AtPiezo Plays an Important Role in Root Cap Mechanotransduction
Abstract
1. Introduction
2. Results
2.1. Identification and Analysis of Piezo Genes in Plants
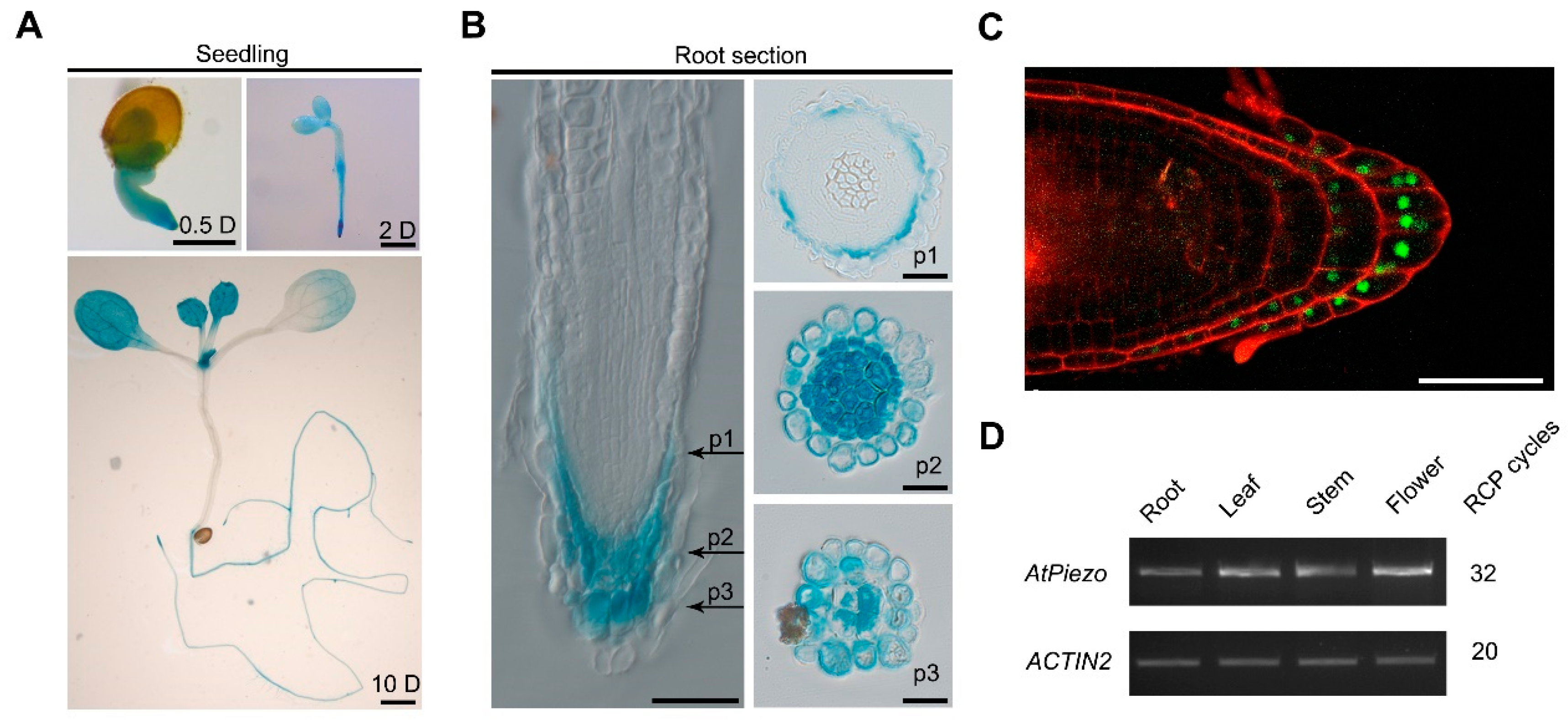
2.2. AtPiezo Is Expressed in Root Cap
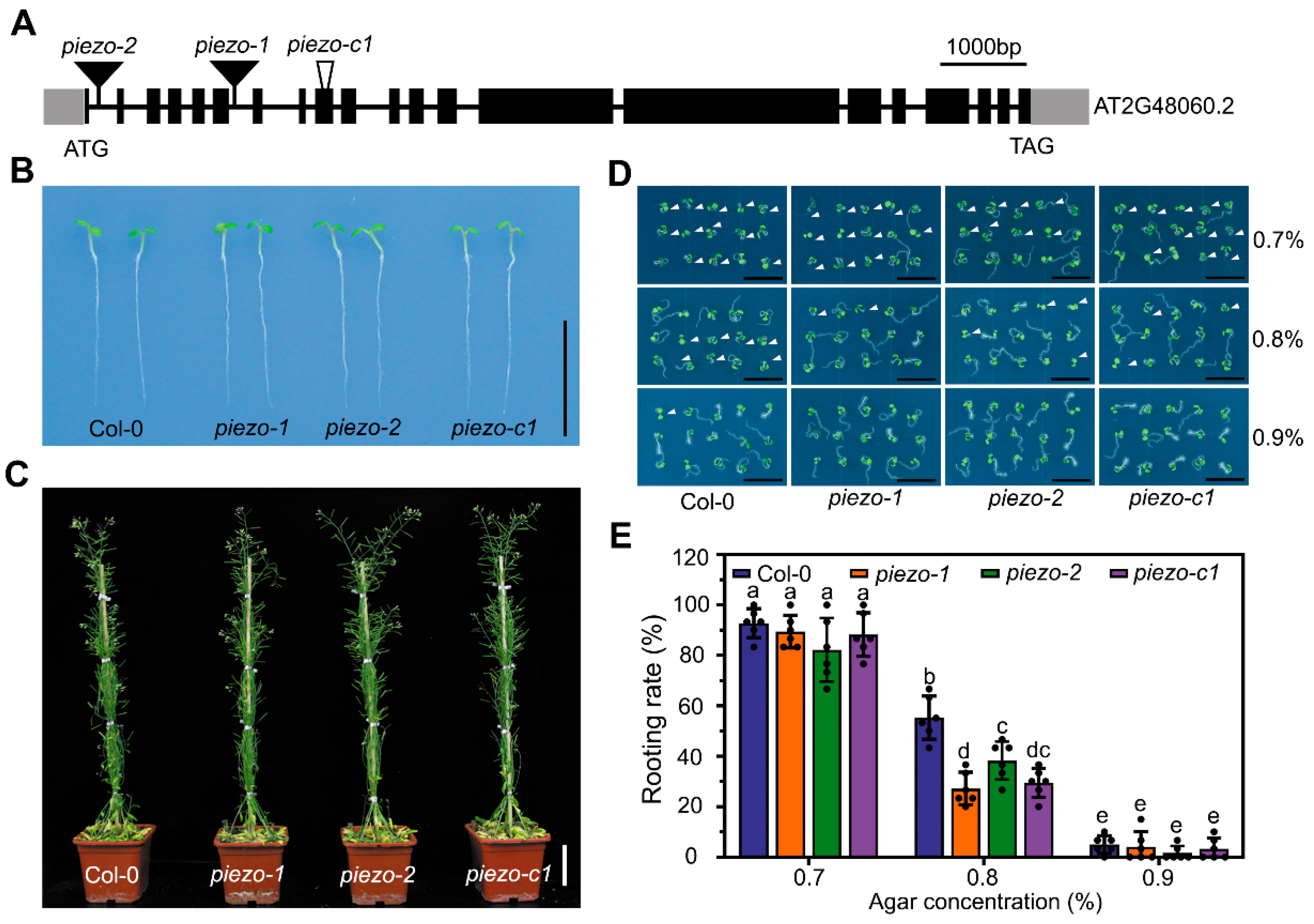
2.3. Atpiezo Mutants Exhibit Reduced Rooting Ability
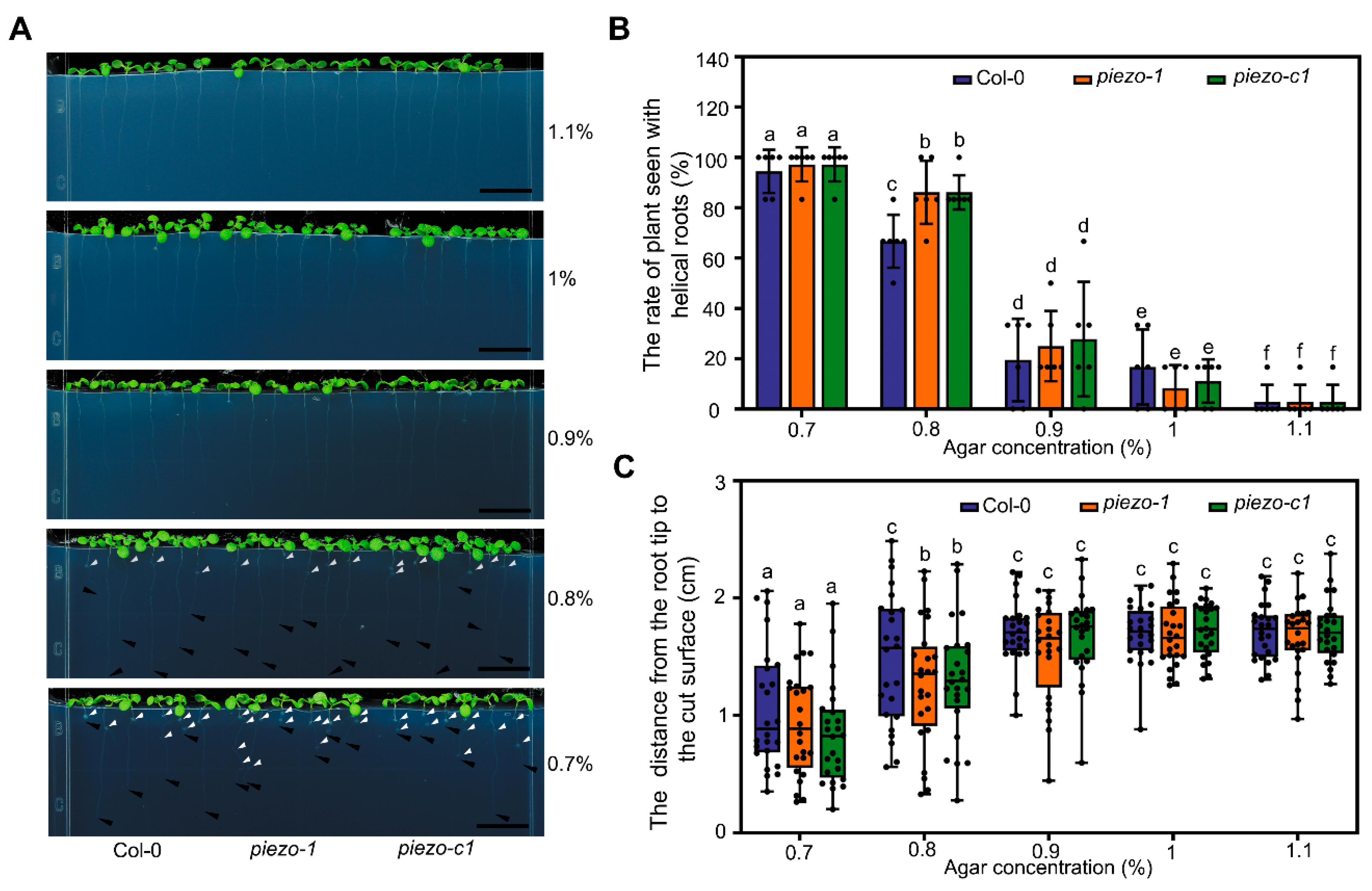
2.4. Atpiezo Mutants Show Altered Growth Status Inside the Medium
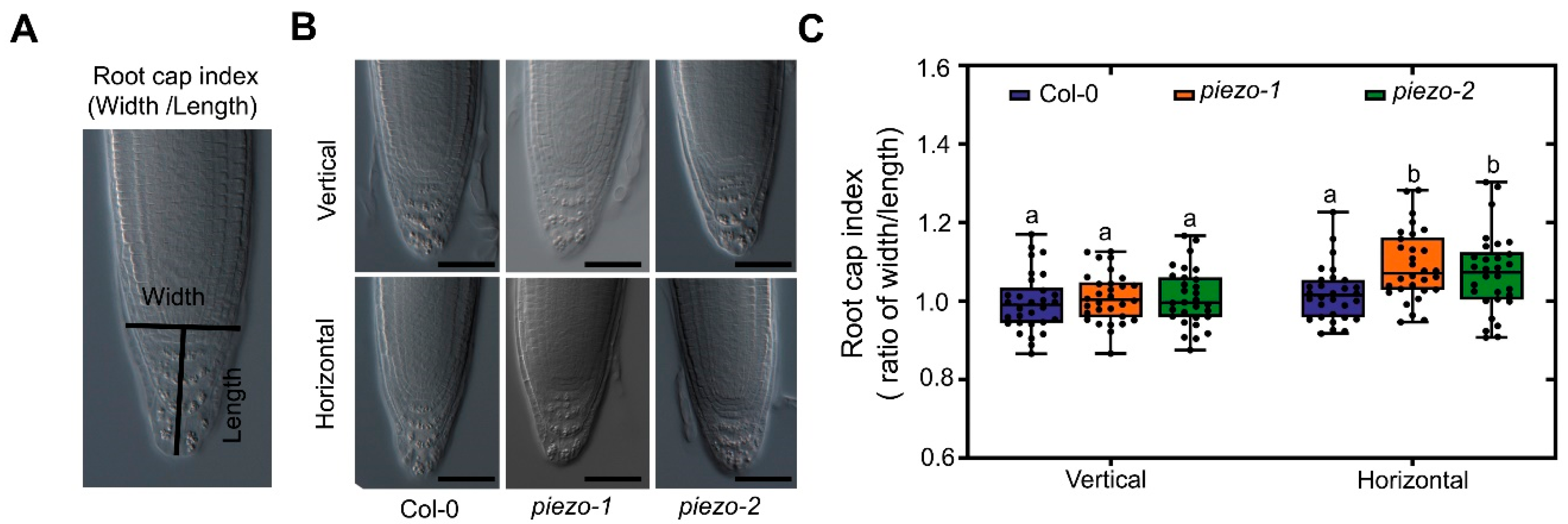
2.5. AtPiezo Affects the Shape of Root Cap in Response to Mechanical Forces
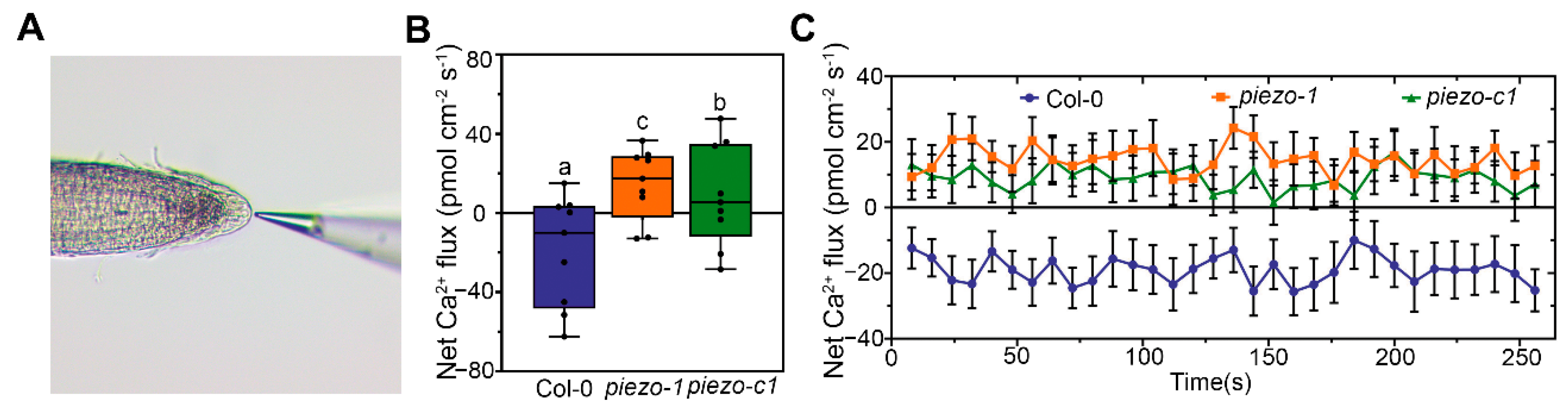
2.6. AtPiezo Change the Ca2+ Flux of Plant Root Cap
3. Discussion
4. Materials and Methods
4.1. Phylogenetic and Sequences Analysis
4.2. Plant Materials and Growth Conditions
4.3. Genotyping Atpiezo Mutants and Cloning AtPiezo
4.4. Plasmid Construction
4.5. Phenotypic Analysis of Root Growth
4.6. GUS Staining and Cross Sectioning
4.7. In Vivo Ca2+ Imaging
4.8. Measurement of the Net Ca2+ Fluxes in Root Cap
4.9. Data Acquisition and Statistics
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Moulia, B.; Coutand, C.; Julien, J.L. Mechanosensitive control of plant growth: Bearing the load, sensing, transducing, and responding. Front. Plant Sci. 2015, 6, 52. [Google Scholar] [CrossRef] [PubMed]
- Hamant, O.; Haswell, E.S. Life behind the wall: Sensing mechanical cues in plants. BMC Biol. 2017, 15, 59. [Google Scholar] [CrossRef] [PubMed]
- Toyota, M.; Gilroy, S. Gravitropism and mechanical signaling in plants. Am. J. Bot. 2013, 100, 111–125. [Google Scholar] [CrossRef] [PubMed]
