Nerve Growth Factor Peptides Bind Copper(II) with High Affinity: A Thermodynamic Approach to Unveil Overlooked Neurotrophin Roles
Abstract
1. Introduction
2. Results
2.1. Protonation Constants
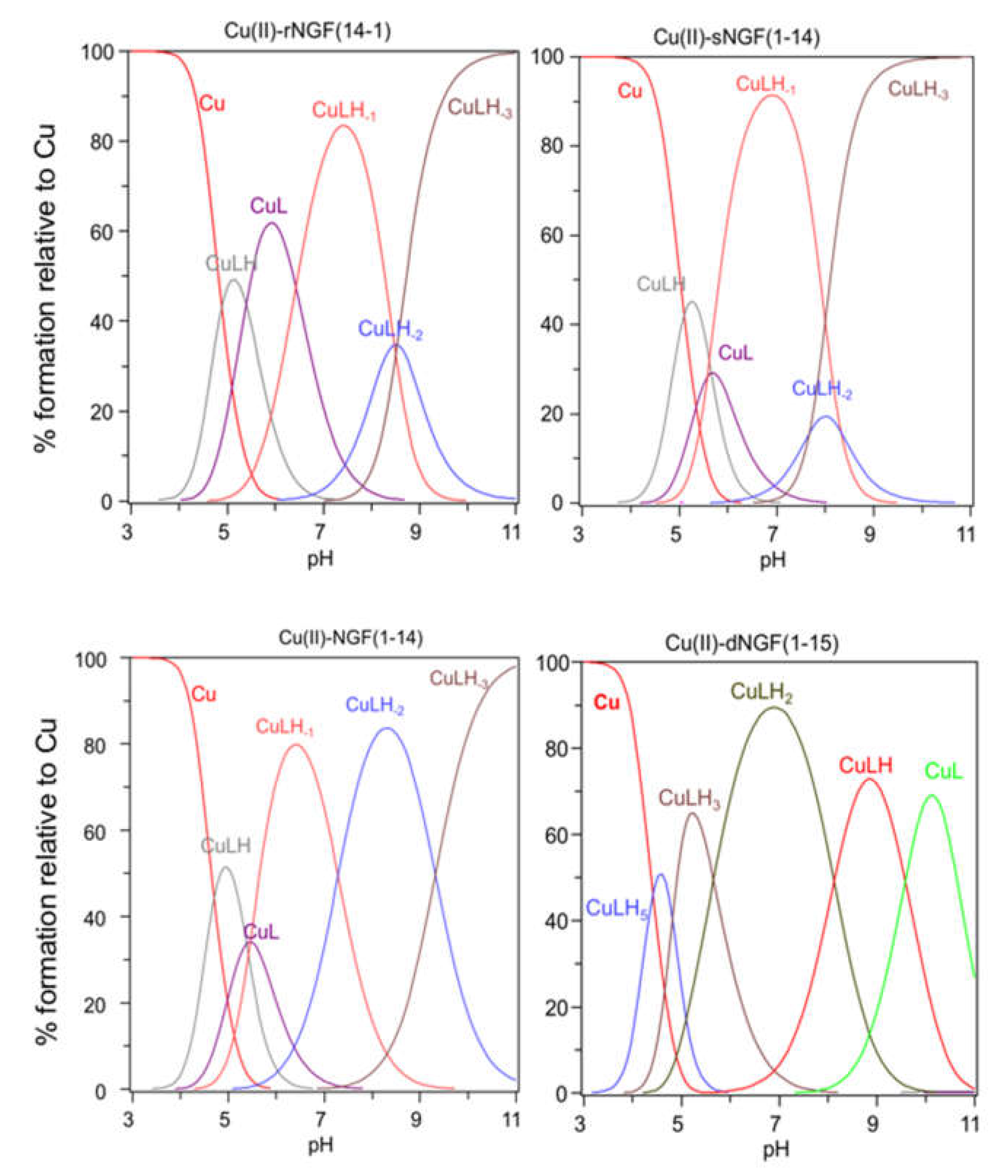
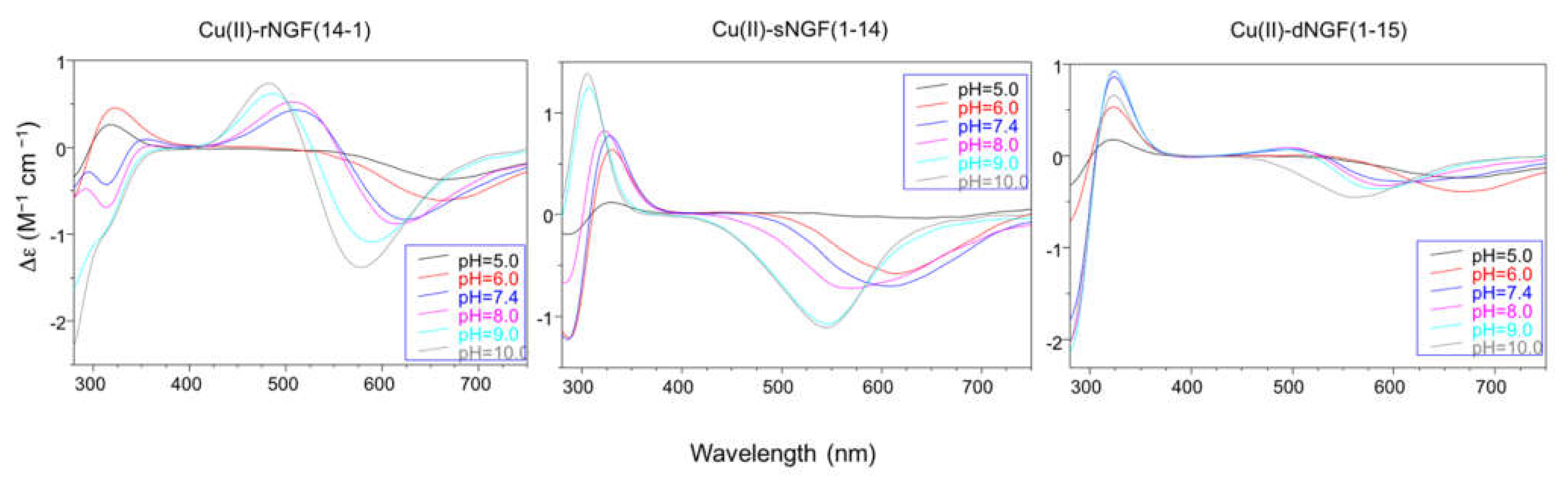
2.2. Speciation and Characterization of Copper-Peptide Complexes
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Chemicals
5.2. Potentiometric Titrations
5.3. UV–Vis and CD Measurements
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Scheiber, I.F.; Mercer, J.F.; Dringen, R. Metabolism and functions of copper in brain. Prog. Neurobiol. 2014, 116, 33–57. [Google Scholar] [CrossRef]
- Ackerman, C.M.; Chang, C.J. Copper signaling in the brain and beyond. J. Biol. Chem. 2018, 293, 4628–4635. [Google Scholar] [CrossRef]
- Lutsenko, S.; Washington-Hughes, C.; Ralle, M.; Schmidt, K. Copper and the brain noradrenergic system. J. Biol. Inorg. Chem. 2019, 24, 1179–1188. [Google Scholar] [CrossRef] [PubMed]
- Grubman, A.; White, A.R. Copper as a key regulator of cell signalling pathways. Expert Rev. Mol. Med. 2014, 16, e11. [Google Scholar] [CrossRef] [PubMed]
- Kardos, J.; Héja, L.; Simon, Á.; Jablonkai, I.; Kovács, R.; Jemnitz, K. Copper signalling: Causes and consequences. Cell Commun. Signal. 2018, 16. [Google Scholar] [CrossRef] [PubMed]
- La Mendola, D.; Giacomelli, C.; Rizzarelli, E. Intracellular Bioinorganic Chemistry and Cross Talk Among Different -Omics. Curr. Top. Med. Chem. 2016, 16, 3103–3130. [Google Scholar] [CrossRef] [PubMed]
- Kardos, J.; Kovács, I.; Hajós, F.; Kálmán, M.; Simonyi, M. Nerve endings from rat brain tissue release copper upon depolarization. A possible role in regulating neuronal excitability. Neurosci. Lett. 1989, 103, 139–144. [Google Scholar] [CrossRef]
- D’Ambrosi, N.; Rossi, L. Copper at synapse: Release, binding and modulation of neurotransmission. Neurochem. Int. 2015, 90, 36–45. [Google Scholar] [CrossRef] [PubMed]
- Kapkaeva, M.R.; Popova, O.V.; Kondratenko, R.V.; Rogozin, P.D.; Genrikhs, E.E.; Stelmashook, E.V.; Skrebitsky, V.G.; Khaspekov, L.G.; Isaev, N.K. Effects of copper on viability and functional properties of hippocampal neurons in vitro. Exp. Toxicol. Pathol. 2017, 69, 259–264. [Google Scholar] [CrossRef]
- Nam, E.; Nam, G.; Lim, M.H. Synaptic Copper, Amyloid-β, and Neurotransmitters in Alzheimer’s Disease. Biochemistry 2020, 59, 15–17. [Google Scholar] [CrossRef]
- Garcia-Osta, A.; Alberini, C.M. Amyloid beta mediates memory formation. Learn. Mem. 2009, 16, 267–272. [Google Scholar] [CrossRef] [PubMed]
- Parihar, M.S.; Brewer, G.J. Amyloid-β as a modulator of synaptic plasticity. J. Alzheimers Dis. 2010, 22, 741–763. [Google Scholar] [CrossRef]
