Abstract
Immune checkpoint inhibitor (ICI) therapies have shown great promise in cancer treatment. However, the intra-heterogeneity is a major barrier to reasonably classifying the potential benefited patients. Comprehensive heterogeneity analysis is needed to solve these clinical issues. In this study, the samples from pan-cancer and independent breast cancer datasets were divided into four tumor immune microenvironment (TIME) subtypes based on tumor programmed death ligand 1 (PD-L1) expression level and tumor-infiltrating lymphocyte (TIL) state. As the combination of the TIL Z score and PD-L1 expression showed superior prediction of response to ICI in multiple data sets compared to other methods, we used the TIL Z score and PD-L1 to classify samples. Therefore, samples were divided by combined TIL Z score and PD-L1 to identify four TIME subtypes, including type I (3.24%), type II (43.24%), type III (6.76%), and type IV (46.76%). Type I was associated with favorable prognosis with more T and DC cells, while type III had the poorest condition and composed a higher level of activated mast cells. Furthermore, TIME subtypes exhibited a distinct genetic and transcriptional feature: type III was observed to have the highest mutation rate (77.92%), while co-mutations patterns were characteristic in type I, and the PD-L1 positive subgroup showed higher carbohydrates, lipids, and xenobiotics metabolism compared to others. Overall, we developed a robust method to classify TIME and analyze the divergence of prognosis, immune cell composition, genomics, and transcriptomics patterns among TIME subtypes, which potentially provides insight for classification of TIME and a referrable theoretical basis for the screening benefited groups in the ICI immunotherapy.
1. Introduction
For the past few years, clinical results revealed that immune checkpoint inhibitor (ICI) treatment, such as programmed death-1 (PD-1) and its ligand 1 (PD-L1) checkpoint blockade, have shown an exhilaratingly long-term effect in a variety of cancer patients and have become a research focus in current tumor immunotherapy [1,2,3]. However, it has been reported that a number of patients showed a low response rate or treatment resistance against the anti-PD-1/PD-L1 checkpoint blockade [4,5,6]. Thus, it is significant to categorize patients into appropriate subpopulation, based on their cellular and molecular characteristics, to elucidate an inner mechanism, resulting in divergence of multi-omics patterns, and to ultimately provide clinical guidance on choosing corresponding treatment strategies for stratifying patients.
The various classifications of population-responding ICIs are mainly attributed to tumor microenvironments (TMEs), especially the composition and quantities of tumor-infiltrating lymphocytes (TILs), as well as numerous factors that independently predict clinical response to ICIs, including PD-L1 expression, tumor mutation burden (TMB), neo-antigen genotype, immune cell exhaustion, and disordered expression levels of cytokines [6,7,8,9,10]. It has been reported that the TIL status in the tumor immune microenvironment (TIME) is positively related to good clinical prognosis and could better predict the response to anti-PD-1/PD-L1 therapies [11,12,13,14]. Considering the inhibitory effect of cancer cells on the function of effector lymphocytes in TIME via immunological checkpoints, such as PD-L1, it is more comprehensive and precise to stratify TIME into different types by combining the two indicators above. Owing to the divergence of TIL status and PD-L1 expression, the immunologic effects of different TIME subtypes can be various, and thus, the corresponding immunotherapeutic strategies can be different. Recent research has described four different subtypes of TIME based on the positive or negative status of TIL and PD-L1 expression, including type I (PD-L1+/TIL+: adaptive immune resistance), type II (PD-L1−/TIL−: immunological ignorance), type III (PD-L1+/TIL−: intrinsic PD-L1 induction), and type IV (PD-L1−/TIL+: Other suppressors) [15,16,17], which may serve as a more systematic biomarker to stratify patients in clinical use of immunotherapy [18,19].
However, there are several issues that need to be addressed. First, most of these studies generally focused on one specific cancer type and classified samples into four subtypes to investigate their molecular characteristics without analyzing the multi-omics discrepancy of four subtypes in pan-cancer [16,20,21]. Second, they merely qualified the PD-L1 expression on the membrane surfaces of tumor cells by immunohistochemistry (IHC) [15,16,17,18,19,20]. However, several studies have reported that tumor cells are able to release a vast of exosomes, containing majority PD-L1, to suppress antitumor immunity rather than merely present PD-L1 on their cell surfaces [22,23]. This discovery may explain the discrepancy of PD-L1 expression between the transcriptomic level and proteomic level and reminds us that exclusive detection of expression of PD-L1 presenting on the membrane surface may have certain limitations. Third, they only evaluated the TIL status according to the CD8+T cell, which was the uppermost effector lymphocyte in TIME, without analyzing other kinds of functional lymphocyte impacts [15,19,20,21,24,25,26,27]. In most big cohort studies of immune-related cancer, researchers only used the expression levels of CD8+ T cell-related genes, such as CD8A or CD8B, to characterize TIL [15,24,25,26,27]. Additionally, they classified different patients into PD-L1 or TIL positive/negative subgroups without illustrating how threshold criteria were set, which was not reasonable for classification or further analysis [15,19,20,21,24,25,26,27]. Thus, the more precise indicator of TIL status, which reflects the interaction among various leukocytes in TIME, needs to be further studied.
In this study, we constructed a new method for classifying TIL states, which are an advanced predictor of responses to ICI. We then stratified patients into four TIME subtypes of 8634 samples overall across 33 cancer types from The Cancer Genome Atlas (TCGA) database, with more optimized classification methods. We analyzed the similarities and differences of distribution of 8 immune cell types in each subtype: T cells, B cells, macrophages, dendritic cells, natural killer cells, mast cells, neutrophils, and eosinophils. We also performed difference analysis of the genomic and transcriptomic level among four subtypes in order to elucidate the mechanism of TIME divergence. Hazard analysis was conducted to identify the impacts of several factors, including our classification patterns on survival statuses. Furthermore, we used 3069 breast cancer patients from the Gene Expression Omnibus (GEO) database for a similar classification study to verify the availability of analysis methods for widespread use. We believe that this stratification of cancer patients sheds light on new approaches to rationally apply the optimal cancer immunotherapeutic strategies for the four different TIME subtypes.
2. Results
2.1. Prognostic Significance of TIL Z Score/PD-L1 to ICI Response Prediction and Stratification of Four TIME Subtypes across Pan-Cancer Types
Five published datasets [28,29,30,31,32] on PD-L1/PD-1 blockade immunotherapy, including pre-treatment transcriptome information and post-treatment clinical response data, were downloaded to evaluate and compare the performance of the TIL Z score and other common indicators, based on CD8A or CD8B expression, in predicting clinical response to ICI. We used the receiver operating characteristic (ROC) curve to measure the true-positive rates against the false-positive rates at various thresholds of the TIL Z score and CD8A and CD8B expression (Figure 1A–C). The results showed that the predictive power of the TIL Z score (AUC = 0.592) was higher than that of CD8A (AUC = 0.575) and CD8B (AUC = 0.552), which suggested that the TIL Z score had a strong robustness to ICI response prediction and was sufficient to characterize TIL. As PD-L1 was also related to ICI response, we assessed the AUC of PD-L1, and the result indicated that the predictive power of PD-L1 (AUC = 0.53) was lower than the TIL Z score (Figure 1D). We then combined PD-L1 expression and the TIL Z score to evaluate their performances. As Figure 1E shows, the combination of PD-L1 and the TIL Z score had a higher AUC (0.64) than others (0.53~0.59), which suggests that this combined index exhibits strong robustness to ICI response prediction (Table 1).
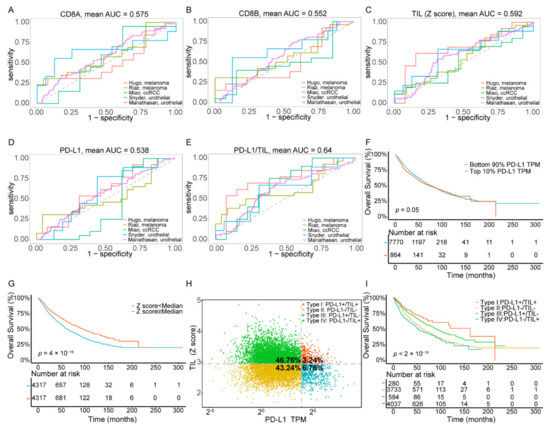
Figure 1.
Combination of the TIL Z score and PD-L1 predicts clinical response to ICI immunotherapy and the stratification of four TIME subtypes across pan-cancer types. (A–E) ROC curves for the performance of CD8A, CD8B, the TIL Z score, PD-L1, and the combined TIL Z score with PD-L1 for predicting anti-PD-1 immunotherapy response in patients who received ICI therapy. (F) Kaplan–Meier survival curves of patients based on PD-L1 expression. (G) Kaplan–Meier survival curves of patients based on the TIL score. (H), The proportions of patients in type I, type II, type III, and type IV. (I) Kaplan–Meier survival curves of patients in type I, type II, type III, and type IV. Abbreviations: TIL: tumor-infiltrating lymphocyte, ICI: immune checkpoint inhibitors, TIME: tumor immune microenvironment.

Table 1.
The sample size statistics and AUC value of different indicators for the immunotherapy research cohort.
We analyzed 8634 tumor samples of 33 cancer types from the TCGA dataset using PD-L1 mRNA expression and the TIL Z-score to classify samples. The value distribution of PD-L1 expression varied according to the cancer types (ranging from 0.03 to 521.31, Figure S1A, Table S2), which reminded us that there may not be one universal definition of positive or negative PD-L1 expression for each cancer type. Thus, we defined PD-L1 subgroups by percentile rather than establishing a definitive cut-off value for PD-L1 expression. The cut-points chosen to define the PD-L1 positive subgroup were the top 10%, 20%, 30%, 40%, and 50% in each independent experiment. We then performed Kaplan–Meier survival analysis on each positive vs. negative PD-L1 group (Figure 1F, Figure S1B). Since patients had the most significant difference in overall survival state (Figure 1F) when the cut-point was set at the top 10%, this threshold was selected to determine the PD-L1 positive subgroup for further analysis.
Analogously, since the distribution of TIL in pan-cancer varied (ranging from 1.69 to 4.86, Figure S1C, Table S2), we classified TIL subgroups by percentile, and the cut-points chosen to define TIL positive group were the same as PD-L1. Our result of Kaplan–Meier survival analysis with a log-rank test showed a significant difference in positive vs. negative TIL groups (Figure 1G, Figure S1D). Here, we selected the top 50% of patients who exhibited the most significant difference in the overall survival state (p value = 4 × 10−16) to determine the TIL positive subgroup for further analysis. Particularly, our results of correlation analysis revealed a weak relationship (Spearman correlation, p value < 2.2× 10−16, R = −0.159) between TIL status and PD-L1 expression (Figure S1E), which indicated that the two indicators were mutually independent.