- Su, S.H.; Gibbs, N.M.; Jancewicz, A.L.; Masson, P.H. Molecular Mechanisms of Root Gravitropism. Curr. Biol. CB 2017, 27, R964–R972. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, M.; Nishimura, T.; Morita, M.T. Gravity sensing and signal conversion in plant gravitropism. J. Exp. Bot. 2019, 70, 3495–3506. [Google Scholar] [CrossRef] [PubMed]
- Chehab, E.W.; Eich, E.; Braam, J. Thigmomorphogenesis: A complex plant response to mechano-stimulation. J. Exp. Bot. 2009, 60, 43–56. [Google Scholar] [CrossRef]
- Braam, J. In touch: Plant responses to mechanical stimuli. New Phytol. 2005, 165, 373–389. [Google Scholar] [CrossRef]
- Wang, K.; Yang, Z.; Qing, D.J.; Ren, F.; Liu, S.C.; Zheng, Q.S.; Liu, J.; Zhang, W.P.; Dai, C.; Wu, M.; et al. Quantitative and functional posttranslational modification proteomics reveals that TREPH1 plays a role in plant touch-delayed bolting. Proc. Natl. Acad. Sci. USA 2018, 115, E10265–E10274. [Google Scholar] [CrossRef]
- Hamilton, E.S.; Schlegel, A.M.; Haswell, E.S. United in diversity: Mechanosensitive ion channels in plants. Annu. Rev. Plant Biol. 2015, 66, 113–137. [Google Scholar] [CrossRef]
- Monshausen, G.B.; Gilroy, S. The exploring root—root growth responses to local environmental conditions. Curr. Opin. Plant Biol. 2009, 12, 766–772. [Google Scholar] [CrossRef]
- Shih, H.W.; Miller, N.D.; Dai, C.; Spalding, E.P.; Monshausen, G.B. The receptor-like kinase FERONIA is required for mechanical signal transduction in arabidopsis seedlings. Curr. Biol. 2014, 24, 1887–1892. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, E.S.; Jensen, G.S.; Maksaev, G.; Katims, A.; Sherp, A.M.; Haswell, E.S. Mechanosensitive channel MSL8 regulates osmotic forces during pollen hydration and germination. Science 2015, 350, 438–441. [Google Scholar] [CrossRef] [PubMed]