- Zimbone, S.; Monaco, I.; Gianì, F.; Pandini, G.; Copani, A.G.; Giuffrida, M.L.; Rizzarelli, E. Amyloid Beta monomers regulate cyclic adenosine monophosphate response element binding protein functions by activating type-1 insulin-like growth factor receptors in neuronal cells. Aging Cell 2018, 17, e12684. [Google Scholar] [CrossRef]
- Naletova, I.; Satriano, C.; Pietropaolo, A.; Gianì, F.; Pandini, G.; Triaca, V.; Amadoro, G.; Latina, V.; Calissano, P.; Travaglia, A.; et al. The Copper(II)-Assisted Connection between NGF and BDNF by Means of Nerve Growth Factor-Mimicking Short Peptides. Cells 2019, 8, 301. [Google Scholar] [CrossRef]
- Levi-Montalcini, R. The nerve growth factor 35 years later. Science 1987, 237, 1154–1162. [Google Scholar] [CrossRef] [PubMed]
- Sofroniew, M.V.; Howe, C.L.; Mobley, W.C. Nerve growth factor signaling, neuroprotection, and neural repair. Annu. Rev. Neurosci. 2001, 24, 1217–1281. [Google Scholar] [CrossRef] [PubMed]
- Conner, J.M.; Franks, K.M.; Titterness, A.K.; Russell, K.; Merrill, D.A.; Christie, B.R.; Sejnowski, T.J.; Tuszynski, M.H. NGF is essential for hippocampal plasticity and learning. J. Neurosci. 2009, 29, 10883–10889. [Google Scholar] [CrossRef]
- Reichardt, L.F. Neurotrophin-regulated signalling pathways. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2006, 361, 1545–1564. [Google Scholar] [CrossRef]
- Riccio, A.; Pierchala, B.A.; Ciarallo, C.L.; Ginty, D.D. An NGF-TrkA-mediated retrograde signal to transcription factor CREB in sympathetic neurons. Science 1997, 277, 1097–1100. [Google Scholar] [CrossRef]
- Travaglia, A.; Arena, G.; Fattorusso, R.; Isernia, C.; La Mendola, D.; Malgieri, G.; Nicoletti, V.G.; Rizzarelli, E. The inorganic perspective of nerve growth factor: Interactions of Cu2+ and Zn2+ with the N-terminus fragment of nerve growth factor encompassing the recognition domain of the TrkA receptor. Chemistry 2011, 17, 3726–3738. [Google Scholar] [CrossRef]
- Pandini, G.; Satriano, C.; Pietropaolo, A.; Gianì, F.; Travaglia, A.; La Mendola, D.; Nicoletti, V.G.; Rizzarelli, E. The Inorganic Side of NGF: Copper(II) and Zinc(II) Affect the NGF Mimicking Signaling of the N-Terminus Peptides Encompassing the Recognition Domain of TrkA Receptor. Front. Neurosci. 2016, 10. [Google Scholar] [CrossRef]
- Mufson, E.J.; Counts, S.E.; Ginsberg, S.D.; Mahady, L.; Perez, S.E.; Massa, S.M.; Longo, F.M.; Ikonomovic, M.D. Nerve Growth Factor Pathobiology during the Progression of Alzheimer’s Disease. Front. Neurosci. 2019, 13. [Google Scholar] [CrossRef] [PubMed]
- Matrone, C.; Ciotti, M.T.; Mercanti, D.; Marolda, R.; Calissano, P. NGF and BDNF signaling control amyloidogenic route and Abeta production in hippocampal neurons. Proc. Natl. Acad. Sci. USA 2008, 105, 13139–13144. [Google Scholar] [CrossRef]
