Human DDX3X Unwinds Japanese Encephalitis and Zika Viral 5′ Terminal Regions
Abstract
1. Introduction
2. Results
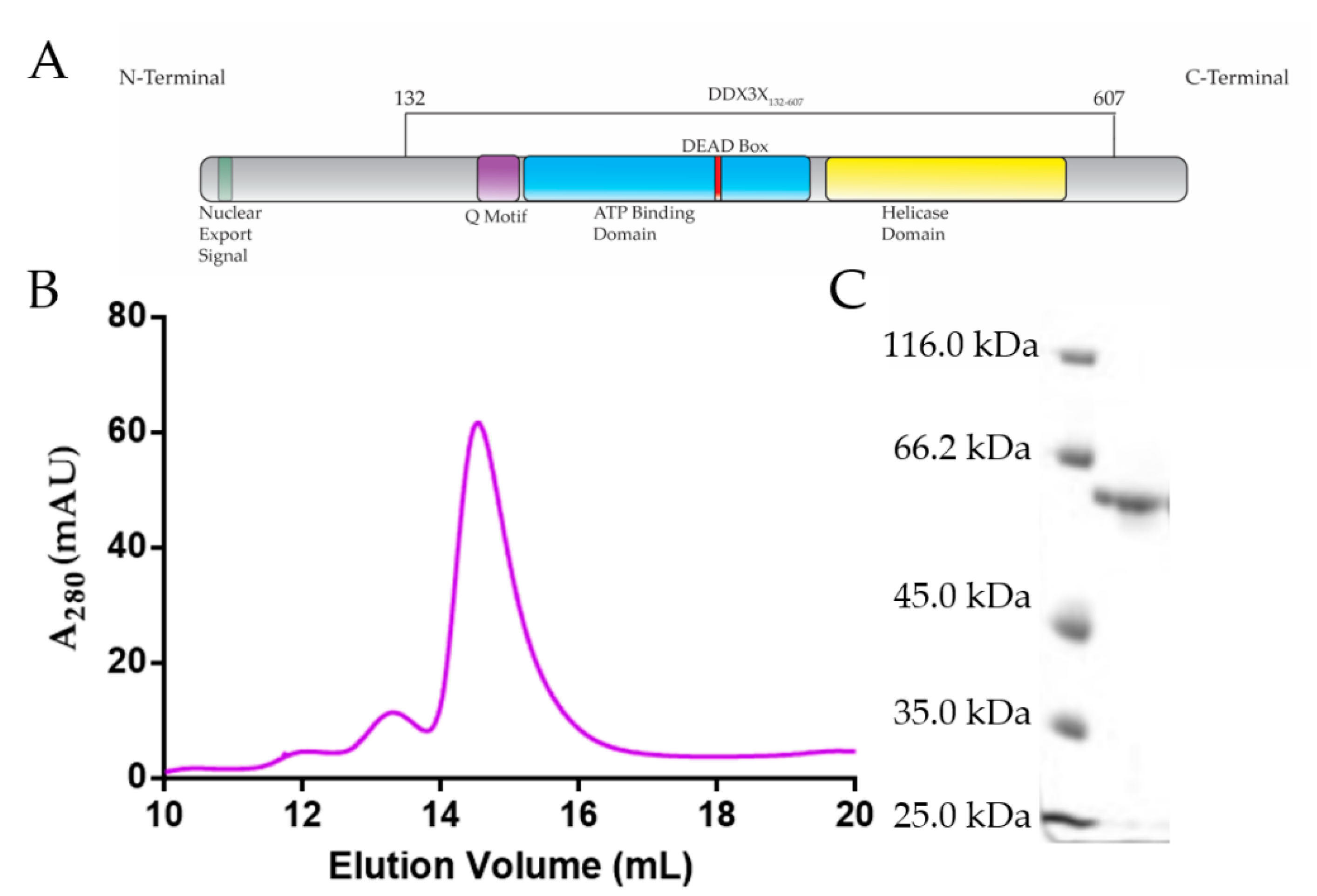
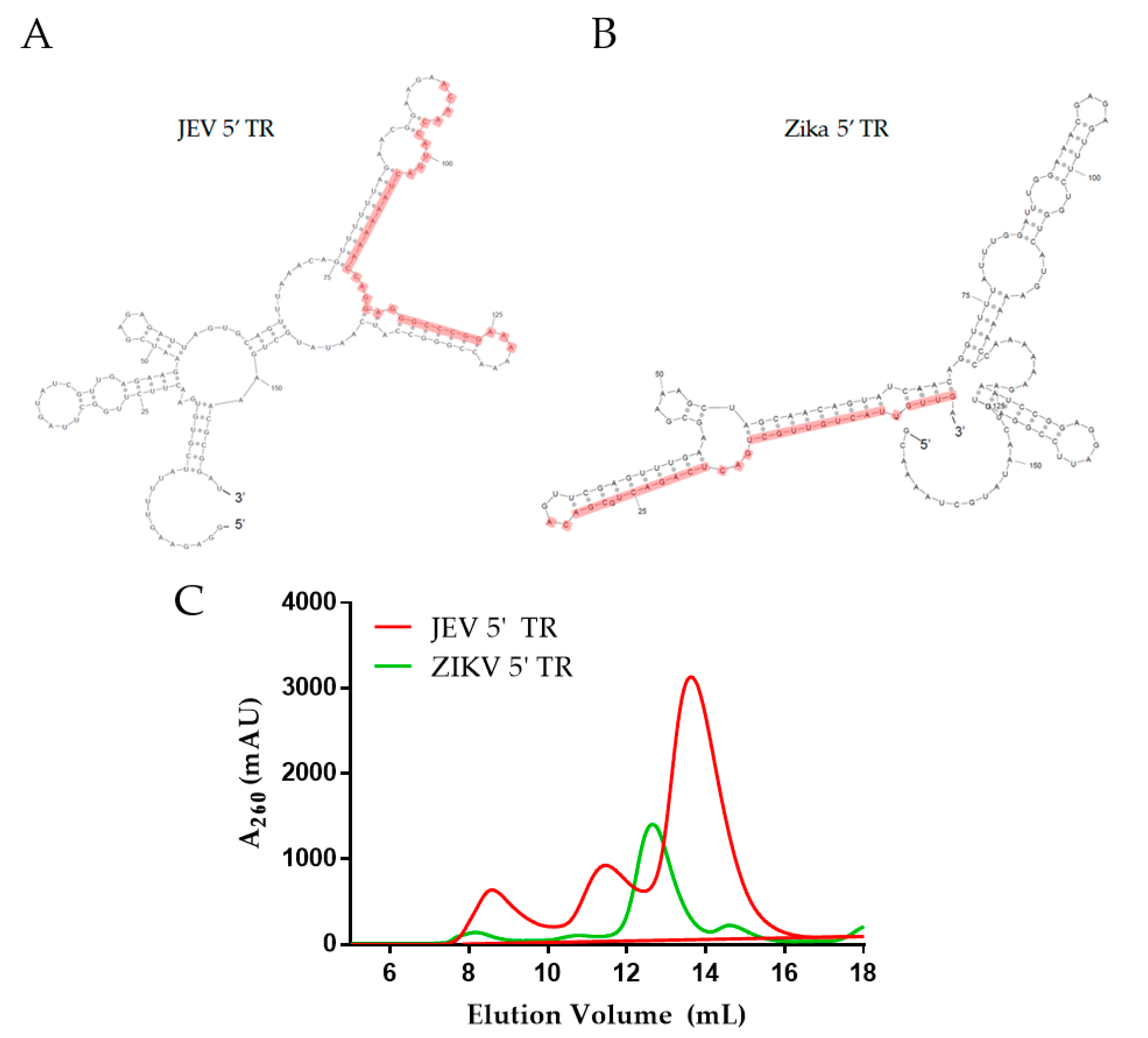
2.1. Purification of DDX3X132–607, JEV 5′ and Zika 5′ TR RNAs
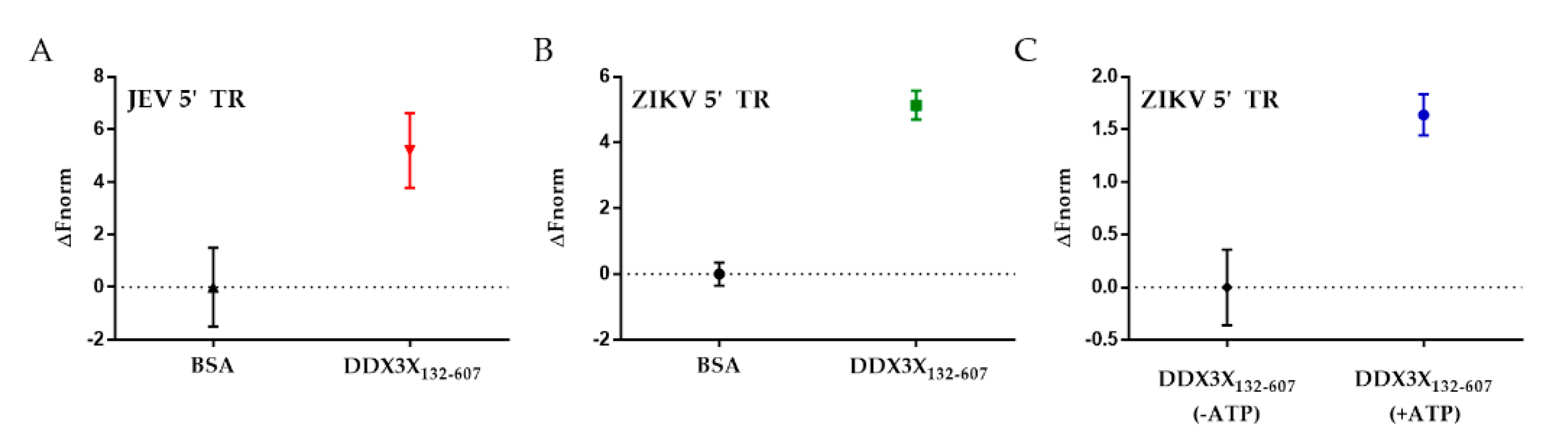
2.2. DDX3X132–607 Binds to 5′ TRs of JEV and ZIKV
2.3. DDX3X132–607 Unwinds 5′ TRs of JEV and ZIKV
3. Discussion
4. Materials and Methods
4.1. Overexpression and Purification of DDX3X132–607
4.2. Preparation of Non-Coding RNAs
- JEV 5′ TR 1–156 (51.5 kDa, 159 nts)
- 2.
- Zika 5′ TR 1–163 (52.3 kDa, 163 nts)
4.3. Fluorescent Labeling of Flaviviral RNA TRs
4.4. RNA–Protein Interaction Studies Using Microscale Thermophoresis
4.5. Helicase Assay
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| MST | microscale thermophoresis |
| SEC | size-exclusion chromatography |
| PAGE | polyacrylamide gel electrophoresis |
| JEV | Japanese encephalitis virus |
| ZIKV | Zika virus |
| TR | terminal regions |
References
- Chambers, T.J.; Monath, T.P. The Flaviviruses: Detection, Diagnosis and Vaccine Development; Elsevier: Amsterdam, The Netherlands, 2003. [Google Scholar]
- Solomon, T.; Kneen, R.; Dung, N.M.; Khanh, V.C.; Thuy, T.T.N.; Day, N.P.; Nisalak, A.; Vaughn, D.W.; White, N.J. Poliomyelitis-like illness due to Japanese encephalitis virus. Lancet 1998, 351, 1094–1097. [Google Scholar] [CrossRef]
- Brasil, P.; Sequeira, P.C.; Freitas, A.D.A.; Zogbi, H.E.; Calvet, G.A.; de Souza, R.V.; Siqueira, A.M.; de Mendonca, M.C.L.; Nogueira, R.M.R.; de Filippis, A.M.B. Guillain-Barré syndrome associated with Zika virus infection. Lancet 2016, 387, 1482. [Google Scholar] [CrossRef]
- Pierson, T.C.; Diamond, M.S. The continued threat of emerging flaviviruses. Nat. Microbiol. 2020, 5, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Fischer, C.; de Oliveira-Filho, E.F.; Drexler, J.F. Viral emergence and immune interplay in flavivirus vaccines. Lancet Infect. Dis. 2020, 20, 15–17. [Google Scholar] [CrossRef]
- Fernandez-Sanles, A.; Rios-Marco, P.; Romero-Lopez, C.; Berzal-Herranz, A. Functional Information Stored in the Conserved Structural RNA Domains of Flavivirus Genomes. Front. Microbiol. 2017, 8, 546. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Epidemic Focus—Flavivirus Epidemics; World Health Organization: Geneva, Switzerland, 2020. [Google Scholar]
