barabadi, M.; Tan, J.L.; Morton, D.A.V.; Frith, J.E.; Lim, R. To Protect and to Preserve: Novel Preservation Strategies for Extracellular Vesicles. Front. Pharmacol. 2018, 9. [Google Scholar] [CrossRef]
- Cheng, Y.; Zeng, Q.; Han, Q.; Xia, W. Effect of PH, Temperature and Freezing-Thawing on Quantity Changes and Cellular Uptake of Exosomes. Protein Cell 2019, 10, 295–299. [Google Scholar] [CrossRef]
- Wu, Y.; Deng, W.; Ii, D.J.K. Exosomes: Improved Methods to Characterize Their Morphology, RNA Content, and Surface Protein Biomarkers. Analyst 2015, 140, 6631–6642. [Google Scholar] [CrossRef]
- Frank, J.; Richter, M.; de Rossi, C.; Lehr, C.-M.; Fuhrmann, K.; Fuhrmann, G. Extracellular Vesicles Protect Glucuronidase Model Enzymes during Freeze-Drying. Sci. Rep. 2018, 8, 12377. [Google Scholar] [CrossRef]
- Lőrincz, Á.M.; Timár, C.I.; Marosvári, K.A.; Veres, D.S.; Otrokocsi, L.; Kittel, Á.; Ligeti, E. Effect of Storage on Physical and Functional Properties of Extracellular Vesicles Derived from Neutrophilic Granulocytes. J. Extracell. Vesicles 2014, 3, 25465. [Google Scholar] [CrossRef]
- Sokolova, V.; Ludwig, A.-K.; Hornung, S.; Rotan, O.; Horn, P.A.; Epple, M.; Giebel, B. Characterisation of Exosomes Derived from Human Cells by Nanoparticle Tracking Analysis and Scanning Electron Microscopy. Colloids Surf. B Biointerfaces 2011, 87, 146–150. [Google Scholar] [CrossRef]
- Kumeda, N.; Ogawa, Y.; Akimoto, Y.; Kawakami, H.; Tsujimoto, M.; Yanoshita, R. Characterization of Membrane Integrity and Morphological Stability of Human Salivary Exosomes. Biol. Pharm. Bull. 2017, 40, 1183–1191. [Google Scholar] [CrossRef]
- Akers, J.C.; Ramakrishnan, V.; Yang, I.; Hua, W.; Mao, Y.; Carter, B.S.; Chen, C.C. Optimizing Preservation of Extracellular Vesicular MiRNAs Derived from Clinical Cerebrospinal Fluid. Cancer Biomark. 2016, 17, 125–132. [Google Scholar] [CrossRef]
- Jin, Y.; Chen, K.; Wang, Z.; Wang, Y.; Liu, J.; Lin, L.; Shao, Y.; Gao, L.; Yin, H.; Cui, C.; et al. DNA in Serum Extracellular Vesicles Is Stable under Different Storage Conditions. BMC Cancer 2016, 16, 753. [Google Scholar] [CrossRef]
- Zhou, H.; Yuen, P.S.T.; Pisitkun, T.; Gonzales, P.A.; Yasuda, H.; Dear, J.W.; Gross, P.; Knepper, M.A.; Star, R.A. Collection, Storage, Preservation, and Normalization of Human Urinary Exosomes for Biomarker Discovery. Kidney Int. 2006, 69, 1471–1476. [Google Scholar] [CrossRef]
- Ge, Q.; Zhou, Y.; Lu, J.; Bai, Y.; Xie, X.; Lu, Z. MiRNA in Plasma Exosome Is Stable under Different Storage Conditions. Molecules 2014, 19, 1568–1575. [Google Scholar] [CrossRef]
- Lv, L.-L.; Cao, Y.; Liu, D.; Xu, M.; Liu, H.; Tang, R.-N.; Ma, K.-L.; Liu, B.-C. Isolation and Quantification of MicroRNAs from Urinary Exosomes/Microvesicles for Biomarker Discovery. Int. J. Biol. Sci. 2013, 9, 1021–1031. [Google Scholar] [CrossRef]
- Linares, R.; Tan, S.; Gounou, C.; Arraud, N.; Brisson, A.R. High-Speed Centrifugation Induces Aggregation of Extracellular Vesicles. J. Extracell. Vesicles 2015, 4, 29509. [Google Scholar] [CrossRef]
- Ban, J.-J.; Lee, M.; Im, W.; Kim, M. Low PH Increases the Yield of Exosome Isolation. Biochem. Biophys. Res. Commun. 2015, 461, 76–79. [Google Scholar] [CrossRef]