As the classification PD-L1 and TIL Z score showed prognostic significance in overall survival of cancer patients, respectively, we further intended to investigate the difference between subtypes in response to ICI treatment. We grouped ICI immunotherapy samples into four TIME subtypes by combining these two predictive indicators, and the result showed that the response rate was higher in type I (40%) and lower in types II and III (28.73%, 29.41%), which indicated that type I samples exhibited a more favorable response rate and may benefit from ICB immunotherapy (Figure S1F, Table S1).
We also grouped all TCGA cancer samples into four TIME subtypes by combining these two predictive indicators (Figure 1H). Among all patients, only 3.24% of the samples were classified as type I (PD-L1+/TIL+), while the proportions of type II (PD-L1−/TIL−), type III (PD-L1+/TIL−), and type IV (PD-L1−/TIL+) were 43.24%, 6.76%, and 46.76%, respectively. Additionally, these proportions were comparable to those reported previously (13.44~54%, 15.4~43.4%, 1~26.20%, 15.4~54.79%, respectively) [14,15,16,17]. The clinical, pathological, cellular, and molecular characteristics of overall cancer cases, according to TIME subtypes, are summarized in Table 2. Kaplan–Meier survival analysis of these four subgroups (Figure 1I) showed that the overall survival of patients within type I was significantly the most favorable, while the patients within type III showed the poorest prognostic condition. Notably, the TIL positive groups (type I and IV) had better survival outcomes than the TIL negative groups (type II and III), which revealed an association between TIL status and increased survival (p value < 2× 10−16).

Table 2.
Clinical, pathological, and molecular characteristics of pan-cancer, according to tumor immune microenvironment subtypes based on programmed death ligand 1 (PD-L1) expression and tumor infiltrating lymphocyte (TIL).
Additionally, the proportion of four TIME types was calculated for a specific cancer type (Figure S1G and Table S2), occurring in different tissues, to display distribution of four subgroups. Notably, thymoma (THYM) was revealed to harbor the highest proportion of type I (55.26%) compared to other cancer types, indicating that tumors derived from lymphoproliferative organs are prone to form a lymphocyte-enriched immune microenvironment. Comparatively, the uterine carcinosarcoma (UCS) was majorly composed of type II (88.24%), while lung squamous cell carcinoma (LUSC) and liver hepatocellular carcinoma (LIHC) processed the highest proportion of type III (31.78%) and type IV (84.13%), respectively. In general, we stratified patients into four subtypes based on their PD-L1 expression and TIL status and proclaimed the distribution divergence of subgroups across pan-cancer. Our results support the hypothesis that combining these two indicators could better predict the prognostic status and the potential response levels of immunotherapy.
2.2. The Composition and Abundance of Lymphocyte among Four Subtypes
Considering the differences of types and abundances of various immune cells would reflect the TIME, and studying the types and content of immune cells in TIME is of a great significance to further reveal immunization surveillance and malignant progression. We used the CIBERSORT tool to classify and estimate the level of immune cell infiltration by the deconvolution algorithm. Here, we divided 22 immune cells into 8 categories: T cells, B cells, macrophages, dendritic cells, natural killer cells, mast cells, neutrophils, and eosinophils. The abundance difference among 8 types of immune cells within four subtypes is shown in Figure 2A and Table S3. Compared to other subtypes, type I (PD-L1+/TIL+) contained a high level of T lymphocytes and DC cells but the lowest proportion of macrophages and mast cells (p values < 0.0001, respectively). In contrast, type II (PD-L1−/TIL−) had the lowest infiltrative levels of T lymphocytes and DCs in four subtypes, whereas its macrophage and mast cells were higher than other subtypes (p value < 0.0001, respectively). The infiltration level of NK cells in type IV (PD-L1−/TIL+) was the highest among four subtypes; however, its infiltration level of T lymphocytes was lower than that of the type I subtype (p value < 0.0001).
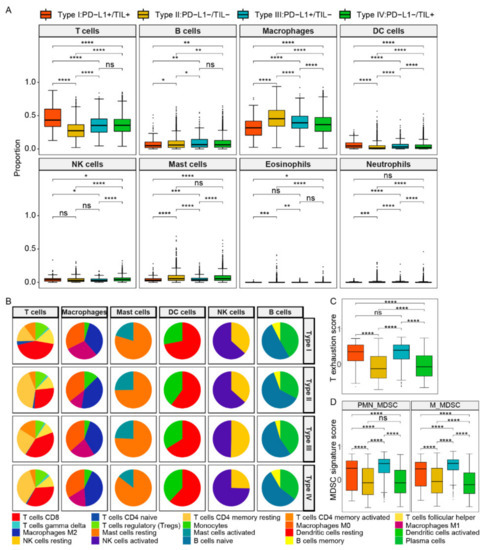
Figure 2.
The composition and abundance of immune cells among four TIME subtypes. (A) The abundance difference among eight types of immune cells within four subtypes. (B) The abundance difference of six main subclass immune cells in each subtype. (C) The T cell exhaustion score between four subtypes. (D) The MDSC signature score between four subtypes. Abbreviations: M_MDSCs: monocytic MDSCs, PMN_MDSCs: polymorphonuclear MDSCs. ****, p < 0.0001; ***, p < 0.001; **, p < 0.01; *, p < 0.05.
We further analyzed the details of immune cells in six main categories within four TIME types (Figure 2B, Table S4). In the T cells category, CD8+ T cells were mainly involved in killing tumor cells, as immune effector activity partially reflected by its content. Type I contained the most CD8+ T cells (43.56%) compared to other subtypes, while type III and type IV were closed to each other (39.43% and 33.81%, respectively), and type II was composed of the least CD8+ T cells (27.44%). The result revealed that better survival of patients may be related to the increased number of CD8 + T cells. The infiltration level of CD4+T memory resting cells in type I (15.03%) and type III (24.86%) were both less than that of type II (41.49%) and type IV (36.15%), yet the infiltration level of CD4+ memory T activated cells in type III was the highest (16.04%). T cells regulatory (Tregs) mainly play a role in suppressing immune cell effects, and their infiltration levels of type I, type II, and type IV were 12.11%, 10.53%, and 11.82%, respectively, which were higher than that of type III (8.05%), but the difference was not obvious. We supposed that Tregs content was not the main contributor to the difference in immune infiltration level.
As for other types of immune cells, the infiltration level of macrophages M2 was similar in type I, type II, and type III, which were 33.88%, 39.88%, and 35.87%, respectively, while that of type IV (45.42%) was higher than the other three types. In addition, the infiltration level of mast cells activated a range from 14.45% to 25.29% in four subtypes, while the TIL positive subgroups (type I/IV) were less than the other two types. The infiltrating level of DC activated cells of type I was the least, while that of type II, type III, and type IV were 39.91%, 33.92%, and 38.02%, respectively. Additionally, the content of NK activated cells in type IV exceeded the other three subtypes, ranging from 49.76% to 74.56%. Notably, the infiltration levels of the subpopulation of B cells were closer in composition among the four subtypes. Additionally, as myeloid-derived suppressor cells (MDSCs) infiltration and the T cell exhaustion state were revealed to be associated with immunosuppression, we further explored the comparison of proportion of MDSCs and the state of T cell exhaustion between the four subtypes. It was observed that the T cell exhaustion score was higher in PD-L1 positive groups (subtype I and subtype III), but there were no significant differences between subtype I and subtype III (Figure 2C). The results showed that the scores of both polymorphonuclear MDSCs (PMN- MDSCs) and monocytic MDSCs (M- MDSCs) were the highest in subtype III (PD-L1+/TIL−), and higher in PD-L1 positive groups compared to negative groups, and higher in TIL negative groups compared to positive groups (Figure 2D).
In general, the TIL positive subgroups that acquired good survival outcomes contained a high proportion of key immune cells, including activated CD8+T cells and NK cells. We speculated that the immunophenotype difference in four subtypes may be due to the abundance difference of these divergent cells.
2.3. Genomics Pattern Discrepancy in Four TIME Subtypes
Here, we investigated the discrepancy of TMB and neoantigen among four subtypes (Figure 3A, Table S5) and we found that type III had a remarkable high somatic mutation burden and neoantigen compared to others (p value < 0.0001). As for type I, type II, and type IV, there were no significant differences of neoantigen, as well as type I and type IV of TMB. We also constructed a 3-dimensional dot plot base on TIL, TMB, and neoantigen and performed linear regression analysis between every two factors (Figure 3B). Notably, a statistically significant correlation between the TMB and the neoantigens number was found (Spearman correlation, R = 0.885, p value < 2.2 × 10−16, Figure 3B). However, there was no significant correlation between TMB and TIL (Spearman correlation, R = −0.084, p = 6.031 × 10−14, Figure 3B) or neoantigen and TIL (Spearman correlation, R = −0.066, p = 4.234 × 10−7, Figure 3B). A correlation between PD-L1 expression and TMB or neoantigen was not found either (Spearman correlation, R = 0.099, p value < 2.2 × 10−16 and R = 0.151, p value < 2.2 × 10−16, respectively) (Figure S2A, Figure 2B).
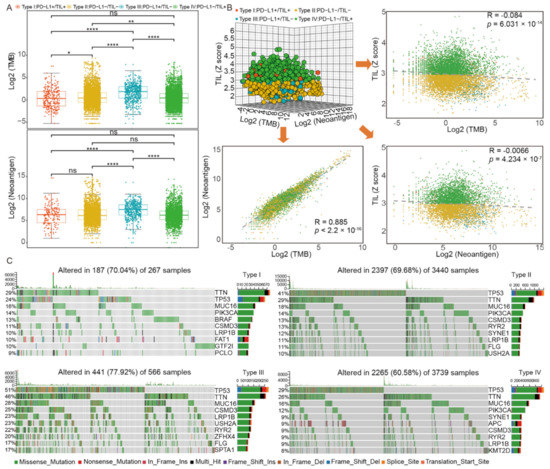
Figure 3.
The genomics pattern discrepancy in four TIME subtypes. (A) The distribution of TMB and neoantigen among four subtypes; (B) correlation analysis among TIL, TMB, and neoantigen; (C) the alteration landscape of somatic variants across four subtypes. ****, p < 0.0001; ***, p < 0.001; **, p < 0.01; *, p < 0.05.