- Basu, D.; Haswell, E.S. The mechanosensitive ion channel MSL10 potentiates responses to cell swelling in Arabidopsis seedlings. Curr. Biol. CB 2020, 30, 2716–2728.e6. [Google Scholar] [CrossRef]
- Nakagawa, Y.; Katagiri, T.; Shinozaki, K.; Qi, Z.; Tatsumi, H.; Furuichi, T.; Kishigami, A.; Sokabe, M.; Kojima, I.; Sato, S.; et al. Arabidopsis plasma membrane protein crucial for Ca2+ influx and touch sensing in roots. Proc. Natl. Acad. Sci. USA 2007, 104, 3639–3644. [Google Scholar] [CrossRef] [PubMed]
- Hattori, T.; Otomi, Y.; Nakajima, Y.; Soga, K.; Wakabayashi, K.; Iida, H.; Hoson, T. MCA1 and MCA2 are involved in the response to hypergravity in arabidopsis hypocotyls. Plants 2020, 9, 590. [Google Scholar] [CrossRef]
- Gobert, A.; Isayenkov, S.; Voelker, C.; Czempinski, K.; Maathuis, F.J. The two-pore channel TPK1 gene encodes the vacuolar K+ conductance and plays a role in K+ homeostasis. Proc. Natl. Acad. Sci. USA 2007, 104, 10726–10731. [Google Scholar] [CrossRef]
- Yuan, F.; Yang, H.; Xue, Y.; Kong, D.; Ye, R.; Li, C.; Zhang, J.; Theprungsirikul, L.; Shrift, T.; Krichilsky, B.; et al. OSCA1 mediates osmotic-stress-evoked Ca2+ increases vital for osmosensing in Arabidopsis. Nature 2014, 514, 367–371. [Google Scholar] [CrossRef]
- Murthy, S.E.; Dubin, A.E.; Whitwam, T.; Jojoa-Cruz, S.; Cahalan, S.M.; Mousavi, S.A.R.; Ward, A.B.; Patapoutian, A. OSCA/TMEM63 are an evolutionarily conserved family of mechanically activated ion channels. eLife 2018, 7, e41844. [Google Scholar] [CrossRef]