- Huang, Y.-J.S.; Higgs, S.; Horne, K.M.; Vanlandingham, D.L. Flavivirus-mosquito interactions. Viruses 2014, 6, 4703–4730. [Google Scholar] [CrossRef] [PubMed]
- Benedict, M.Q.; Levine, R.S.; Hawley, W.A.; Lounibos, L.P. Spread of the tiger: Global risk of invasion by the mosquito Aedes albopictus. Vector Borne Zoonotic Dis. 2007, 7, 76–85. [Google Scholar] [CrossRef] [PubMed]
- Reinhold, J.M.; Lazzari, C.R.; Lahondère, C. Effects of the environmental temperature on Aedes aegypti and Aedes albopictus mosquitoes: A review. Insects 2018, 9, 158. [Google Scholar] [CrossRef] [PubMed]
- Baig, S.; Fox, K.K.; Jee, Y.; O’Connor, P.; Hombach, J.; Wang, S.A.; Hyde, T.; Fischer, M.; Hills, S.L. Japanese encephalitis surveillance and immunization—Asia and the Western Pacific, 2012. MMWR Morb. Mortal. Wkly. Rep. 2013, 62, 658. [Google Scholar]
- Campbell, G.L.; Hills, S.L.; Fischer, M.; Jacobson, J.A.; Hoke, C.H.; Hombach, J.M.; Marfin, A.A.; Solomon, T.; Tsai, T.F.; Tsu, V.D. Estimated global incidence of Japanese encephalitis: A systematic review. Bull. World Health Organ. 2011, 89, 766–774. [Google Scholar] [CrossRef]
- Dubischar, K.L.; Kadlecek, V.; Sablan, B.; Borja-Tabora, C.F.; Gatchalian, S.; Eder-Lingelbach, S.; Kiermayr, S.; Spruth, M.; Westritschnig, K. Immunogenicity of the inactivated Japanese encephalitis virus vaccine IXIARO in children from a Japanese encephalitis virus-endemic region. Pediatric Infect. Dis. J. 2017, 36, 898–904. [Google Scholar] [CrossRef] [PubMed]
- Van Gessel, Y.; Klade, C.S.; Putnak, R.; Formica, A.; Krasaesub, S.; Spruth, M.; Cena, B.; Tungtaeng, A.; Gettayacamin, M.; Dewasthaly, S. Correlation of protection against Japanese encephalitis virus and JE vaccine (IXIARO®) induced neutralizing antibody titers. Vaccine 2011, 29, 5925–5931. [Google Scholar] [CrossRef] [PubMed]
- Samarasekera, U.; Triunfol, M. Concern over Zika virus grips the world. Lancet 2016, 387, 521–524. [Google Scholar] [CrossRef]
- World Health Organization. Zika Situation Report; World Health Organization: Geneva, Switzerland, 2016. [Google Scholar]