We sought to investigate the alteration landscape of somatic variants across four subtypes and the specific tumor mutation spectrum, the frequencies of which varied in the top 10 mutant genes. Generally, we found that the patients of type III had the highest altered rate (77.92%) among four subtypes. As shown in Figure 3C and Table S6, five gene mutations were found in all four subtypes: TTN (29%, 29%, 46%, and 26%, respectively), TP53 (24%, 41%, 51%, and 29%, respectively), LRP1B (10%, 11%, 23%, and 9%, respectively), MUC16 (16%, 18%, 28%, and 16%, respectively), and CSMD3 (11%, 13%, 25%, and 9%, respectively). Specifically, compared to others, tumors of type III acquired the highest mutation rate of these five genes. We then investigated the unique mutated genes of each subtype: BRAF (13%), FAT1 (10%), GTF2I (10%), and PCLO (9%) in type I, ZFHX4 (20%) and SPTA1 (17%) in type III, APC (9%) and KMT2D (9%) in type IV, and no unique characteristics of type II mutated genes. We further investigated the relationship between TIL and gene mutation. The result indicated that there was a statistically significant difference of TIL status between the TP53 mutation subgroup and the wild-type subgroup (p value < 0.0001, Figure S2C) and the proportion of patients who were TIL positive in the wild-type subgroup was higher than that of the mutation subgroup (Figure S2D).
Considering that many mutated genes were co-occurring or displayed strong exclusiveness, we then explored the potential different somatic interactions among four subtypes to expound their mutation pattern (Figure S2E). The interaction of these genes with oncogenes suggests a close relationship to cancer occurrence and development. In type I, specific interaction patterns were found: GTF2I and BRAF mutations were both significantly mutually exclusive to other gene mutations (p value < 0.01, respectively), while the other mutations co-occurred more obviously. However, in type II, type III, and type IV, most of the gene mutations were significantly co-occurring (p value < 0.01), except for TP53 with SYNE1 in type II and PIK3CA with TP53 in type IV. We further evaluated and identified oncogenes in each subtype (Figure S2F, Table S7). Type IV owned the most oncogenes (16 in total) compared to the other three subtypes, and the most common oncogene KRAS mutation appeared across all four subtypes. Of these oncogenes, three of them (GTF2I, BRAF, and PIK3CA) had relatively higher mutated frequencies in type I compared to the other three subtypes. In addition, the BRAF mutation subgroup had a higher proportion of patients who were TIL positive (p value < 0.0001, Figure S2G, Figure S2H). However, the difference of TIL levels between the HRAS mutation subgroups and wild-type subgroup was not found, though HRAS mutation was uniquely identified in the TIL positive subgroup (type I/IV) (p value = 0.78, Figure S2I). Furthermore, different PD-L1 expression between the IDH-1 mutation subgroup and wild-type subgroup was statistically significant (p value < 0.0001, Figure S2J). In conclusion, the specific somatic mutation spectrum of each subtype could help us accurately classify patients into such subgroups.
2.4. Transcriptomics Pattern Discrepancy in Four TIME Subtypes
Understanding the divergence of immunomodulators (IM) expression and state is critical to descript transcriptomics patterns of each subtype. We thus examined the IM gene expression, as well as copy number variation (CNV), including amplification and deletion (Figure 4A, Table S8). In general, the gene expression differences of IMs across immune subtypes were not significant. Thereinto, PD-L1 positive subgroups (type I/III) presented similar states in co-inhibitor, ligand, receptor, and other modulators, as their gene expression levels were largely higher than PD-L1 negative groups (type II/III). For copy number alterations, type I generally showed low frequency amplification and deletion of IM genes, except for IM genes PDCD1LG2 and CD274 (PD-L1), which amplified a higher frequency, and noticeably, these genes had the highest frequencies in type III. Additionally, CD28, VTCN1, PDCD1, CTLA4, and ICOS had higher frequency deletion in type III as well. We found that the PD-L1 expression level in PDCD1LG2 and CD274 copy number amplification subgroups were higher than that of non-amplification subgroups (p value < 0.0001, Figure S3A,B, respectively), but PDCD1 or CTLA4 subgroups suggested opposite conclusions (p value < 0.01 & < 0.0001, Figure S3C,D, respectively). In conclusion, these marked divergences in IM genes clarified the perspective of PD-L1 subgroups referring molecular patterns discrepancy, which may be reflective of the immunomodulator state of the TIME in patients.
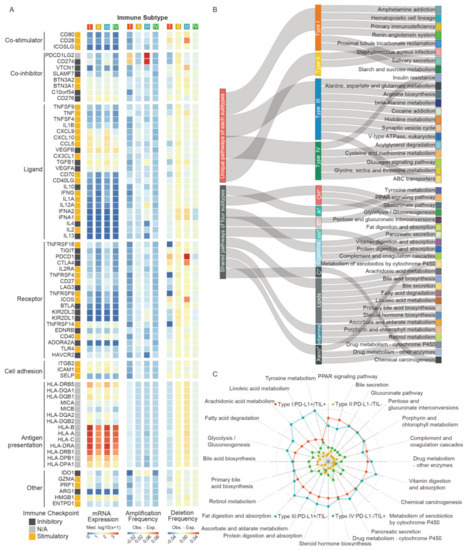
Figure 4.
The transcriptomic pattern discrepancy in four TIME subtypes. (A) The immunomodulators gene expression and copy number variation for each subtype. (B) The shared and unique pathway features for each subtype. (C) The distinct difference weight score of pathways in each group. Abbreviations: CH*: carbohydrates, A*: Amino acid, E*: Endocrine, Im*: Immune, C*: Cancer, Xeno*: Xenobiotics.
To reveal the key deregulated pathways occurring in each subtype, we analyzed different gene expression and calculated gene scores based on log fold changes values by comparing samples within one subtype with the other three integrated samples. Magnitude of pathway dysregulation was calculated by gene scores and assigning scores, based on the enrichment pathways of different expressed genes (DEGs) from The Kyoto Encyclopedia of Genes and Genomes (KEGG). As shown in the result, four TIME subtypes exhibited common signatures but maintained some unique features of their own (Figure 4B). Type I exhibited six unique pathways, including amphetamine addiction, hematopoietic cell lineage, primary immunodeficiency, renin-angiotensin system, salivary secretion, starch, and sucrose metabolism. Proximal tubule bicarbonate reclamation and staphylococcus aureus infection were the only unique pathways activated in type II. Notably, the most common pathways showed in type III were metabolic-related processes, such as alanine, aspartate, and glutamate metabolism, arginine biosynthesis, and ABC transporters. The specific pathway terms in type IV were also different, such as the glucagon signaling pathway and cysteine and methionine metabolism. We deemed that dysregulation of unique pathways in each subtype suggested different TIME signatures and potential differential sensitivity, providing the fundamentals of theoretical mechanism research for therapeutic intervention. We also determined the distinct difference weight scores of pathways in each subtype, which indicate enrichment degree and differential status of DEGs (Figure 4C, Table S9). With few exceptions (e.g., immune system, carcinogenic process), there was significant enrichment in metabolic genes that were frequently shared across all subtypes, but to a different degree. Overall, type I and type III (PD-L1+) harbored higher pathway scores than the other two types. Specifically, type III exhibited the highest score of pathways, except for PPAR signaling, bile secretion, and complement and coagulation cascades, while some were consistent with type I. Compared to type II and type IV (PD-L1−), these pathways in type IV changed more dramatically.
We then analyzed the expression distributions of cytokines and cytotoxic-related genes in each subtype found that the gene expression of immuno-activation cytokines (IFNG, TNF, IL12A, and IL12B), immuno-suppressive cytokines (TGFB1, IL6, and IL10), and cytolytic factors (GZMB and PRF1) were largely higher in type I and type III (PD-L1+) than the other two PD-L1 negative subtypes (Figure S3E, Table S10), which may indicate that PDL1 expression is involved in regulating immune balance. Moreover, there were also differences in cytokines observed among types I and III or typed II and IV. TGFB1, IL6, and IL10 were higher in TIL negative groups compared to TIL positive groups, which indicated the potential immunosuppressive effects brought by these cytokines. However, IFNG, TNF, and IL12A were also higher in type III compared to type I and higher in type II compared to type IV, suggesting the complexity of the immune microenvironment (Figure S3E, Table S10). Moreover, it emerged that the tumor vasculature itself constituted an important barrier to T cells. We analyzed the association between angiogenesis-related growth factors, as well as their receptors with TIL subtypes. We found that expression of EDN1, EDNRA, VEGFB, KDR, and FLT1 were higher in TIL negative groups; to be specific, they were higher in type III compared to type I and higher in type II compared to type IV (Figure S3F,G). Additionally, the correlation analysis showed the gene expression of growth factors and receptors were negatively correlated with the TIL score; EDNRA especially exhibited a higher negative correlation coefficient with the TIL score (Figure S3H). These results further suggested the adverse effects of tumor vasculature disorder on TIL.
2.5. Hazard Analysis for Multiple Omics Factors across Four TIME Subtypes
Significant variables (p value < 0.05) of the univariate analysis were into entered a multivariate Cox model. In the model, we examined several factors, including age, gender, tumor stage, TIL status (overall and specific cell types), TMB, neoantigen level, TP53, BRAF, and IDH1 mutation state, copy number variation of PD-L1, PDCD1, and CTLA4, and immuno-activating/suppressive cytokines and cytolytic activity (Figure 5, Table 3). We found that positive TIL was associated with a good prognosis and higher overall survival (Hazard Ratio (HR): 0.846; 95% CI: 0.734–0.975; p value = 0.02). In contrast, high Macrophage M2 and activated mast cells were associated with significantly higher overall mortality and were not conducive to survival (HR: 1.244; 95% CI: 1.079–1.434; p value = 0.0026 and HR: 1.242; 95% CI: 1.044–1.477; p value = 0.015, respectively). Furthermore, an advanced tumor stage, such as stage IV (HR: 3.406; 95% CI: 2.787–4.163; p value < 2 × 10−16) and stage III (HR: 1.874; 95% CI: 1.546–2.272; p value = 1.69 × 10−10), a high level of immuno-suppressive cytokines (HR: 1.165; 95% CI: 1.001–1.356; p value = 0.048), and TP53 mutation (HR: 1.322; 95% CI:1.138–1.535; p value = 0.000255) were all associated with poorer overall survival.
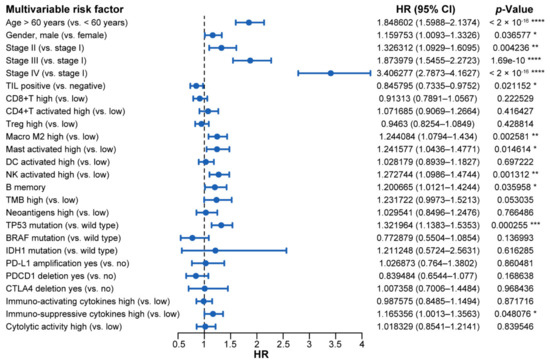
Figure 5.