- Deja-Muylle, A.; Parizot, B.; Motte, H.; Beeckman, T. Exploiting natural variation in root system architecture via genome-wide association studies. J. Exp. Bot. 2020, 71, 2379–2389. [Google Scholar] [CrossRef]
- Bengough, A.G.; McKenzie, B.M.; Hallett, P.D.; Valentine, T.A. Root elongation, water stress, and mechanical impedance: A review of limiting stresses and beneficial root tip traits. J. Exp. Bot. 2011, 62, 59–68. [Google Scholar] [CrossRef]
- Bengough, A.G. Root elongation is restricted by axial but not by radial pressures: So what happens in field soil? Plant Soil 2012, 360, 15–18. [Google Scholar] [CrossRef]
- Potocka, I.; Szymanowska-Pulka, J. Morphological responses of plant roots to mechanical stress. Ann. Bot. 2018, 122, 711–723. [Google Scholar] [CrossRef] [PubMed]
- Kumpf, R.P.; Nowack, M.K. The root cap: A short story of life and death. J. Exp. Bot. 2015, 66, 5651–5662. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N.; Iyer-Pascuzzi, A.S. Shedding the last layer: Mechanisms of root cap cell release. Plants 2020, 9, 308. [Google Scholar] [CrossRef]
- Iijima, M.; Barlow, P.W.; Bengough, A.G. Root cap structure and cell production rates of maize (Zea mays) roots in compacted sand. New Phytol. 2003, 160, 127–134. [Google Scholar] [CrossRef]
- Coste, B.; Mathur, J.; Schmidt, M.; Earley, T.J.; Ranade, S.; Petrus, M.J.; Dubin, A.E.; Patapoutian, A. Piezo1 and Piezo2 are essential components of distinct mechanically activated cation channels. Science 2010, 330, 55–60. [Google Scholar] [CrossRef]