- Canu, N.; Pagano, I.; La Rosa, L.R.; Pellegrino, M.; Ciotti, M.T.; Mercanti, D.; Moretti, F.; Sposato, V.; Triaca, V.; Petrella, C.; et al. Association of TrkA and APP Is Promoted by NGF and Reduced by Cell Death-Promoting Agents. Front. Mol. Neurosci. 2017, 10. [Google Scholar] [CrossRef]
- Canu, N.; Amadoro, G.; Triaca, V.; Latina, V.; Sposato, V.; Corsetti, V.; Severini, C.; Ciotti, M.T.; Calissano, P. The Intersection of NGF/TrkA Signaling and Amyloid Precursor Protein Processing in Alzheimer’s Disease Neuropathology. Int. J. Mol. Sci. 2017, 18, 1319. [Google Scholar] [CrossRef] [PubMed]
- Su, R.; Su, W.; Jiao, Q. NGF protects neuroblastoma cells against β-amyloid-induced apoptosis via the Nrf2/HO-1 pathway. FEBS Open Bio 2019, 9, 2063–2071. [Google Scholar] [CrossRef]
- Sáez, E.T.; Pehar, M.; Vargas, M.R.; Barbeito, L.; Maccioni, R.B. Production of nerve growth factor by beta-amyloid-stimulated astrocytes induces p75NTR-dependent tau hyperphosphorylation in cultured hippocampal neurons. J. Neurosci. Res. 2006, 84, 1098–1106. [Google Scholar] [CrossRef] [PubMed]
- Ejaz, H.W.; Wang, W.; Lang, M. Copper Toxicity Links to Pathogenesis of Alzheimer’s Disease and Therapeutics Approaches. Int. J. Mol. Sci. 2020, 21, 7660. [Google Scholar] [CrossRef]
- Kaden, D.; Bush, A.I.; Danzeisen, R.; Bayer, T.A.; Multhaup, G. Disturbed copper bioavailability in Alzheimer’s disease. Int. J. Alzheimers Dis. 2011, 2011. [Google Scholar] [CrossRef]
- Barnham, K.J.; Bush, A.I. Metals in Alzheimer’s and Parkinson’s diseases. Curr. Opin. Chem. Biol. 2008, 12, 222–228. [Google Scholar] [CrossRef]
- Lovell, M.A.; Robertson, J.D.; Teesdale, W.J.; Campbell, J.L.; Markesbery, W.R. Copper, iron and zinc in Alzheimer’s disease senile plaques. J. Neurol. Sci. 1998, 158, 47–52. [Google Scholar] [CrossRef]
- Bagheri, S.; Squitti, R.; Haertlé, T.; Siotto, M.; Saboury, A.A. Role of Copper in the Onset of Alzheimer’s Disease Compared to Other Metals. Front. Aging Neurosci. 2018, 9. [Google Scholar] [CrossRef]
- Exley, C.; House, E.; Polwart, A.; Esiri, M.M. Brain burdens of aluminum, iron, and copper and their relationships with amyloid-β pathology in 60 human brains. J. Alzheimers Dis. 2012, 31, 725–730. [Google Scholar] [CrossRef]
- Scholefield, M.; Church, S.J.; Xu, J.; Patassini, S.; Roncaroli, F.; Hooper, N.M.; Unwin, R.D.; Cooper, G.J.S. Widespread Decreases in Cerebral Copper Are Common to Parkinson’s Disease Dementia and Alzheimer’s Disease Dementia. Front. Aging Neurosci. 2021, 13. [Google Scholar] [CrossRef] [PubMed]
- Squitti, R.; Siotto, M.; Polimanti, R. Low-copper diet as a preventive strategy for Alzheimer’s disease. Neurobiol. Aging 2014, 35 (Suppl. 2), S40–S50. [Google Scholar] [CrossRef]