The forest plot of prognostic values for multivariable cox proportional hazard regression models. Abbreviations: HR: hazard ratio; CI: confidence interval; TIL: tumor infiltrating lymphocyte; TMB: tumor mutation burden. ****, p < 0.0001; ***, p < 0.001; **, p < 0.01; *, p < 0.05.

Table 3.
Univariate and multivariate cox proportional hazards analysis for overall survival in pan-cancer patients.
2.6. Validation in GEO Dataset
To further validate the widespread use of this classification method based on PD-L1 and TIL level, we performed similar analysis at a public mRNA expression dataset (GSE96058) containing sufficiently large numbers of breast cancer samples (n = 3069) deposited in GEO. As before, we set the intervals that define PD-L1 and TIL positive to multiple percentiles: top 10%, 20%, 30%, 40%, and 50%. We then performed the Kaplan–Meier survival analysis log-rank test and found that, when PD-L1 and TIL positive were in the top 10% (p value = 0.009) and top 50% (p value = 0.032), respectively, the difference of the overall survival curve was the most significant, which was consistent with the results of TCGA dataset analysis, indicating that the thresholds we took were appropriate (Figure 6A,B, Figure S4A,B). We further grouped the GEO samples into four TIME subtypes based on the combination of PD-L1 and TIL, as previously described. The difference of overall survival curve of the four subtypes was statistically significant (p value = 0.015), the prognosis condition of type III was poorest, and the survival outcomes of the TIL positive groups (type I and IV) were better than the TIL negative groups (type II and III), which were similar to the results of TCGA dataset analysis, but the prognosis condition of type I was not the most favorable, unlike the TCGA dataset analysis (Figure S5A). Among all patients in GEO validation, the proportions of type I, type II, type III, and type IV were 3.68%, 43.66%, 6.32%, and 46.34%, respectively, which was similar to the results of the TCGA cohort (Figure S5B).
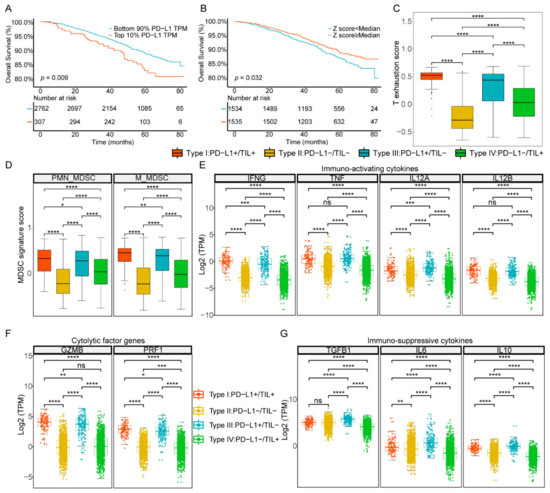
Figure 6.
Stratification of four TIME subtypes in the GEO database. (A) Survival analysis of positive vs. negative PD-L1 groups. (B) Survival analysis of positive vs. negative TIL groups. (C) The T cell exhaustion score between four subtypes. (D) The MDSCs signature score between four subtypes. (E–G) The gene expression distributions of cytokines and cytolysis factors in each subtype. ****, p < 0.0001; ***, p < 0.001; **, p < 0.01; *, p < 0.05.
As before, we used the CIBERSORT tool to classify and evaluate the infiltration level of immune cells. The abundance difference among eight types of immune cells within four subtypes was shown in Figure S5C and Table S11. Analogously, type I (PD-L1+/TIL+) contained the highest level of T lymphocytes and the lowest proportion of macrophages and mast cells (p values < 0.0001, respectively), and type II (PD-L1−/TIL−) had the lowest infiltrative levels of T lymphocytes and the highest level of macrophage and mast cells (p value < 0.0001, respectively), and the infiltration level of T lymphocytes of type IV was lower than that of the type I subtype (p value < 0.001). However, there was no significant difference in the abundance of DC cells among the four TIME subtypes.
The proportion of 20 immune cell types classified into 6 main cell types among the 4 TIME subtypes was shown in Figure S5D and Table S12. Type I contained the most CD8+ T cells (37.82%) compared to other subtypes, while type II was composed of the least CD8+ T cells (29.39%) and type III and type IV were closed to each other (34.83% and 31.91%, respectively). The infiltration level of CD4+T memory resting cells in type I (43.69%) and type III (48.32%) were both less than that of type II (57.34%) and type IV (55.23%). The infiltration levels of T cells regulatory (Tregs) among type I, type II, type III, and type IV were 8.44%, 5.88%, 7.42%, and 6.64%, respectively, the difference of which was not obvious. The infiltration level of macrophages M2 of type II (64.74%) and type IV (62.46%) were higher than type I (47.91%) and type III (52.55%). The infiltration levels of mast cells of type I (2.8%) and type IV (4.8%), which belong to TIL positive subgroups, were lower than type II (6.15%) and type III (7.92%). The infiltration levels of the B cells subpopulation were closer in composition among the four subtypes. As above, the proportions of cell types among the four types were similar to those in the results of the TCGA dataset. We also explored the comparison of proportion of MDSCs and the state of T cell exhaustion between four subtypes in the GEO dataset. It was observed that the T cell exhaustion score was higher in PD-L1 positive groups (subtype I and subtype III) but higher in subtype I compared to subtype III. The results showed that the scores of both PMN-MDSCs and M-MDSCs were higher in PD-L1 positive groups compared to negative groups and higher in TIL positive groups compared to negative groups (Figure 6C,D).
As in the TCGA dataset analysis, the expression levels of immuno-activation cytokines (IFNG, TNF, IL12A, and IL12B), immune-suppressive cytokines (VEGFA, TGFB1, IL6, and IL10) and cytolytic factors (GZMB and PRF1) were higher in the PD-L1 positive subtypes (type I and type III) than in the PD-L1 negative subtypes (type II and type IV) (Figure 6E–G, Table S13). Consistent with the TCGA results, we found that the expression of growth factors and their receptors were higher in TIL negative groups (Figure S5E,F). Additionally, the correlation analysis showed the gene expressions of growth factors and receptors were also negatively correlated with the TIL score (Figure S5G). In general, GEO dataset results showed a similar pattern to that of the TCGA dataset, no matter the classification of PD-L1 and TIL, the composition of immune cells, or the expression of transcriptome, indicating the reliability of our results and universality of the classification method.
3. Discussion
PD-L1, as an immune checkpoint, is generally upregulated in TIME and promotes immune escape of tumor cells [33,34]. As a main target of immunotherapy, PD-L1 immunoblockade therapy brings great benefits to many patients, but its clinical application still has certain limitations. For example, many studies have found that the PD-L1 expression state is not directly correlated to the response rate or immunotherapy prognosis in different cancer types [35,36]. In this study, using a large scale of TCGA pan-cancer datasets, we systematically investigated the distribution of PD-L1 expression and TIL status, examined their prognostic impacts, and stratified 8634 patients into four subtypes across 33 cancer types by combining these two factors. We also used the GEO breast cancer dataset to validate our findings and found analogous conclusions. Although a positive correlation between PD-L1 expression and CD8+T cells was reported by previous researches [37,38], our results showed that TIL status was independent of PD-L1 expression, which allowed a further reasonable classification. Overall survival analysis illustrated that patients in TIL+ groups (type I and type IV) had better prognostic outcomes than that in TIL− groups (type II and type III), which were consistent with the prognostic outcome of TIL alone. Type I has a higher survival rate than that of type IV, suggesting that the prognostic outcome of PD-L1+/TIL+ subtypes was better than that of PD-L1−/TIL+ results, which is inconsistent with some previous studies [20], since only CD8+ T cells were considered as TILs in their research. Notably, the lower proportion of PD-L1 positive subtypes (type I and III) that was revealed by our study may imply a relative low proportion of patients who would potentially benefit from PD-L1 immunosuppressor. In particular, the distribution of four subtypes varied among the 33 cancer types, which inspired us to consider that different immunotherapy strategies should be adopted for different cancer types, even different patients with the same kind of cancer, to achieve precise treatment effect [20].
The TIME is a bidirectional, dynamic, and intricate interaction network between tumor cells and non-malignant cells, including immune cells and stromal cells [11,39]. Among them, owing to the difference of types and abundance of various immune cells, the formation of different TIME types could guide the tumor occurrence, development, and even transfer patterns. Therefore, analyzing the type and abundance of immune cells in corresponding subtypes of TIME is of great significance for further revealing the molecular mechanism of tumorigenesis and malignant progression [40,41]. Our results show that CD8+T cells and DC cells in type I were richer than the other three subtypes. We believe that the higher CD8+T cell infiltration level may endow type I patients with higher immunity, since the cytolytic activity-related gene GZMB and PRF1 expressions were also higher in type I, as shown in transcriptome analysis, thus giving a more promising prognostic effect. The proportion of T cells of type IV was lower than that of type I, while its content of NK-activated cells was higher than that of type I. We hypothesize that the tumor killing effect of type IV patients is more dependent on NK cells. The intrinsic mechanism of different subtypes in recruiting T cells and NK cells, particularly the presence of PD-L1, remains to be elucidated. T cell exhaustion state was higher in PD-L1 positive groups, which further suggest the strong association between PD-L1 signals and T cell exhaustion. Of immune cells that exert immunosuppressive effects, Treg cells were not responsible for differences in immune microenvironment, but TIL negative groups had higher rates of MDSCs compared to the positive subtypes, as well as the relatively high proportions of M2 macrophage. Therefore, we reasoned that MDSCs and M2 macrophage were important factors to prevent T cell infiltration, and the difference of immune microenvironment in different subtypes is mainly reflected by a relative abundance of CD8+ T cells, MDSCs, and M2 macrophage [42,43].
Previous research has reported that TMB and neoantigen were associated with better immunotherapy effect, but its predictive effect has a limited effect on certain cancers, such as non-small cell lung cancer and colorectal cancer [44,45]. Our results reveal a significant correlation between TMB and neoantigens, but the relationships between TIL and TMB or neoantigen were not found. Therefore, we expect that high TMB or neoantigen would not primarily lead to high levels of immune infiltration, which remind us that novel and robust factors predicting the immunotherapy effect for various cancer should be further discovered. We also investigated the mutation landscape of high frequency foreach subtypes. For high frequency mutated genes of specific subtype, gene BRAF in type I encodes a protein belonging to the RAF family of serine/threonine protein kinases, which have been identified in various cancers [46]. Some research has reported that BRAF V600E mutation would sustain IFN-γ inducible PD-L1 expression by coactivating STAT1 and increasing protein translation and is associated with high levels of PD-L1 expression [47,48,49,50], and the patients with BRAF mutations appeared to benefit from monotherapy with PD-L1 inhibitors, which is consistent with results of the present study, to some extent. APC gene in type IV encodes a tumor suppressor protein that acts as an antagonist of the Wnt signaling pathway, which was involved in other processes, including cell migration and adhesion, transcriptional activation, and apoptosis [51]. However, we did not retrieve any reports concerning the relationship between this gene mutation and PD-L1 expression. As for oncogenes for each subtype, we found that KRAS mutation was the most common oncogene, while some studies reported that PD-L1 expression was upregulated by KRAS G12D mutation and KRAS mutations could serve as a potential predictor of anti-PD-1/PD-L1 immunotherapy [52,53]. In general, gene mutation spectrums present genomics divergence among four subtypes, and, in the future, highly specific targeted drugs for different patients need to be used to maximize the therapeutic effect, and the combination of targeted therapy and immunotherapy will be a promising treatment.