- Kim, S.E.; Coste, B.; Chadha, A.; Cook, B.; Patapoutian, A. The role of Drosophila Piezo in mechanical nociception. Nature 2012, 483, 209–212. [Google Scholar] [CrossRef]
- Li, J.; Hou, B.; Tumova, S.; Muraki, K.; Bruns, A.; Ludlow, M.J.; Sedo, A.; Hyman, A.J.; McKeown, L.; Young, R.S.; et al. Piezo1 integration of vascular architecture with physiological force. Nature 2014, 515, 279–282. [Google Scholar] [CrossRef]
- Pathak, M.M.; Nourse, J.L.; Tran, T.; Hwe, J.; Arulmoli, J.; Le, D.T.; Bernardis, E.; Flanagan, L.A.; Tombola, F. Stretch-activated ion channel Piezo1 directs lineage choice in human neural stem cells. Proc. Natl. Acad. Sci. USA 2014, 111, 16148–16153. [Google Scholar] [CrossRef]
- Ranade, S.S.; Woo, S.H.; Dubin, A.E.; Moshourab, R.A.; Wetzel, C.; Petrus, M.; Mathur, J.; Begay, V.; Coste, B.; Mainquist, J.; et al. Piezo2 is the major transducer of mechanical forces for touch sensation in mice. Nature 2014, 516, 121–330. [Google Scholar] [CrossRef]
- Woo, S.H.; Lukacs, V.; de Nooij, J.C.; Zaytseva, D.; Criddle, C.R.; Francisco, A.; Jessell, T.M.; Wilkinson, K.A.; Patapoutian, A. Piezo2 is the principal mechanotransduction channel for proprioception. Nat. Neurosci. 2015, 18, 1756–1762. [Google Scholar] [CrossRef] [PubMed]
- Gudipaty, S.A.; Lindblom, J.; Loftus, P.D.; Redd, M.J.; Edes, K.; Davey, C.F.; Krishnegowda, V.; Rosenblatt, J. Mechanical stretch triggers rapid epithelial cell division through Piezo1. Nature 2017, 543, 118–121. [Google Scholar] [CrossRef] [PubMed]
- Ge, J.; Li, W.; Zhao, Q.; Li, N.; Chen, M.; Zhi, P.; Li, R.; Gao, N.; Xiao, B.; Yang, M. Architecture of the mammalian mechanosensitive Piezo1 channel. Nature 2015, 527, 64–69. [Google Scholar] [CrossRef] [PubMed]