- Behzadfar, L.; Abdollahi, M.; Sabzevari, O.; Hosseini, R.; Salimi, A.; Naserzadeh, P.; Sharifzadeh, M.; Pourahmad, J. Potentiating role of copper on spatial memory deficit induced by beta amyloid and evaluation of mitochondrial function markers in the hippocampus of rats. Metallomics 2017, 9, 969–980. [Google Scholar] [CrossRef] [PubMed]
- Squitti, R.; Ventriglia, M.; Gennarelli, M.; Colabufo, N.A.; El Idrissi, I.G.; Bucossi, S.; Mariani, S.; Rongioletti, M.; Zanetti, O.; Congiu, C.; et al. Non-Ceruloplasmin Copper Distincts Subtypes in Alzheimer’s Disease: A Genetic Study of ATP7B Frequency. Mol. Neurobiol. 2017, 54, 671–681. [Google Scholar] [CrossRef]
- Zhao, J.; Shi, Q.; Tian, H.; Li, Y.; Liu, Y.; Xu, Z.; Robert, A.; Liu, Q.; Meunier, B. TDMQ20, a Specific Copper Chelator, Reduces Memory Impairments in Alzheimer’s Disease Mouse Models. ACS Chem. Neurosci. 2021, 12, 140–149. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Yin, Y.L.; Liu, X.Z.; Shen, P.; Zheng, Y.G.; Lan, X.R.; Lu, C.B.; Wang, J.Z. Current understanding of metal ions in the pathogenesis of Alzheimer’s disease. Transl. Neurodegener. 2020, 9. [Google Scholar] [CrossRef]
- Stefaniak, E.; Pushie, M.J.; Vaerewyck, C.; Corcelli, D.; Griggs, C.; Lewis, W.; Kelley, E.; Maloney, N.; Sendzik, M.; Bal, W.; et al. Exploration of the Potential Role for Aβ in Delivery of Extracellular Copper to Ctr1. Inorg. Chem. 2020, 59, 16952–16966. [Google Scholar] [CrossRef]
- Stefaniak, E.; Bal, W. CuII Binding Properties of N-Truncated Aβ Peptides: In Search of Biological Function. Inorg. Chem. 2019, 58, 13561–13577. [Google Scholar] [CrossRef] [PubMed]
- Alies, B.; Bijani, C.; Sayen, S.; Guillon, E.; Faller, P.; Hureau, C. Copper coordination to native N-terminally modified versus full-length amyloid-β: Second-sphere effects determine the species present at physiological pH. Inorg. Chem. 2012, 51, 12988–13000. [Google Scholar] [CrossRef] [PubMed]
- Arena, G.; Pappalardo, G.; Sovago, I.; Rizzarelli, E. Copper(II) interaction with amyloid-β: Affinity and speciation. Coord. Chem. Rev. 2012, 256, 3–12. [Google Scholar] [CrossRef]
- Lyros, E.; Ragoschke-Schumm, A.; Kostopoulos, P.; Sehr, A.; Backens, M.; Kalampokini, S.; Decker, Y.; Lesmeister, M.; Liu, Y.; Reith, W.; et al. Normal brain aging and Alzheimer’s disease are associated with lower cerebral pH: An in vivo histidine 1H-MR spectroscopy study. Neurobiol. Aging 2020, 87, 60–69. [Google Scholar] [CrossRef] [PubMed]
- Decker, Y.; Németh, E.; Schomburg, R.; Chemla, A.; Fülöp, L.; Menger, M.D.; Liu, Y.; Fassbender, K. Decreased pH in the aging brain and Alzheimer’s disease. Neurobiol. Aging 2021, 101, 40–49. [Google Scholar] [CrossRef]
- Tóth, M.O.; Menyhárt, Á.; Frank, R.; Hantosi, D.; Farkas, E.; Bari, F. Tissue Acidosis Associated with Ischemic Stroke to Guide Neuroprotective Drug Delivery. Biology 2020, 9, 460. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.; Chang, P.T. Acidic pH promotes the formation of toxic fibrils from beta-amyloid peptide. Brain Res. 2001, 893, 287–291. [Google Scholar] [CrossRef]