- Carteaux, G.; Maquart, M.; Bedet, A.; Contou, D.; Brugières, P.; Fourati, S.; Cleret de Langavant, L.; de Broucker, T.; Brun-Buisson, C.; Leparc-Goffart, I. Zika virus associated with meningoencephalitis. N. Engl. J. Med. 2016, 374, 1595–1596. [Google Scholar] [CrossRef] [PubMed]
- Cauchemez, S.; Besnard, M.; Bompard, P.; Dub, T.; Guillemette-Artur, P.; Eyrolle-Guignot, D.; Salje, H.; Van Kerkhove, M.D.; Abadie, V.; Garel, C. Association between Zika virus and microcephaly in French Polynesia, 2013–2015: A retrospective study. Lancet 2016, 387, 2125–2132. [Google Scholar] [CrossRef]
- Gladwyn-Ng, I.; Cordón-Barris, L.; Alfano, C.; Creppe, C.; Couderc, T.; Morelli, G.; Thelen, N.; America, M.; Bessières, B.; Encha-Razavi, F. Stress-induced unfolded protein response contributes to Zika virus–associated microcephaly. Nat. Neurosci. 2018, 21, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Mécharles, S.; Herrmann, C.; Poullain, P.; Tran, T.-H.; Deschamps, N.; Mathon, G.; Landais, A.; Breurec, S.; Lannuzel, A. Acute myelitis due to Zika virus infection. Lancet 2016, 387, 1481. [Google Scholar] [CrossRef]
- Nascimento, O.J.; da Silva, I.R. Guillain–Barré syndrome and Zika virus outbreaks. Curr. Opin. Neurol. 2017, 30, 500–507. [Google Scholar] [CrossRef]
- Oliveira Melo, A.; Malinger, G.; Ximenes, R.; Szejnfeld, P.; Alves Sampaio, S.; Bispo de Filippis, A. Zika virus intrauterine infection causes fetal brain abnormality and microcephaly: Tip of the iceberg? Ultrasound Obstet. Gynecol. 2016, 47, 6–7. [Google Scholar] [CrossRef]
- Schwartzmann, P.V.; Ramalho, L.N.; Neder, L.; Vilar, F.C.; Ayub-Ferreira, S.M.; Romeiro, M.F.; Takayanagui, O.M.; Dos Santos, A.C.; Schmidt, A.; Figueiredo, L.T. Zika Virus Meningoencephalitis in an Immunocompromised Patient. Mayo Clin. Proc. 2017, 92, 460–466. [Google Scholar] [CrossRef]
- Devhare, P.; Meyer, K.; Steele, R.; Ray, R.B.; Ray, R. Zika virus infection dysregulates human neural stem cell growth and inhibits differentiation into neuroprogenitor cells. Cell Death Dis. 2017, 8, e3106. [Google Scholar] [CrossRef] [PubMed]
- Broutet, N.; Krauer, F.; Riesen, M.; Khalakdina, A.; Almiron, M.; Aldighieri, S.; Espinal, M.; Low, N.; Dye, C. Zika virus as a cause of neurologic disorders. N. Engl. J. Med. 2016, 374, 1506–1509. [Google Scholar] [CrossRef] [PubMed]