The divergence of transcriptomic patterns between PD-L1 positive groups and PD-L1 negative groups demonstrated that difference of IM gene expression pattern might attribute to a PD-L1-related pathway, while this assumption needs further confirmation. In the unique pathway studies, the association of immune types with signaling pathways was investigated based on RNA expression data of DEGs. Type I was associated with hematopoietic cell lineage, which could be contributed to CD4+T cells, suggesting a fundamental role of TIL in hematopoiesis through the secretion of cytokines or interferon [54]. It is the arginine biosynthesis pathway that mainly draws our attention to type III, whose concentrations impact the metabolic fitness directly and T cells capacity, which are crucial for anti-tumor functionality in TIME, as previously reported [55]. Arginine biosynthesis is more active in the cancer cells of type III and indicate that lack of arginine, because of weak competition in immune cells, may lead to energy depletion and less TIL in local TIME. Combined with previous research, our results suggest that TIL is associated with multiple biological states, such as genesis of blood cells and synthesis and metabolism of amino acid in TIME. Other TIL-related factors need further confirmation.
The common pathway analysis revealed distinguishing patterns of activity shared by four subtypes. Surprisingly, type I and type III exhibited higher scores in most shared metabolism pathways, suggesting that PD-L1 high expression is more likely associated with metabolic alternations in TIME. The previous study discovered an unexpected role for PD-L1 in regulating tumor cell metabolism in the D42m1-T3 mice model. Specifically, PD-L1 could enhance the glycolysis of tumor cells by association with some signaling proteins, such as mTOR [56]. Combined with our study, higher PD-L1 expression may affect certain energy metabolism in tumor cells and thus weaken the nutrient intake of immune cells due to competition in type I and type III, compared to type II and type IV. Xenobiotics was metabolized by cytochrome P450, which could be induced by aryl hydrocarbon receptor (AHR) activation [57]. Type I and type III exhibit a more active xenobiotics metabolism, while type II and type IV show less, indicating that xenobiotics metabolism may influence PD-L1 expression through AHR signaling in TIME [58]. More work is required to determine how PD-L1 signals and the accurate connection between PD-L1 and metabolic pathways or biological processes in tumor and immune cells. Moreover, malignant cells can deprive glucose in TIME, thus blocking effective anticancer immunity, as glucose is used by T cells, NK cells, macrophages, and DCs to support their effector functions [56,59]. Glycolysis was shown to regulate TIL on account of metabolic competition in the tumor microenvironment, which can blunt Ca2+ signaling, glycolytic capacity, and cytokine production of TILs because glucose consumption by tumors metabolically restricts T cells [59,60]. Our research shows that cancer cells in type III (PD-L1+/TIL−) had more active glycolysis, suggesting that glycolysis is vital to TIL and is affected by PD-L1 expression. In summary, PD-L1 positive subtype (type I/III) and PD-L1 negative subtype (type II/IV) harbor distinct alterations in cell metabolism pathways, while the TIL subtypes have minor differences, and it seems that there are more potential connections between PD-L1 and TIME metabolism. These results may catalyze a better understanding of the role of immune cells’ altered metabolism in anti-cancer ability and provide novel means to stratify patients based on metabolic features and immunological status. Moreover, gene expression of endothelial-related growth factors and receptors were lower in TIL positive groups, which suggest that endothelial tumors and disorganized vasculature establish the barrier preventing T cell infiltration into tumors [61].
Hazards analysis identified several reliable indicators for evaluation of clinical treatment effects, except for common factors present, such as age, gender, and tumor stage. Most of the multivariable prognostic factors, such as macrophages M2, activated mast cells, TP53 mutation, and immuno-suppressive cytokines expression, are unfavorable for survival by promoting the occurrence and development of tumors. In particular, TIL is implied to reduce the risk of death and is considered a good prognostic factor in cancer patients. In addition, consideration of the combination of more factors may improve sensitivity or specificity of clinical diagnosis and treatment.
4. Materials and Methods
4.1. Data Collection and Preprocessing
Immunotherapy dataset: Pre-treatment transcriptome information and post-treatment clinical response data from the five datasets of previous studies [28,29,30,31,32], whose patients received anti-PD-1 or anti-PD-L1 immunotherapy and were downloaded to evaluate the power of CD8A, CD8B, the TIL Z score, PD-L1, and the PD-L1/TIL Z score to predict clinical response to ICIs.
TCGA dataset: We acquired available level-3 data published by TCGA, including 8634 samples with available survival information of 33 cancer types. Genomic somatic mutation data, copy number variation (CNV) data, mRNA expression data, and clinical information of each sample were downloaded from the GDC Data Portal (https://portal.gdc.cancer.gov, accessed on 30 April 2019).
GEO dataset: A public mRNA high throughput sequencing dataset (GSE96058), containing sufficiently large numbers of breast cancer samples (n = 3069) deposited in GEO, was used to construct the validation cohort. The expressing matrix of mRNA plus clinical metadata were downloaded from GEO. Clinical metadata were used for Kaplan– Meier overall survival analysis, and mRNA expression profiles, which were constructed by GPL11154 of the Illumina HiSeq 2000 platform, were presented as fragments per kilobase of exon model per million mapped fragments (FPKM) and were transformed into TPM for transcriptome analysis.
4.2. Tumor-Infiltrating Lymphocyte Z Score
We calculated a comprehensive TIL score for each sample by applying an algorithmically optimized method, which was based on the expression of representative genes or gene sets of single samples from 26 determinants, consisting of 20 single factors (classified in MHC molecules, immunoinhibitors, and immunostimulators) and 6 immune cell types (activated CD4+ T cells, activated CD8+ T cells, effector memory CD4+ T cells, effector memory CD8+ T cells, Tregs, and MDSCs). The calculation was conducted through R code, developed by Charoentong et al. [42], and the source codes are available (https://github.com/mui-icbi/Immunophenogram, accessed on 20 May 2019). The RNA expression matrix was transformed into log2 (TPM+1) values and used as an input to calculate the comprehensive score of TILs. The result file generated by algorithm operation contained an average Z score and immunophenoscore (IPS); therefore, the Z score was selected as a TIL comprehensive score for further research.
4.3. TIME Subtypes and Immune Cells Proportion
According to previous reports regarding the four TIME types [5], we stratified PD-L1 expression level and the TIL Z score into positive and negative groups: type I, PD-L1 positive with TIL positive; type II, PD-L1 negative with TIL negative; type III, PD-L1 positive with TIL negative; and type IV, PD-L1 negative with TIL positive, with a cut-off value of 90 percentile and median value, respectively. Additionally, a deconvolution approach [62], CIBERSORT, was applied to calculate the proportion of 22 immune cell types (https://cibersort.stanford.edu, accessed on 3 June 2019).
4.4. Genomic Analysis
The resulting data, consisting of detected somatic variants, was stored in mutation annotation format (MAF), and R package “Maftools” was used to summarize, analyze, annotate, and visualize MAF files in an efficient manner [63]. To evaluate TMB across samples, multiple somatic mutations, including nonsynonymous mutations, insertion-deletion mutations, and silent mutations, were counted and summated, with the exome size of 38 Mb, while germline mutations without somatic mutations were excluded [8]. The neoantigen number (n = 5,798) was evaluated by Vésteinn Thorsson et al. [64].
The data of amplification, deletion, and neutral status within a CNV threshold, recorded as “1”, “−1”, and “0”, respectively, was calculated by Gistic 2.0 [65]. mRNA expression profiles, which were constructed by the Illumina HiSeq V2 platform, were presented as counts and were transformed into transcripts per million (TPM) for analysis. Genes with multiple probes were represented by mean values of probes.
4.5. Differential Gene Analysis and Pathway Score Analysis
In this part, each subtype was determined as an experimental group in turns, while others were set as control groups, and R package “edgeR” was subsequently performed on differential gene expression analysis [66,67]. Differential gene lists were identified with statistical significance (|log2FC| ≥ 2, FDR < 0.05). Pathways were downloaded from the Kyoto Encyclopedia of Genes and Genomes (KEGG), and Metascape (http://metascape.org, accessed on 14 July 2019) was performed on gene annotation and functional enrichment analysis with a significant threshold (p value < 0.01, enriched genes number ≥ 3) [68]. Gene and pathway scores were calculated via Python 3.7.1. Gene scores were computed by differential gene lists of each subtype in order to calculate which single pathway scores contributed to shared pathways. The calculation processes of each pathway score for type k (k = I, II, III, IV) was described as follows.
For each gene, the mean log2FC score was firstly calculated across 4 subtypes:
where log2FCk is the log2 fold change score of type k (k = I, II, III, IV). If there is no such gene in the differential gene list of this subtype, then .
Gene score of type k was then determined by an equation:
Mean log2FC score is the result of Equation (1).
Finally, the pathway score of type k was calculated:
where t∑ (Gene score) represents the total gene score at the same pathway of type k and is the total gene score of that pathway from all subtypes. At the end, we visualized the result by using Power-BI (https://powerbi.microsoft.com) to plot a radar map of thr pathway score.
4.6. Gene Set Variation Analysis (GSVA) Score of Gene Expression Signature
To compare the difference in the proportion of T cell exhaustion and MDSCs between four TIME subtypes, we used the gene set obtained for previous studies [69,70] to calculate the GSVA score across four subtypes.
4.7. Survival Analysis
Univariate and multivariate logistic regression analyses were performed to determine significant factors of clinicopathologic characteristics. Patients who lacked follow-up or death time were pre-excluded when performing survival analyses. For categorical variables, such as TIME subtypes, PD-L1 expression status, and TIL status, a prognostic condition was estimated via Kaplan –Meier plots with a log-rank test and Cox proportional hazards regression analysis. Survival times were determined in months, from initial pathological diagnosis to death, or the last time the patient was known to be alive. p values less than 0.05 were considered statistically significant.