- Grasso, G.; Magrì, A.; Bellia, F.; Pietropaolo, A.; La Mendola, D.; Rizzarelli, E. The copper(II) and zinc(II) coordination mode of HExxH and HxxEH motif in small peptides: The role of carboxylate location and hydrogen bonding network. J. Inorg. Biochem. 2014, 130, 92–102. [Google Scholar] [CrossRef]
- Rajković, S.; Kállay, C.; Serényi, R.; Malandrinos, G.; Hadjiliadis, N.; Sanna, D.; Sóvágó, I. Complex formation processes of terminally protected peptides containing two or three histidyl residues. Characterization of the mixed metal complexes of peptides. Dalton Trans. 2008, 37, 5059–5071. [Google Scholar] [CrossRef]
- Kállay, C.; Várnagy, K.; Micera, G.; Sanna, D.; Sóvágó, I. Copper(II) complexes of oligopeptides containing aspartyl and glutamyl residues. Potentiometric and spectroscopic studies. J. Inorg. Biochem. 2005, 99, 1514–1525. [Google Scholar] [CrossRef]
- La Mendola, D.; Magrì, A.; Campagna, T.; Campitiello, M.A.; Raiola, L.; Isernia, C.; Hansson, O.; Bonomo, R.P.; Rizzarelli, E. A doppel alpha-helix peptide fragment mimics the copper(II) interactions with the whole protein. Chemistry 2010, 16, 6212–6223. [Google Scholar] [CrossRef]
- Karavelas, T.; Malandrinos, G.; Hadjiliadis, N.; Mlynarz, P.; Kozlowski, H.; Barsan, M.; Butler, I. Coordination properties of Cu(II) and Ni(II) ions towards the C-terminal peptide fragment -TYTEHA- of histone H4. Dalton Trans. 2008, 9, 1215–1223. [Google Scholar] [CrossRef]
- Kowalik-Jankowska, T.; Ruta, M.; Wiśniewska, K.; Lankiewicz, L. Coordination abilities of the 1-16 and 1-28 fragments of beta-amyloid peptide towards copper(II) ions: A combined potentiometric and spectroscopic study. J. Inorg. Biochem. 2003, 95, 270–282. [Google Scholar] [CrossRef]
- Damante, C.A.; Osz, K.; Nagy, Z.; Pappalardo, G.; Grasso, G.; Impellizzeri, G.; Rizzarelli, E.; Sóvágó, I. The metal loading ability of beta-amyloid N-terminus: A combined potentiometric and spectroscopic study of copper(II) complexes with beta-amyloid(1-16), its short or mutated peptide fragments, and its polyethylene glycol (PEG)-ylated analogue. Inorg. Chem. 2008, 47, 9669–9683. [Google Scholar] [CrossRef] [PubMed]
- Magrì, A.; Munzone, A.; Peana, M.; Medici, S.; Zoroddu, M.A.; Hansson, O.; Satriano, C.; Rizzarelli, E.; La Mendola, D. Coordination Environment of Cu(II) Ions Bound to N-Terminal Peptide Fragments of Angiogenin Protein. Int. J. Mol. Sci. 2016, 17, 1240. [Google Scholar] [CrossRef]
- Kállay, C.; Nagy, Z.; Várnagy, K.; Malandrinos, G.; Hadjiliadis, N.; Sóvágó, I. Thermodynamic and structural characterization of the copper(II) complexes of peptides containing both histidyl and aspartyl residues. Bioinorg. Chem. Appl. 2007, 2007. [Google Scholar] [CrossRef]