- Mlakar, J.; Korva, M.; Tul, N.; Popović, M.; Poljšak-Prijatelj, M.; Mraz, J.; Kolenc, M.; Resman Rus, K.; Vesnaver Vipotnik, T.; Fabjan Vodušek, V. Zika virus associated with microcephaly. N. Engl. J. Med. 2016, 374, 951–958. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Hammack, C.; Ogden, S.C.; Wen, Z.; Qian, X.; Li, Y.; Yao, B.; Shin, J.; Zhang, F.; Lee, E.M. Zika virus infects human cortical neural progenitors and attenuates their growth. Cell Stem Cell 2016, 18, 587–590. [Google Scholar] [CrossRef]
- Li, C.; Xu, D.; Ye, Q.; Hong, S.; Jiang, Y.; Liu, X.; Zhang, N.; Shi, L.; Qin, C.-F.; Xu, Z. Zika virus disrupts neural progenitor development and leads to microcephaly in mice. Cell Stem Cell 2016, 19, 120–126. [Google Scholar] [CrossRef]
- Li, H.; Saucedo-Cuevas, L.; Shresta, S.; Gleeson, J.G. The neurobiology of Zika virus. Neuron 2016, 92, 949–958. [Google Scholar] [CrossRef]
- Russo, F.B.; Jungmann, P.; Beltrão-Braga, P.C.B. Zika infection and the development of neurological defects. Cellular Microbial. 2017, 19, e12744. [Google Scholar] [CrossRef]
- Barzon, L.; Pacenti, M.; Franchin, E.; Lavezzo, E.; Trevisan, M.; Sgarabotto, D.; Palu, G. Infection dynamics in a traveller with persistent shedding of Zika virus RNA in semen for six months after returning from Haiti to Italy, January 2016. Eurosurveillance 2016, 21. [Google Scholar] [CrossRef]
- Foy, B.D.; Kobylinski, K.C.; Chilson Foy, J.L.; Blitvich, B.J.; Travassos da Rosa, A.; Haddow, A.D.; Lanciotti, R.S.; Tesh, R.B. Probable non-vector-borne transmission of Zika virus, Colorado, USA. Emerg. Infect. Dis. 2011, 17, 880–882. [Google Scholar] [CrossRef]
- Paul, L.M.; Carlin, E.R.; Jenkins, M.M.; Tan, A.L.; Barcellona, C.M.; Nicholson, C.O.; Michael, S.F.; Isern, S. Dengue virus antibodies enhance Zika virus infection. Clin. Transl. Immunol. 2016, 5, e117. [Google Scholar] [CrossRef]
- Myers, T.R.; McCarthy, N.L.; Panagiotakopoulos, L.; Omer, S.B. Estimation of the Incidence of Guillain-Barre Syndrome During Pregnancy in the United States. Open Forum Infect. Dis. 2019, 6, ofz071. [Google Scholar] [CrossRef] [PubMed]