4.8. Statistical Analysis
R package “pROC” was used to plot the rate of response at various threshold settings of CD8A, CD8B, or the TIL Z score for generating the receiver operator characteristic (ROC) curve. Spearman rank correlation analysis was applied to compute the statistical significance of two continuous variables, which were exemplified as TMB, neoantigens, the TIL Z score, PD-L1 expression, and so on. One-way analysis of variance or a Wilcoxon rank sum test was applied for significance of differences between continuous values, which were listed as the immune cells proportion, tumor mutation burden, number of neoantigens, gene expression, such IFNG expression, and so on. Comparison of proportion according to categorical variables was performed using Pearson’s Chi-square test or the Fisher exact test. p values less than 0.05 were considered statistically significant.
5. Conclusions
In the current study, we developed a more robust method for classifying TIME subtypes at the big data analysis level and studied their characteristics shaping their corresponding microenvironments. It is noteworthy that the performance in the prognosis and prediction of the response to ICI immunotherapy of our method is superior to previous methods used in previous research. Considering the effectiveness, our classification method exhibits a better performance, which provides a potential option for clinical research and applications.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/ijms22105158/s1. Figure S1: Based on survival analysis of positive vs. negative PD-L1 or TIL subgroups to classify samples. (A) The value distribution of PD-L1 expression across 33 cancer types. (B) Survival analysis of positive vs. negative PD-L1 subgroups in each cut-point. (C) The value distribution of TIL status across 33 cancer types. (D) Survival analysis of positive vs. negative TIL subgroups in each cut-point. (E) Correlation relationship between TIL status and PD-L1 expression. (F) Response rate to ICI immunotherapy of four TIME subtypes. (G) The proportions of 4 TIME subtypes across 33 cancer types. Figure S2: Genomic characterization between four subtypes. (A) The correlation between tumor mutation burden and PD-L1 expression. (B) The correlation between neoantigens and PD-L1 expression. (C) Difference in TIL between TP53 mutation and wild type. (D) The samples proportion of TIL+ and TIL− between TP53 mutation and wild type. (E) Somatic mutational interactions among 4 subtypes. (F) The oncogene pattern in each subtype. (G) Difference in TIL between BRAF mutation and wild type. (H) The samples proportion of TIL+ and TIL− between BRAF mutation and wild type. (I) Difference in TIL between HRAS mutation and wild type. (J) Difference in PD-L1 expression between IDH1 mutation and wild type. ****, p < 0.0001; ***, p < 0.001; **, p < 0.01; *, p < 0.05. Figure S3: The transcriptomic patterns discrepancy in four TIME subtypes. (A) Difference in PD-L1 expression between PDCD1LG2 amplification and not amplification. (B) Difference in PD-L1 expression between PD-L1 amplification and not amplification. (C) Difference in PD-L1 expression between PDCD1 deletion and not deletion. (D) Difference in PD-L1 expression between CTLA4 deletion and not deletion. (E) The gene expression distributions of cytokines and cytolysis factors in each subtype. (F) The gene expression distributions of growth factors and receptors in each subtype. (G) The gene expression distributions of growth factors and receptors between TIL positive and TIL negative samples. (H) The correlation coefficient between the TIL score and expression of growth factors, as well as receptors. ****, p < 0.0001; ***, p < 0.001; **, p < 0.01; *, p < 0.05. Figure S4: Survival analysis of positive vs. negative PD-L1 or TIL subgroups in the validation cohort from the Gene Expression Omnibus database. (A) Survival analysis of positive vs. negative PD-L1 subgroups in each cut-point. (B) Survival analysis of positive vs. negative TIL subgroups in each cut-point. Figure S5: The composition and abundance of immune cell types and expression distribution among four TIME subtypes in validation cohort from the Gene Expression Omnibus database. (A) Survival analysis of type I, type II, type III, and type IV. (B) The proportions of patients in type I, type II, type III, and type IV. (C) The abundance difference among 8 types of immune cells within 4 subtypes. (D) The abundance difference of 6 main subclass in each subtype. (E) The gene expression distributions of growth factors and receptors in each subtype. (F) The gene expression distributions of growth factors and receptors between TIL positive and TIL negative samples. (G) The correlation coefficient between the TIL score and expression of growth factors, as well as receptors. ****, p < 0.0001; ***, p < 0.001; **, p < 0.01; *, p < 0.05. Table S1: The response rate to ICI immunotherapy among four subtypes. Table S2: Statistical data of PD-L1 TPM, TIL (Z score), and subtypes proportion across pan-cancer. Table S3: Statistical data of 8 immune cell proportion across the four subtypes of tumor immune microenvironment. Table S4: The relative proportion of 20 immune cell subtypes across the four subtypes of tumor immune microenvironment. Table S5: Statistical data of the log2 value of TMB and the log2 value of neoantigens across the four subtypes of tumor immune microenvironment. Table S6: Mutation rate of the top 10 genes across the four subtypes of tumor immune microenvironment. Table S7: The drive gene detected of the four subtypes of tumor immune microenvironment. Table S8: Statistical data of 75 immune related genes across the four subtypes of tumor immune microenvironment. Table S9: The pathway scores of the shared pathways across the four subtypes of tumor immune microenvironment. Table S10: Statistical data of the specific gene TPM across the four subtypes of tumor immune microenvironment. Table S11: Statistical data of 8 immune cell proportion across the four subtypes of tumor immune microenvironment in the validation cohort from the Gene Expression Omnibus database. Table S12: The relative proportion of 20 immune cell subtypes across the four subtypes of tumor immune microenvironment in the validation cohort from the Gene Expression Omnibus database. Table S13: Statistical data of the specific gene TPM across the four subtypes of tumor immune microenvironment in the validation cohort from the Gene Expression Omnibus database.
Author Contributions
Conceptualization, K.H., M.H., and H.D.; data analysis, K.H. and J.C.; literature survey, J.Q.; illustrations, K.H.; writing—original draft preparation, K.H. and M.H.; writing—review and editing, J.W., S.L., and H.D; funding acquisition, H.D. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Key R&D Program of China (2018YFC0910201), the Key R&D Program of Guangdong Province (2019B020226001) and the Science and the Technology Planning Project of Guangzhou (201704020176).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data is contained within the article or supplementary material.
Acknowledgments
This work was supported by the National Key R&D Program of China (2018YFC0910201), the Key R&D Program of Guangdong Province (2019B020226001), and the Science and the Technology Planning Project of Guangzhou (201704020176).
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| ICI | Immune checkpoint inhibitors; |
| TIME | Tumor immune microenvironment; |
| PD-1 | Programmed death1; |
| PD-L1 | Programmed death ligand 1; |
| TCGA | The Cancer Genome Atlas; |
| TIL | Tumor-infiltrating lymphocyte; |
| TMB | Tumor mutation burden; |
| IHC | Immunohistochemistry; |
| THYM | Thymoma; |
| UCS | Uterine carcinosarcoma; |
| LUSC | Lung squamous cell carcinoma; |
| LIHC | Liver hepatocellular carcinoma; |
| TP53 | Tumor protein 53; |
| TTN | Titin; |
| LRP1B | LDL receptor related protein 1B; |
| CSMD3 | CUB and Sushi Multiple Domains 3; |
| BRAF | B-Raf Proto-Oncogene; |
| FAT1 | FAT Atypical Cadherin 1; |
| GTF2I | General Transcription Factor Iii; |
| PCLO | Piccolo; |
| ZFHX4 | Zinc Finger Homeobox 4; |
| SPTA1 | Spectrin alpha, erythrocytic 1; |
| APC | Adenomatous polyposis coli; |
| KMT2D | Lysine methyltransferase 2D; |
| PIK3CA | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha; |
| PDCD1LG2 | Programmed Cell Death 1 Ligand 2; |
| VTCN1 | V-set domain containing T cell activation inhibitor 1; |
| PDCD1 | Programmed Cell Death 1; |
| CTLA4 | Cytotoxic T-lymphocyte-associated protein 4; |
| DEG | Different expressed gene; |
| KEGG | Kyoto Encyclopedia of Genes and Genomes; |
| IFNG | Interferon gamma; |
| TNF | Tumor necrosis factor; |
| TNFA | Tumor Necrosis Factor Alpha; |
| IL6 | Interleukin 6; |
| IL12 | Interleukin 12; |
| IL12A | Interleukin 12A; |
| IL12B | Interleukin 12B; |
| IL10 | Interleukin 10; |
| GZMB | Granzyme B; |