- La Mendola, D.; Magrì, A.; Hansson, Ö.; Bonomo, R.P.; Rizzarelli, E. Copper(II) complexes with peptide fragments encompassing the sequence 122-130 of human doppel protein. J. Inorg. Biochem. 2009, 103, 758–765. [Google Scholar] [CrossRef] [PubMed]
- Matera, A.; Brasuń, J.; Cebrat, M.; Świątek-Kozłowska, J. The role of the histidine residue in the coordination abilities of peptides with a multi-histidine sequence towards copper(II) ions. Polyhedron 2008, 27, 1539–1555. [Google Scholar] [CrossRef]
- Zoroddu, M.A.; Kowalik-Jankowska, T.; Medici, S.; Peana, M.; Kozlowski, H. Copper(II) binding to Cap43 protein fragments. Dalton Trans. 2008, 44, 6127–6134. [Google Scholar] [CrossRef]
- Pontecchiani, F.; Simonovsky, E.; Wieczorek, R.; Barbosa, N.; Rowinska-Zyrek, M.; Potocki, S.; Remelli, M.; Miller, Y.; Kozlowski, H. The unusual binding mechanism of Cu(II) ions to the poly-histidyl domain of a peptide found in the venom of an African viper. Dalton Trans. 2014, 43, 16680–16689. [Google Scholar] [CrossRef] [PubMed]
- Pietropaolo, A.; Muccioli, L.; Zannoni, C.; La Mendola, D.; Maccarrone, G.; Pappalardo, G.; Rizzarelli, E. Unveiling the role of histidine and tyrosine residues on the conformation of the avian prion hexarepeat domain. J. Phys. Chem. B 2008, 112, 5182–5188. [Google Scholar] [CrossRef]
- Bataille, M.; Formicka-Kozlowska, G.; Kozlowski, H.; Pettit, L.D.; Steel, I. The L-proline residue as a ‘break-point’ in the co-ordination of metal–peptide systems. J. Chem. Soc. Chem. Commun. 1984, 231–232. [Google Scholar] [CrossRef]
- Magrì, A.; Tabbì, G.; Breglia, R.; De Gioia, L.; Fantucci, P.; Bruschi, M.; Bonomo, R.P.; La Mendola, D. Copper ion interaction with the RNase catalytic site fragment of the angiogenin protein: An experimental and theoretical investigation. Dalton Trans. 2017, 46, 8524–8538. [Google Scholar] [CrossRef]
- La Mendola, D.; Arena, G.; Pietropaolo, A.; Satriano, C.; Rizzarelli, E. Metal ion coordination in peptide fragments of neurotrophins: A crucial step for understanding the role and signaling of these proteins in the brain. Coord. Chem. Rev. 2021, 435. [Google Scholar] [CrossRef]
- Murphy, J.M.; Powell, B.A.; Brumaghim, J.L. Stability constants of bio-relevant, redox-active metals with amino acids: The challenges of weakly binding ligands. Coord. Chem. Rev. 2020, 412. [Google Scholar] [CrossRef]
- Tõugu, V.; Karafin, A.; Palumaa, P. Binding of zinc(II) and copper(II) to the full-length Alzheimer’s amyloid-beta peptide. J. Neurochem. 2008, 104, 1249–1259. [Google Scholar] [CrossRef] [PubMed]