- Stettler, K.; Beltramello, M.; Espinosa, D.A.; Graham, V.; Cassotta, A.; Bianchi, S.; Vanzetta, F.; Minola, A.; Jaconi, S.; Mele, F.; et al. Specificity, cross-reactivity, and function of antibodies elicited by Zika virus infection. Science 2016, 353, 823–826. [Google Scholar] [CrossRef] [PubMed]
- Cleaves, G.R.; Dubin, D.T. Methylation status of intracellular dengue type 2 40 S RNA. Virology 1979, 96, 159–165. [Google Scholar] [CrossRef]
- Brinton, M.A.; Fernandez, A.V.; Dispoto, J.H. The 3′-nucleotides of flavivirus genomic RNA form a conserved secondary structure. Virology 1986, 153, 113–121. [Google Scholar] [CrossRef]
- Wengler, G.; Wengler, G.; Gross, H.J. Studies on Virus-Specific Nucleic-Acids Synthesized in Vertebrate and Mosquito Cells Infected with Flaviviruses. Virology 1978, 89, 423–437. [Google Scholar] [CrossRef]
- Gritsun, T.S.; Gould, E.A. Origin and evolution of 3′UTR of flaviviruses: Long direct repeats as a basis for the formation of secondary structures and their significance for virus transmission. Adv. Virus Res. 2007, 69, 203–248. [Google Scholar]
- Thurner, C.; Witwer, C.; Hofacker, I.L.; Stadler, P.F. Conserved RNA secondary structures in Flaviviridae genomes. J. Gen. Virol. 2004, 85 Pt 5, 1113–1124. [Google Scholar] [CrossRef]
- Hahn, C.S.; Hahn, Y.S.; Rice, C.M.; Lee, E.; Dalgarno, L.; Strauss, E.G.; Strauss, J.H. Conserved elements in the 3′ untranslated region of flavivirus RNAs and potential cyclization sequences. J. Mol. Biol. 1987, 198, 33–41. [Google Scholar] [CrossRef]
- Alvarez, D.E.; Lodeiro, M.F.; Luduena, S.J.; Pietrasanta, L.I.; Gamarnik, A.V. Long-range RNA-RNA interactions circularize the dengue virus genome. J. Virol. 2005, 79, 6631–6643. [Google Scholar] [CrossRef]
- Li, X.D.; Deng, C.L.; Yuan, Z.M.; Ye, H.Q.; Zhang, B. Different Degrees of 5′-to-3′ DAR Interactions Modulate Zika Virus Genome Cyclization and Host-Specific Replication. J. Virol. 2020, 94. [Google Scholar] [CrossRef]
- De Borba, L.; Villordo, S.M.; Iglesias, N.G.; Filomatori, C.V.; Gebhard, L.G.; Gamarnik, A.V. Overlapping local and long-range RNA-RNA interactions modulate dengue virus genome cyclization and replication. J. Virol. 2015, 89, 3430–3437. [Google Scholar] [CrossRef] [PubMed]
- Khromykh, A.A.; Meka, H.; Guyatt, K.J.; Westaway, E.G. Essential role of cyclization sequences in flavivirus RNA replication. J. Virol. 2001, 75, 6719–6728. [Google Scholar] [CrossRef] [PubMed]