- Beech, D.J.; Xiao, B. Piezo channel mechanisms in health and disease. J. Physiol. 2018, 596, 965–967. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Tong, X.; Liu, S.Y.; Chai, L.X.; Zhu, F.F.; Zhang, X.P.; Zou, J.Z.; Wang, X.B. Genetic analysis of a Piezo-like protein suppressing systemic movement of plant viruses in Arabidopsis thaliana. Sci. Rep. 2019, 9, 3187. [Google Scholar] [CrossRef] [PubMed]
- Saotome, K.; Murthy, S.E.; Kefauver, J.M.; Whitwam, T.; Patapoutian, A.; Ward, A.B. Structure of the mechanically activated ion channel Piezo1. Nature 2018, 554, 481–486. [Google Scholar] [CrossRef]
- Yan, J.; Wang, B.; Zhou, Y.; Hao, S. Resistance from agar medium impacts the helical growth of Arabidopsis primary roots. J. Mech. Behav. Biomed. Mater. 2018, 85, 43–50. [Google Scholar] [CrossRef]
- Moreno-Risueno, M.A.; Van Norman, J.M.; Moreno, A.; Zhang, J.; Ahnert, S.E.; Benfey, P.N. Oscillating gene expression determines competence for periodic Arabidopsis root branching. Science 2010, 329, 1306–1311. [Google Scholar] [CrossRef]
- Wu, J.; Lewis, A.H.; Grandl, J. Touch, Tension, and transduction—The function and regulation of piezo ion channels. Trends Biochem. Sci. 2017, 42, 57–71. [Google Scholar] [CrossRef]
- Frachisse, J.M.; Thomine, S.; Allain, J.M. Calcium and plasma membrane force-gated ion channels behind development. Curr. Opin. Plant Biol. 2020, 53, 57–64. [Google Scholar] [CrossRef]
- Kurusu, T.; Kuchitsu, K.; Nakano, M.; Nakayama, Y.; Iida, H. Plant mechanosensing and Ca2+ transport. Trends Plant Sci. 2013, 18, 227–233. [Google Scholar] [CrossRef]