- Paredes-Rosan, C.A.; Valencia, D.E.; Barazorda-Ccahuana, H.L.; Aguilar-Pineda, J.A.; Gómez, B. Amyloid beta oligomers: How pH influences over trimer and pentamer structures? J. Mol. Model. 2020, 26, 1–8. [Google Scholar] [CrossRef]
- Liao, Q.; Owen, M.C.; Bali, S.; Barz, B.; Strodel, B. Aβ under stress: The effects of acidosis, Cu2+-binding, and oxidation on amyloid β-peptide dimers. Chem. Commun. 2018, 54, 7766–7769. [Google Scholar] [CrossRef]
- Amorini, A.M.; Bellia, F.; Di Pietro, V.; Giardina, B.; La Mendola, D.; Lazzarino, G.; Sortino, S.; Tavazzi, B.; Rizzarelli, E.; Vecchio, G. Synthesis and antioxidant activity of new homocarnosine beta-cyclodextrin conjugates. Eur. J. Med. Chem. 2007, 42, 910–920. [Google Scholar] [CrossRef]
- Gans, P.; Sabatini, A.; Vacca, A. Investigation of equilibria in solution. Determination of equilibrium constants with the HYPERQUAD suite of programs. Talanta 1996, 43, 1739–1753. [Google Scholar] [CrossRef]
- Alderighi, L.; Gans, P.; Ienco, A.; Peters, D.; Sabatini, A.; Vacca, A. Hyperquad simulation and speciation (HySS): A utility program for the investigation of equilibria involving soluble and partially soluble species. Coord. Chem. Rev. 1999, 184, 311–318. [Google Scholar] [CrossRef]
- Bellia, F.; La Mendola, D.; Maccarrone, G.; Mineo, P.; Vitalini, D.; Scamporrino, E.; Sortino, S.; Vecchio, G.; Rizzarelli, E. Copper(II) complexes with β-cyclodextrin–homocarnosine conjugates and their antioxidant activity. Inorg. Chim. Acta 2007, 360, 945–954. [Google Scholar] [CrossRef]
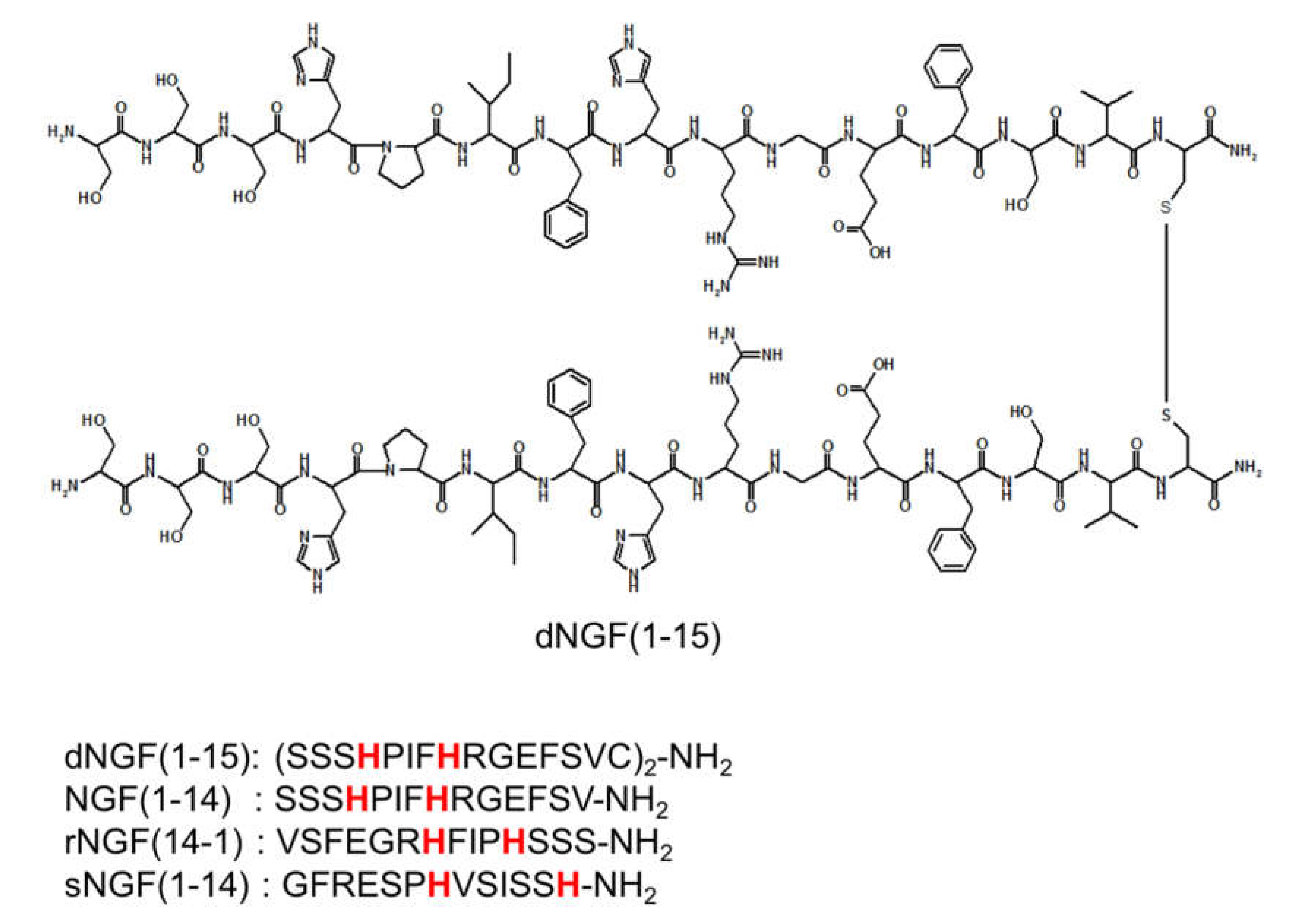
| Species | NGF(1-14) b | rNGF(14-1) | sNGF(1-14) | dNGF(1-15) |
|---|---|---|---|---|
| LH | 7.56 | 7.82 (2) | 7.85 (3) | 7.65 (5) |
| LH2 | 14.13 | 14.51 (2) | 14.65 (3) | 15.35 (2) |
| LH3 | 20.14 | 20.60 (2) | 20.87 (3) | - |
| LH4 | 24.44 | 24.88 (2) | 25.11 (4) | 28.77 (6) |
| LH5 | - | 35.12 (4) | ||
| LH6 | - | 41.31 (4) | ||
| LH7 | - | 46.62 (4) | ||
| LH8 | - | 50.75 (4) | ||
| pK COO- | 4.13 | 4.28 | 4.23 | 4.13 |
| pK COO- | - | - | - | 5.28 |
| pK His | 6.01 | 6.09 | 6.22 | 6.21 |
| pK His | 6.57 | 6.69 | 6.81 | 6.35 |
| pK His (×2) | - | - | - | (6.71 × 2) |
| pK NH2 | 7.56 | 7.82 | 7.85 | 7.65 |
| pK NH2 | - | 7.70 |
| Species (pqr) b | logβpqr NGF(1-14)c | logβpqr rNGF(14-1) | logβpqr sNGF(1-14) | logβpqr dNGF(1-15) |
|---|---|---|---|---|
| CuLH5 | - | - | - | 41.18 (3) |
| CuLH3 | - | - | - | 31.56 (2) |
| CuLH2 | - | - | - | 25.91 (5) |
| CuLH | 14.09 | 14.15 (1) | 13.97 (4) | 17.79 (5) |
| CuL | 8.72 | 8.77 (1) | 8.32 (5) | 8.21 (4) |
| CuLH-1 | 3.27 | 2.33 (2) | −2.74 (3) | - |
| CuLH-2 | −4.02 | −6.15 (4) | −5.58 (8) | - |
| CuLH-3 | −13.34 | −14.67 (2) | −13.27 (4) | - |
| pK (n/m) | ||||
| pK (5/3) | - | - | - | 4.81 × 2 |
| pK (3/2) | - | - | - | 5.65 |
| pK (2/1) | - | - | - | 8.12 |
| pK (1/0) | 5.37 | 5.38 | 5.65 | 9.57 |
| pK (0/−1) | 5.45 | 6.43 | 5.57 | - |