- Friebe, P.; Shi, P.Y.; Harris, E. The 5′ and 3′ downstream AUG region elements are required for mosquito-borne flavivirus RNA replication. J. Virol. 2011, 85, 1900–1905. [Google Scholar] [CrossRef] [PubMed]
- Chambers, T.J.; Hahn, C.S.; Galler, R.; Rice, C.M. Flavivirus genome organization, expression, and replication. Annu. Rev. Microbiol. 1990, 44, 649–688. [Google Scholar] [CrossRef]
- Tingting, P.; Caiyun, F.; Zhigang, Y.; Pengyuan, Y.; Zhenghong, Y. Subproteomic analysis of the cellular proteins associated with the 3′ untranslated region of the hepatitis C virus genome in human liver cells. Biochem. Biophys. Res. Commun. 2006, 347, 683–691. [Google Scholar] [CrossRef]
- Harris, D.; Zhang, Z.; Chaubey, B.; Pandey, V.N. Identification of cellular factors associated with the 3′-nontranslated region of the hepatitis C virus genome. Mol. Cell. Proteom. 2006, 5, 1006–1018. [Google Scholar] [CrossRef]
- Li, C.; Ge, L.L.; Li, P.P.; Wang, Y.; Dai, J.J.; Sun, M.X.; Huang, L.; Shen, Z.Q.; Hu, X.C.; Ishag, H.; et al. Cellular DDX3 regulates Japanese encephalitis virus replication by interacting with viral un-translated regions. Virology 2014, 449, 70–81. [Google Scholar] [CrossRef]
- Li, C.; Ge, L.L.; Li, P.P.; Wang, Y.; Sun, M.X.; Huang, L.; Ishag, H.; Di, D.D.; Shen, Z.Q.; Fan, W.X.; et al. The DEAD-box RNA helicase DDX5 acts as a positive regulator of Japanese encephalitis virus replication by binding to viral 3′ UTR. Antivir. Res. 2013, 100, 487–499. [Google Scholar] [CrossRef]
- Ward, A.M.; Bidet, K.; Yinglin, A.; Ler, S.G.; Hogue, K.; Blackstock, W.; Gunaratne, J.; Garcia-Blanco, M.A. Quantitative mass spectrometry of DENV-2 RNA-interacting proteins reveals that the DEAD-box RNA helicase DDX6 binds the DB1 and DB2 3′ UTR structures. RNA Biol. 2011, 8, 1173–1186. [Google Scholar] [CrossRef]
- Dong, Y.; Yang, J.; Ye, W.; Wang, Y.; Miao, Y.; Ding, T.; Xiang, C.; Lei, Y.; Xu, Z. LSm1 binds to the Dengue virus RNA 3′ UTR and is a positive regulator of Dengue virus replication. Int. J. Mol. Med. 2015, 35, 1683–1689. [Google Scholar]
- Gomila, R.C.; Martin, G.W.; Gehrke, L. NF90 binds the dengue virus RNA 3′ terminus and is a positive regulator of dengue virus replication. PLoS ONE 2011, 6, e16687. [Google Scholar] [CrossRef] [PubMed]