- Sabapathy, T.; Helmerhorst, E.; Bottomley, S.; Babaeff, S.; Munyard, K.; Newsholme, P.; Mamotte, C.D. Use of Virus-like Particles as a Native Membrane Model to Study the Interaction of Insulin with the Insulin Receptor. Biochim. Biophys. Acta BBA-Biomembr. 2019, 1861, 1204–1212. [Google Scholar] [CrossRef]
- Meckes, D.G.; Raab-Traub, N. Microvesicles and Viral Infection. J. Virol. 2011, 85, 12844–12854. [Google Scholar] [CrossRef]
- Xin, L.; Yuan, Y.-W.; Liu, C.; Zhou, L.-Q.; Liu, L.; Zhou, Q.; Li, S.-H. Preparation of Internalizing RGD-Modified Recombinant Methioninase Exosome Active Targeting Vector and Antitumor Effect Evaluation. Dig. Dis. Sci. 2020. [Google Scholar] [CrossRef]
- Park, O.; Choi, E.S.; Yu, G.; Kim, J.Y.; Kang, Y.Y.; Jung, H.; Mok, H. Efficient Delivery of Tyrosinase Related Protein-2 (TRP2) Peptides to Lymph Nodes Using Serum-Derived Exosomes. Macromol. Biosci. 2018, 18, 1800301. [Google Scholar] [CrossRef]
- Haney, M.J.; Klyachko, N.L.; Zhao, Y.; Gupta, R.; Plotnikova, E.G.; He, Z.; Patel, T.; Piroyan, A.; Sokolsky, M.; Kabanov, A.V.; et al. Exosomes as Drug Delivery Vehicles for Parkinson’s Disease Therapy. J. Control. Release 2015, 207, 18–30. [Google Scholar] [CrossRef]
- Mattera, V.S.; Gerber, P.P.; Glisoni, R.; Ostrowski, M.; Verstraeten, S.V.; Pasquini, J.M.; Correale, J.D. Extracellular Vesicles Containing the Transferrin Receptor as Nanocarriers of Apotransferrin. J. Neurochem. 2020, 155, 327–338. [Google Scholar] [CrossRef]
- Gao, X.; Ran, N.; Dong, X.; Zuo, B.; Yang, R.; Zhou, Q.; Moulton, H.M.; Seow, Y.; Yin, H. Anchor Peptide Captures, Targets, and Loads Exosomes of Diverse Origins for Diagnostics and Therapy. Sci. Transl. Med. 2018, 10. [Google Scholar] [CrossRef]
- Corso, G.; Heusermann, W.; Trojer, D.; Görgens, A.; Steib, E.; Voshol, J.; Graff, A.; Genoud, C.; Lee, Y.; Hean, J.; et al. Systematic Characterization of Extracellular Vesicle Sorting Domains and Quantification at the Single Molecule–Single Vesicle Level by Fluorescence Correlation Spectroscopy and Single Particle Imaging. J. Extracell. Vesicles 2019, 8, 1663043. [Google Scholar] [CrossRef]
- Jafari, D.; Shajari, S.; Jafari, R.; Mardi, N.; Gomari, H.; Ganji, F.; Moghadam, M.F.; Samadikuchaksaraei, A. Designer Exosomes: A New Platform for Biotechnology Therapeutics. BioDrugs 2020. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reinsalu, O.; Samel, A.; Niemeister, E.; Kurg, R. MAGEA4 Coated Extracellular Vesicles Are Stable and Can Be Assembled In Vitro. Int. J. Mol. Sci. 2021, 22, 5208. https://doi.org/10.3390/ijms22105208
Reinsalu O, Samel A, Niemeister E, Kurg R. MAGEA4 Coated Extracellular Vesicles Are Stable and Can Be Assembled In Vitro. International Journal of Molecular Sciences. 2021; 22(10):5208. https://doi.org/10.3390/ijms22105208
Chicago/Turabian StyleReinsalu, Olavi, Anneli Samel, Elen Niemeister, and Reet Kurg. 2021. "MAGEA4 Coated Extracellular Vesicles Are Stable and Can Be Assembled In Vitro" International Journal of Molecular Sciences 22, no. 10: 5208. https://doi.org/10.3390/ijms22105208
APA StyleReinsalu, O., Samel, A., Niemeister, E., & Kurg, R. (2021). MAGEA4 Coated Extracellular Vesicles Are Stable and Can Be Assembled In Vitro. International Journal of Molecular Sciences, 22(10), 5208. https://doi.org/10.3390/ijms22105208