| pK (−1/−2) | 7.29 | 8.48 | 8.33 | - |
| pK (−2/−3) | 9.30 | 8.52 | 7.69 | - |
| Peptide | pH | UV-vis λ (nm) (ε (M−1 cm−1)) | CD λ (nm) (Δε (M−1 cm−1) |
|---|---|---|---|
| rNGF(14-1) | 5 | 640 (64) | 280 (−0.30); 316 (+0.20); 670 (−0.27) |
| 6 | 625 (144) | 280 (−0.40); 323 (+0.33); 671 (−0.44) | |
| 7.4 | 609 (178) | 280 (−0.30); 314 (−0.22); 352 (+0.07); 508 (+0.31); 625 (−0.60) | |
| 9 | 532 (194) | 280 (−1.30); 484 (+0.46); 589 (−0.79) | |
| 10 | 522 (232) | 280 (−1.80); 483 (+0.54); 577 (−1.00) | |
| sNGF(1-14) | 5 | 670 (40) | 289 (−0.20); 328 (+0.12); 647 (−0.04) |
| 6 | 617 (94) | 287 (−1.34); 328 (+0.68); 617 (−0.59) | |
| 7.4 | 603 (102) | 288 (−1.38); 329 (+0.82); 603 (−0.72) | |
| 9-10 | 522 (141) | 306 (+1.47); 544 (−1.13) | |
| dNGF(1-15) | 5 | 635 (50) | 280 (−0.34); 321 (+0.19); 669 (−0.24) |
| 6 | 612 (70) | 280 (−0.40); 323 (+0.33); 671 (−0.44) | |
| 7.4 | 569 (94) | 280 (−1.80); 324 (+0.92); 504 (+0.08); 611 (−0.30) | |
| 8 | 561 (104) | 280 (−2.04); 323 (+0.98); 496 (+0.11); 586 (−0.34) | |
| 9 | 554 (119) | 280 (−2.15); 324 (+0.98); 496 (+0.08); 587 (−0.38) | |
| 10 | 530 (138) | 280 (−2.14); 324 (+0.71); 563 (−0.46) |
| Peptide | pH | |
|---|---|---|
| NGF(1-14) | 7.4 | 2.5 × 10−11 |
| dNGF(1-15) | 7.4 | 4.2 × 10−11 |
| rNGF(14-1) | 7.4 | 6.5 × 10−10 |
| sNGF(1-14) | 7.4 | 2.7 × 10−10 |
| Aβ1-16-PEG a | 7.4 | 1.1 × 10−10 |
| hCtr1-14 b | 7.4 | 1.0 × 10−13 |
| NGF(1-14) | 5.5 | 4.0 × 10−6 |
| dNGF(1-15) | 5.5 | 1.9 × 10−7 |
| rNGF(14-1) | 5.5 | 1.4 × 10−5 |
| sNGF(1-14) | 5.5 | 3.8 × 10−5 |
| Aβ1-16-PEG | 5.5 | 2.5 × 10−7 |
| hCtr1-14 | 5.5 | 7.4 × 10−8 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Magrì, A.; La Mendola, D.; Rizzarelli, E. Nerve Growth Factor Peptides Bind Copper(II) with High Affinity: A Thermodynamic Approach to Unveil Overlooked Neurotrophin Roles. Int. J. Mol. Sci. 2021, 22, 5085. https://doi.org/10.3390/ijms22105085
Magrì A, La Mendola D, Rizzarelli E. Nerve Growth Factor Peptides Bind Copper(II) with High Affinity: A Thermodynamic Approach to Unveil Overlooked Neurotrophin Roles. International Journal of Molecular Sciences. 2021; 22(10):5085. https://doi.org/10.3390/ijms22105085
Chicago/Turabian StyleMagrì, Antonio, Diego La Mendola, and Enrico Rizzarelli. 2021. "Nerve Growth Factor Peptides Bind Copper(II) with High Affinity: A Thermodynamic Approach to Unveil Overlooked Neurotrophin Roles" International Journal of Molecular Sciences 22, no. 10: 5085. https://doi.org/10.3390/ijms22105085
APA StyleMagrì, A., La Mendola, D., & Rizzarelli, E. (2021). Nerve Growth Factor Peptides Bind Copper(II) with High Affinity: A Thermodynamic Approach to Unveil Overlooked Neurotrophin Roles. International Journal of Molecular Sciences, 22(10), 5085. https://doi.org/10.3390/ijms22105085