- Ward, A.M.; Calvert, M.E.; Read, L.R.; Kang, S.; Levitt, B.E.; Dimopoulos, G.; Bradrick, S.S.; Gunaratne, J.; Garcia-Blanco, M.A. The Golgi associated ERI3 is a Flavivirus host factor. Sci. Rep. 2016, 6, 34379. [Google Scholar] [CrossRef] [PubMed]
- Lei, Y.; Huang, Y.; Zhang, H.; Yu, L.; Zhang, M.; Dayton, A. Functional interaction between cellular p100 and the dengue virus 3′ UTR. J. Gen. Virol. 2011, 92 Pt 4, 796–806. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Nomaguchi, M.; Padmanabhan, R.; Markoff, L. Specific requirements for elements of the 5′ and 3′ terminal regions in flavivirus RNA synthesis and viral replication. Virology 2008, 374, 170–185. [Google Scholar] [CrossRef]
- Meier-Stephenson, V.; Mrozowich, T.; Pham, M.; Patel, T.R. DEAD-box helicases: The Yin and Yang roles in viral infections. Biotechnol. Genet. Eng. Rev. 2018, 34, 3–32. [Google Scholar] [CrossRef]
- Ng, W.C.; Soto-Acosta, R.; Bradrick, S.S.; Garcia-Blanco, M.A.; Ooi, E.E. The 5′ and 3′ Untranslated Regions of the Flaviviral Genome. Viruses 2017, 9, 137. [Google Scholar] [CrossRef]
- Leitao, A.L.; Costa, M.C.; Enguita, F.J. Unzippers, Resolvers and Sensors: A Structural and Functional Biochemistry Tale of RNA Helicases. Int. J. Mol. Sci. 2015, 16, 2269–2293. [Google Scholar] [CrossRef]
- Linder, P.; Jankowsky, E. From unwinding to clamping—The DEAD box RNA helicase family. Nat. Rev. Mol. Cell Biol. 2011, 12, 505–516. [Google Scholar] [CrossRef]
- Pyle, A.M. Translocation and unwinding mechanisms of RNA and DNA helicases. Annu. Rev. Biophys. 2008, 37, 317–336. [Google Scholar] [CrossRef]
- Jankowsky, E. RNA helicases at work: Binding and rearranging. Trends Biochem. Sci. 2011, 36, 19–29. [Google Scholar] [CrossRef]
- Chao, C.H.; Chen, C.M.; Cheng, P.L.; Shih, J.W.; Tsou, A.P.; Lee, Y.H. DDX3, a DEAD box RNA helicase with tumor growth-suppressive property and transcriptional regulation activity of the p21waf1/cip1 promoter, is a candidate tumor suppressor. Cancer Res. 2006, 66, 6579–6588. [Google Scholar] [CrossRef] [PubMed]