| PRF1 | Perforin-1; |
| KRAS | Kirsten ras; |
| VEGFA | Vascular endothelial growth factor A; |
| TGFB1 | Transforming growth factor beta 1; |
| HRAS | HRas proto-oncogene; |
| IDH1 | Isocitrate dehydrogenase (NADP(+)) 1; |
| POLE | DNA polymerase epsilon; |
| POLD1 | DNA polymerase delta 1; |
| MUC16 | Mucin 16; |
| RYR2 | Ryanodine receptor 2; |
| SYNE1 | Spectrin repeat containing nuclear envelope protein 1; |
| FLG | Filaggrin; |
| USH2A | Usherin; |
| CDKN2A | Cyclin dependent kinase inhibitor 2A; |
| MB21D2 | Mab-21 domain containing 2; |
| NDUFA13 | NADH:ubiquinone oxidoreductase subunit A13; |
| DGCR6L | DiGeorge syndrome critical region gene 6 like; |
| S100A1 | S100 calcium binding protein A1; |
| IAPP | Islet amyloid polypeptide; |
| SLC3A2 | Solute carrier family 3 member 2; |
| KLF3 | Kruppel like factor 3; |
| GNG12 | G protein subunit gamma 12; |
| NRAS | NRAS proto-oncogene; |
| RAB9B | RAB9B, member RAS oncogene family; |
| SH3BGRL2 | SH3 domain binding glutamate rich protein like 2; |
| TNP1 | Transition protein 1; |
| RPL22 | Ribosomal protein L22; |
| MRPL22 | Mitochondrial ribosomal protein L22; |
| CBLN3 | Cerebellin 3 precursor; |
| PAIP2 | Poly(A) binding protein interacting protein 2; |
| SEC61B | SEC61 translocon subunit beta; |
| DBI | Diazepam binding inhibitor; |
| GNA11 | G protein subunit alpha 11; |
| ARHGAP1 | Rho GTPase activating protein 1. |
References
- Havel, J.J.; Chowell, D.; Chan, T.A. The evolving landscape of biomarkers for checkpoint inhibitor immunotherapy. Nat. Rev. Cancer 2019, 19, 133–150. [Google Scholar] [CrossRef]
- Sunshine, J.; Taube, J.M. PD-1/PD-L1 inhibitors. Curr. Opin. Pharmacol. 2015, 23, 32–38. [Google Scholar] [CrossRef]
- Smyth, M.J.; Ngiow, S.F.; Ribas, A.; Teng, M.W. Combination cancer immunotherapies tailored to the tumour microenvironment. Nat. Rev. Clin. Oncol. 2016, 13, 143–158. [Google Scholar] [CrossRef] [PubMed]
- Braun, D.A.; Burke, K.P.; Van Allen, E.M. Genomic Approaches to Understanding Response and Resistance to Immunotherapy. Clin. Cancer Res. 2016, 22, 5642–5650. [Google Scholar] [CrossRef]
- Balar, A.V.; Weber, J.S. PD-1 and PD-L1 antibodies in cancer: Current status and future directions. Cancer Immunol. Immunother. 2017, 66, 551–564. [Google Scholar] [CrossRef]
- Chen, D.S.; Mellman, I. Elements of cancer immunity and the cancer-immune set point. Nature 2017, 541, 321–330. [Google Scholar] [CrossRef]
- Gasser, S.; Lim, L.H.K.; Cheung, F.S.G. The role of the tumour microenvironment in immunotherapy. Endocr. Relat. Cancer 2017, 24, T283–T295. [Google Scholar] [CrossRef]
- Chalmers, Z.R.; Connelly, C.F.; Fabrizio, D.; Gay, L.; Ali, S.M.; Ennis, R.; Schrock, A.; Campbell, B.; Shlien, A.; Chmielecki, J.; et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 2017, 9, 34. [Google Scholar] [CrossRef] [PubMed]
- Rizvi, N.A.; Hellmann, M.D.; Snyder, A.; Kvistborg, P.; Makarov, V.; Havel, J.J.; Lee, W.; Yuan, J.; Wong, P.; Ho, T.S.; et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015, 348, 124–128. [Google Scholar] [CrossRef] [PubMed]
- Schumacher, T.N.; Schreiber, R.D. Neoantigens in cancer immunotherapy. Science 2015, 348, 69–74. [Google Scholar] [CrossRef]
- Fridman, W.H.; Pages, F.; Sautes-Fridman, C.; Galon, J. The immune contexture in human tumours: Impact on clinical outcome. Nat. Rev. Cancer 2012, 12, 298–306. [Google Scholar] [CrossRef]
- Bremnes, R.M.; Al-Shibli, K.; Donnem, T.; Sirera, R.; Al-Saad, S.; Andersen, S.; Stenvold, H.; Camps, C.; Busund, L.T. The role of tumor-infiltrating immune cells and chronic inflammation at the tumor site on cancer development, progression, and prognosis: Emphasis on non-small cell lung cancer. J. Thorac. Oncol. 2011, 6, 824–833. [Google Scholar] [CrossRef]
- Gibney, G.T.; Weiner, L.M.; Atkins, M.B. Predictive biomarkers for checkpoint inhibitor-based immunotherapy. Lancet Oncol. 2016, 17, e542–e551. [Google Scholar] [CrossRef]
- Jin, Z.; Yoon, H.H. The promise of PD-1 inhibitors in gastro-esophageal cancers: Microsatellite instability vs. PD-L1. J. Gastrointest. Oncol. 2016, 7, 771–788. [Google Scholar] [CrossRef] [PubMed]
- Teng, M.W.; Ngiow, S.F.; Ribas, A.; Smyth, M.J. Classifying Cancers Based on T-cell Infiltration and PD-L1. Cancer Res. 2015, 75, 2139–2145. [Google Scholar] [CrossRef]
- Taube, J.M.; Anders, R.A.; Young, G.D.; Xu, H.; Sharma, R.; McMiller, T.L.; Chen, S.; Klein, A.P.; Pardoll, D.M.; Topalian, S.L.; et al. Colocalization of inflammatory response with B7-h1 expression in human melanocytic lesions supports an adaptive resistance mechanism of immune escape. Sci. Transl. Med. 2012, 4, 127ra37. [Google Scholar] [CrossRef]
- Sznol, M.; Chen, L. Antagonist antibodies to PD-1 and B7-H1 (PD-L1) in the treatment of advanced human cancer. Clin. Cancer Res. 2013, 19, 1021–1034. [Google Scholar] [CrossRef] [PubMed]
- Sanmamed, M.F.; Chen, L. A Paradigm Shift in Cancer Immunotherapy: From Enhancement to Normalization. Cell 2018, 175, 313–326. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, L. Classification of Advanced Human Cancers Based on Tumor Immunity in the MicroEnvironment (TIME) for Cancer Immunotherapy. JAMA Oncol. 2016, 2, 1403–1404. [Google Scholar] [CrossRef] [PubMed]
- Yagi, T.; Baba, Y.; Ishimoto, T.; Iwatsuki, M.; Miyamoto, Y.; Yoshida, N.; Watanabe, M.; Baba, H. PD-L1 Expression, Tumor-infiltrating Lymphocytes, and Clinical Outcome in Patients With Surgically Resected Esophageal Cancer. Ann. Surg. 2019, 269, 471–478. [Google Scholar] [CrossRef]
- Lin, Z.; Gu, J.; Cui, X.; Huang, L.; Li, S.; Feng, J.; Liu, B.; Zhou, Y. Deciphering Microenvironment of NSCLC based on CD8 + TIL Density and PD-1/PD-L1 Expression. J. Cancer 2019, 10, 211–222. [Google Scholar] [CrossRef] [PubMed]
- Ricklefs, F.L.; Alayo, Q.; Krenzlin, H.; Mahmoud, A.B.; Speranza, M.C.; Nakashima, H.; Hayes, J.L.; Lee, K.; Balaj, L.; Passaro, C.; et al. Immune evasion mediated by PD-L1 on glioblastoma-derived extracellular vesicles. Sci. Adv. 2018, 4, eaar2766. [Google Scholar] [CrossRef] [PubMed]
- Poggio, M.; Hu, T.; Pai, C.C.; Chu, B.; Belair, C.D.; Chang, A.; Montabana, E.; Lang, U.E.; Fu, Q.; Fong, L.; et al. Suppression of Exosomal PD-L1 Induces Systemic Anti-tumor Immunity and Memory. Cell 2019, 177, 414–427.e13. [Google Scholar] [CrossRef] [PubMed]
- Ock, C.Y.; Keam, B.; Kim, S.; Lee, J.S.; Kim, M.; Kim, T.M.; Jeon, Y.K.; Kim, D.W.; Chung, D.H.; Heo, D.S. Pan-Cancer Immunogenomic Perspective on the Tumor Microenvironment Based on PD-L1 and CD8 T-Cell Infiltration. Clin. Cancer Res. 2016, 22, 2261–2270. [Google Scholar] [CrossRef]
- Chen, S.; Zhang, N.; Shao, J.; Wang, T.; Wang, X. Multi-omics Perspective on the Tumor Microenvironment based on PD-L1 and CD8 T-Cell Infiltration in Urothelial Cancer. J. Cancer 2019, 10, 697–707. [Google Scholar] [CrossRef]
- Kondou, R.; Iizuka, A.; Nonomura, C.; Miyata, H.; Ashizawa, T.; Nagashima, T.; Ohshima, K.; Urakami, K.; Kusuhara, M.; Yamaguchi, K.; et al. Classification of tumor microenvironment immune types based on immune response-associated gene expression. Int. J. Oncol. 2019, 54, 219–228. [Google Scholar] [CrossRef]
- Wang, Y.; Zhu, C.; Song, W.; Li, J.; Zhao, G.; Cao, H. PD-L1 Expression and CD8(+) T Cell Infiltration Predict a Favorable Prognosis in Advanced Gastric Cancer. J. Immunol. Res. 2018, 2018, 4180517. [Google Scholar] [CrossRef]
- Hugo, W.; Zaretsky, J.M.; Sun, L.; Song, C.; Moreno, B.H.; Hu-Lieskovan, S.; Berent-Maoz, B.; Pang, J.; Chmielowski, B.; Cherry, G.; et al. Genomic and Transcriptomic Features of Response to Anti-PD-1 Therapy in Metastatic Melanoma. Cell 2016, 165, 35–44. [Google Scholar] [CrossRef]
- Snyder, A.; Nathanson, T.; Funt, S.A.; Ahuja, A.; Buros Novik, J.; Hellmann, M.D.; Chang, E.; Aksoy, B.A.; Al-Ahmadie, H.; Yusko, E.; et al. Contribution of systemic and somatic factors to clinical response and resistance to PD-L1 blockade in urothelial cancer: An exploratory multi-omic analysis. PLoS Med. 2017, 14, e1002309. [Google Scholar] [CrossRef]
- Miao, D.; Margolis, C.A.; Gao, W.H.; Voss, M.H.; Li, W.; Martini, D.J.; Norton, C.; Bosse, D.; Wankowicz, S.M.; Cullen, D.; et al. Genomic correlates of response to immune checkpoint therapies in clear cell renal cell carcinoma. Science 2018, 359, 801–805. [Google Scholar] [CrossRef]
- Riaz, N.; Havel, J.J.; Makarov, V.; Desrichard, A.; Urba, W.J.; Sims, J.S.; Hodi, F.S.; Martin-Algarra, S.; Mandal, R.; Sharfman, W.H.; et al. Tumor and Microenvironment Evolution during Immunotherapy with Nivolumab. Cell 2017, 171, 934–949. [Google Scholar] [CrossRef] [PubMed]
- Mariathasan, S.; Turley, S.J.; Nickles, D.; Castiglioni, A.; Yuen, K.; Wang, Y.; Kadel, E.E., III; Koeppen, H.; Astarita, J.L.; Cubas, R.; et al. TGFbeta attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature 2018, 554, 544–548. [Google Scholar] [CrossRef]