- Johnson, M.; Zaretskaya, I.; Raytselis, Y.; Merezhuk, Y.; McGinnis, S.; Madden, T.L. NCBI BLAST: A better web interface. Nucleic Acids Res. 2008, 36, W5–W9. [Google Scholar] [CrossRef]
- UniProt, C. UniProt: A hub for protein information. Nucleic Acids Res. 2015, 43, D204–D212. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Moller, S.; Croning, M.D.R.; Apweiler, R. Evaluation of methods for the prediction of membrane spanning regions. Bioinformatics 2001, 17, 646–653. [Google Scholar] [CrossRef]
- Kall, L.; Krogh, A.; Sonnhammer, E.L.L. Advantages of combined transmembrane topology and signal peptide prediction-the Phobius web server. Nucleic Acids Res. 2007, 35, W429–W432. [Google Scholar] [CrossRef]
- Schultz, J.; Copley, R.R.; Doerks, T.; Ponting, C.P.; Bork, P. SMART: A web-based tool for the study of genetically mobile domains. Nucleic Acids Res. 2000, 28, 231–234. [Google Scholar] [CrossRef]
- Omasits, U.; Ahrens, C.H.; Muller, S.; Wollscheid, B. Protter: Interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics 2014, 30, 884–886. [Google Scholar] [CrossRef]
- Herrero, J.; Muffato, M.; Beal, K.; Fitzgerald, S.; Gordon, L.; Pignatelli, M.; Vilella, A.J.; Searle, S.M.; Amode, R.; Brent, S.; et al. Ensembl comparative genomics resources. Database: The journal of biological databases and curation 2016. Database 2016, 2016, bav096. [Google Scholar] [CrossRef]
- Wang, Z.P.; Xing, H.L.; Dong, L.; Zhang, H.Y.; Han, C.Y.; Wang, X.C.; Chen, Q.J. Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome Biol. 2015, 16, 144. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Zhang, C.; Gong, F.C.; Lambert, G.M.; Galbraith, D.W. Cell type-specific characterization of nuclear DNA contents within complex tissues and organs. Plant Methods 2005, 1, 7. [Google Scholar] [CrossRef]
- Shao, Q.; Gao, Q.; Lhamo, D.; Zhang, H.; Luan, S. Two glutamate- and pH-regulated Ca2+ channels are required for systemic wound signaling in Arabidopsis. Sci. Signal. 2020, 13. [Google Scholar] [CrossRef]
- Sun, J.; Chen, S.; Dai, S.; Wang, R.; Li, N.; Shen, X.; Zhou, X.; Lu, C.; Zheng, X.; Hu, Z.; et al. NaCl-induced alternations of cellular and tissue ion fluxes in roots of salt-resistant and salt-sensitive poplar species. Plant Physiol. 2009, 149, 1141–1153. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fang, X.; Liu, B.; Shao, Q.; Huang, X.; Li, J.; Luan, S.; He, K. AtPiezo Plays an Important Role in Root Cap Mechanotransduction. Int. J. Mol. Sci. 2021, 22, 467. https://doi.org/10.3390/ijms22010467
Fang X, Liu B, Shao Q, Huang X, Li J, Luan S, He K. AtPiezo Plays an Important Role in Root Cap Mechanotransduction. International Journal of Molecular Sciences. 2021; 22(1):467. https://doi.org/10.3390/ijms22010467
Chicago/Turabian StyleFang, Xianming, Beibei Liu, Qianshuo Shao, Xuemei Huang, Jia Li, Sheng Luan, and Kai He. 2021. "AtPiezo Plays an Important Role in Root Cap Mechanotransduction" International Journal of Molecular Sciences 22, no. 1: 467. https://doi.org/10.3390/ijms22010467
APA StyleFang, X., Liu, B., Shao, Q., Huang, X., Li, J., Luan, S., & He, K. (2021). AtPiezo Plays an Important Role in Root Cap Mechanotransduction. International Journal of Molecular Sciences, 22(1), 467. https://doi.org/10.3390/ijms22010467