- Dong, H.; Zhu, G.; Tamada, K.; Chen, L. B7-H1, a third member of the B7 family, co-stimulates T-cell proliferation and interleukin-10 secretion. Nat. Med. 1999, 5, 1365–1369. [Google Scholar] [CrossRef] [PubMed]
- Butte, M.J.; Keir, M.E.; Phamduy, T.B.; Sharpe, A.H.; Freeman, G.J. Programmed death-1 ligand 1 interacts specifically with the B7-1 costimulatory molecule to inhibit T cell responses. Immunity 2007, 27, 111–122. [Google Scholar] [CrossRef]
- Lee, J.S.; Ruppin, E. Multiomics Prediction of Response Rates to Therapies to Inhibit Programmed Cell Death 1 and Programmed Cell Death 1 Ligand 1. JAMA Oncol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Topalian, S.L.; Drake, C.G.; Pardoll, D.M. Immune checkpoint blockade: A common denominator approach to cancer therapy. Cancer Cell 2015, 27, 450–461. [Google Scholar] [CrossRef]
- Nowicki, T.S.; Akiyama, R.; Huang, R.R.; Shintaku, I.P.; Wang, X.; Tumeh, P.C.; Singh, A.; Chmielowski, B.; Denny, C.; Federman, N.; et al. Infiltration of CD8 T Cells and Expression of PD-1 and PD-L1 in Synovial Sarcoma. Cancer Immunol. Res. 2017, 5, 118–126. [Google Scholar] [CrossRef]
- Zhang, X.C.; Wang, J.; Shao, G.G.; Wang, Q.; Qu, X.; Wang, B.; Moy, C.; Fan, Y.; Albertyn, Z.; Huang, X.; et al. Comprehensive genomic and immunological characterization of Chinese non-small cell lung cancer patients. Nat. Commun. 2019, 10, 1772. [Google Scholar] [CrossRef]
- Binnewies, M.; Roberts, E.W.; Kersten, K.; Chan, V.; Fearon, D.F.; Merad, M.; Coussens, L.M.; Gabrilovich, D.I.; Ostrand-Rosenberg, S.; Hedrick, C.C.; et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat. Med. 2018, 24, 541–550. [Google Scholar] [CrossRef]
- Li, B.; Li, T.; Pignon, J.C.; Wang, B.; Wang, J.; Shukla, S.A.; Dou, R.; Chen, Q.; Hodi, F.S.; Choueiri, T.K.; et al. Landscape of tumor-infiltrating T cell repertoire of human cancers. Nat. Genet. 2016, 48, 725–732. [Google Scholar] [CrossRef]
- Gentles, A.J.; Newman, A.M.; Liu, C.L.; Bratman, S.V.; Feng, W.; Kim, D.; Nair, V.S.; Xu, Y.; Khuong, A.; Hoang, C.D.; et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat. Med. 2015, 21, 938–945. [Google Scholar] [CrossRef] [PubMed]
- Charoentong, P.; Finotello, F.; Angelova, M.; Mayer, C.; Efremova, M.; Rieder, D.; Hackl, H.; Trajanoski, Z. Pan-cancer Immunogenomic Analyses Reveal Genotype-Immunophenotype Relationships and Predictors of Response to Checkpoint Blockade. Cell Rep. 2017, 18, 248–262. [Google Scholar] [CrossRef]
- Baert, T.; Vankerckhoven, A.; Riva, M.; Van Hoylandt, A.; Thirion, G.; Holger, G.; Mathivet, T.; Vergote, I.; Coosemans, A. Myeloid Derived Suppressor Cells: Key Drivers of Immunosuppression in Ovarian Cancer. Front. Immunol. 2019, 10, 1273. [Google Scholar] [CrossRef]
- Samstein, R.M.; Lee, C.H.; Shoushtari, A.N.; Hellmann, M.D.; Shen, R.; Janjigian, Y.Y.; Barron, D.A.; Zehir, A.; Jordan, E.J.; Omuro, A.; et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat. Genet. 2019, 51, 202–206. [Google Scholar] [CrossRef]
- Yarchoan, M.; Hopkins, A.; Jaffee, E.M. Tumor Mutational Burden and Response Rate to PD-1 Inhibition. N. Engl. J. Med. 2017, 377, 2500–2501. [Google Scholar] [CrossRef]
- Davies, H.; Bignell, G.R.; Cox, C.; Stephens, P.; Edkins, S.; Clegg, S.; Teague, J.; Woffendin, H.; Garnett, M.J.; Bottomley, W.; et al. Mutations of the BRAF gene in human cancer. Nature 2002, 417, 949–954. [Google Scholar] [CrossRef] [PubMed]
- Wasylecka-Juszczynska, M.M.; Gorniak, P.; Szydlowski, M.; Polak, A.; Juszczynski, P. BRAF V600E mutation in melanoma sustains IFN-gamma inducible PD-L1 expression by coactivating STAT1 and increasing protein translation. Ann. Oncol. 2018, 29, viii463–viii464. [Google Scholar] [CrossRef]
- Dudnik, E.; Peled, N.; Nechushtan, H.; Wollner, M.; Onn, A.; Agbarya, A.; Moskovitz, M.; Keren, S.; Popovits-Hadari, N.; Urban, D.; et al. BRAF Mutant Lung Cancer: Programmed Death Ligand 1 Expression, Tumor Mutational Burden, Microsatellite Instability Status, and Response to Immune Check-Point Inhibitors. J. Thorac. Oncol. 2018, 13, 1128–1137. [Google Scholar] [CrossRef]
- Feng, D.; Qin, B.; Pal, K.; Sun, L.; Dutta, S.; Dong, H.; Liu, X.; Mukhopadhyay, D.; Huang, S.; Sinicrope, F.A. BRAF(V600E)-induced, tumor intrinsic PD-L1 can regulate chemotherapy-induced apoptosis in human colon cancer cells and in tumor xenografts. Oncogene 2019, 38, 6752–6766. [Google Scholar] [CrossRef] [PubMed]
- Gibney, G.T.; Tang, S.J.; Poorman, K.; Olszanski, A.J.; Eisenberg, B.L.; Mehmi, I.; Farma, J.M.; In, G.K.; Amin, A.; Rapisuwon, S.; et al. Associations of age, PD-L1 status, BRAF mutation and tumor mutational burden (TMB) in advanced melanoma. J. Clin. Oncol. 2018, 36. [Google Scholar] [CrossRef]
- Aoki, K.; Taketo, M.M. Adenomatous polyposis coli (APC): A multi-functional tumor suppressor gene. J. Cell Sci. 2007, 120, 3327–3335. [Google Scholar] [CrossRef] [PubMed]
- Scheel, A.H.; Ansen, S.; Schultheis, A.M.; Scheffler, M.; Fischer, R.N.; Michels, S.; Hellmich, M.; George, J.; Zander, T.; Brockmann, M.; et al. PD-L1 expression in non-small cell lung cancer: Correlations with genetic alterations. Oncoimmunology 2016, 5, e1131379. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.Y.; Zhong, W.Z.; Zhang, X.C.; Su, J.; Xie, Z.; Liu, S.Y.; Tu, H.Y.; Chen, H.J.; Sun, Y.L.; Zhou, Q.; et al. Potential Predictive Value of TP53 and KRAS Mutation Status for Response to PD-1 Blockade Immunotherapy in Lung Adenocarcinoma. Clin. Cancer Res. 2017, 23, 3012–3024. [Google Scholar] [CrossRef] [PubMed]
- Riether, C.; Schurch, C.M.; Ochsenbein, A.F. Regulation of hematopoietic and leukemic stem cells by the immune system. Cell Death Differ. 2015, 22, 187–198. [Google Scholar] [CrossRef]
- Geiger, R.; Rieckmann, J.C.; Wolf, T.; Basso, C.; Feng, Y.; Fuhrer, T.; Kogadeeva, M.; Picotti, P.; Meissner, F.; Mann, M.; et al. L-Arginine Modulates T Cell Metabolism and Enhances Survival and Anti-tumor Activity. Cell 2016, 167, 829–842.e13. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.H.; Qiu, J.; O’Sullivan, D.; Buck, M.D.; Noguchi, T.; Curtis, J.D.; Chen, Q.; Gindin, M.; Gubin, M.M.; van der Windt, G.J.; et al. Metabolic Competition in the Tumor Microenvironment Is a Driver of Cancer Progression. Cell 2015, 162, 1229–1241. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Locasale, J.W.; Bielas, J.H.; O’Sullivan, J.; Sheahan, K.; Cantley, L.C.; Vander Heiden, M.G.; Vitkup, D. Heterogeneity of tumor-induced gene expression changes in the human metabolic network. Nat. Biotechnol. 2013, 31, 522–529. [Google Scholar] [CrossRef]
- Wang, G.Z.; Zhang, L.; Zhao, X.C.; Gao, S.H.; Qu, L.W.; Yu, H.; Fang, W.F.; Zhou, Y.C.; Liang, F.; Zhang, C.; et al. The Aryl hydrocarbon receptor mediates tobacco-induced PD-L1 expression and is associated with response to immunotherapy. Nat. Commun. 2019, 10, 1125. [Google Scholar] [CrossRef]
- Badur, M.G.; Metallo, C.M. Reverse engineering the cancer metabolic network using flux analysis to understand drivers of human disease. Metab. Eng. 2018, 45, 95–108. [Google Scholar] [CrossRef]
- O’Neill, L.A.; Pearce, E.J. Immunometabolism governs dendritic cell and macrophage function. J. Exp. Med. 2016, 213, 15–23. [Google Scholar] [CrossRef]
- Motz, G.T.; Santoro, S.P.; Wang, L.P.; Garrabrant, T.; Lastra, R.R.; Hagemann, I.S.; Lal, P.; Feldman, M.D.; Benencia, F.; Coukos, G. Tumor endothelium FasL establishes a selective immune barrier promoting tolerance in tumors. Nat. Med. 2014, 20, 607–615. [Google Scholar] [CrossRef] [PubMed]
- Newman, A.M.; Liu, C.L.; Green, M.R.; Gentles, A.J.; Feng, W.; Xu, Y.; Hoang, C.D.; Diehn, M.; Alizadeh, A.A. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 2015, 12, 453–457. [Google Scholar] [CrossRef]
- Mayakonda, A.; Lin, D.C.; Assenov, Y.; Plass, C.; Koeffler, H.P. Maftools: Efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018, 28, 1747–1756. [Google Scholar] [CrossRef]
- Mermel, C.H.; Schumacher, S.E.; Hill, B.; Meyerson, M.L.; Beroukhim, R.; Getz, G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011, 12, R41. [Google Scholar] [CrossRef]
- Thorsson, V.; Gibbs, D.L.; Brown, S.D.; Wolf, D.; Bortone, D.S.; Ou Yang, T.H.; Porta-Pardo, E.; Gao, G.F.; Plaisier, C.L.; Eddy, J.A.; et al. The Immune Landscape of Cancer. Immunity 2018, 48, 812–830.e14. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- McCarthy, D.J.; Chen, Y.; Smyth, G.K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucl. Acids Res. 2012, 40, 4288–4297. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef]
- Veglia, F.; Sanseviero, E.; Gabrilovich, D.I. Myeloid-derived suppressor cells in the era of increasing myeloid cell diversity. Nat. Rev. Immunol. 2021. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.Y.; Zhang, Y.Y.; Zheng, L.T.; Zheng, C.H.; Song, J.T.; Zhang, Q.M.; Kang, B.X.; Liu, Z.Z.R.; Jin, L.; Xing, R.; et al. Global characterization of T cells in non-small-cell lung cancer by single-cell sequencing. Nat. Med. 2018, 24, 978–985. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).