- Ariumi, Y.; Kuroki, M.; Abe, K.; Dansako, H.; Ikeda, M.; Wakita, T.; Kato, N. DDX3 DEAD-box RNA helicase is required for hepatitis C virus RNA replication. J. Virol. 2007, 81, 13922–13926. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Ji, X. The mechanism of RNA duplex recognition and unwinding by DEAD-box helicase DDX3X. Nat. Commun. 2019, 10, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Feng, T.; Pan, W.; Shi, X.; Dai, J. DEAD-box RNA helicase DDX3X inhibits DENV replication via regulating type one interferon pathway. Biochem. Biophys. Res. Commun. 2015, 456, 327–332. [Google Scholar] [CrossRef]
- Brai, A.; Martelli, F.; Riva, V.; Garbelli, A.; Fazi, R.; Zamperini, C.; Pollutri, A.; Falsitta, L.; Ronzini, S.; Maccari, L. DDX3X helicase inhibitors as a new strategy to fight the West Nile virus infection. J. Med. Chem. 2019, 62, 2333–2347. [Google Scholar] [CrossRef]
- Brinton, M.A. Replication cycle and molecular biology of the West Nile virus. Viruses 2013, 6, 13–53. [Google Scholar] [CrossRef]
- Brinton, M.A.; Basu, M. Functions of the 3′ and 5′ genome RNA regions of members of the genus Flavivirus. Virus Res. 2015, 206, 108–119. [Google Scholar] [CrossRef]
- Reuten, R.; Patel, T.R.; McDougall, M.; Rama, N.; Nikodemus, D.; Gibert, B.; Delcros, J.-G.; Prein, C.; Meier, M.; Metzger, S. Structural decoding of netrin-4 reveals a regulatory function towards mature basement membranes. Nat. Commun. 2016, 7, 1–17. [Google Scholar] [CrossRef]
- Ferens, F.G.; Patel, T.R.; Oriss, G.; Court, D.A.; Stetefeld, J. A Cholesterol Analog Induces an Oligomeric Reorganization of VDAC. Biophys. J. 2019, 116, 847–859. [Google Scholar] [CrossRef]
- Wienken, C.J.; Baaske, P.; Rothbauer, U.; Braun, D.; Duhr, S. Protein-binding assays in biological liquids using microscale thermophoresis. Nat. Commun. 2010, 1, 100. [Google Scholar] [CrossRef]
- Garbelli, A.; Beermann, S.; Di Cicco, G.; Dietrich, U.; Maga, G. A motif unique to the human DEAD-box protein DDX3 is important for nucleic acid binding, ATP hydrolysis, RNA/DNA unwinding and HIV-1 replication. PLoS ONE 2011, 6, e19810. [Google Scholar] [CrossRef] [PubMed]
- Nelson, C.; Mrozowich, T.; Park, S.; D’souza, S.; Henrickson, A.; Vigar, J.; Wieden, H.-J.; Owens, R.; Demeler, B.; Patel, T.R. Human DDX17 unwinds Rift Valley Fever Virus Non-coding RNAs. Int. J. Mol. Sci. 2020, 22, 54. [Google Scholar] [CrossRef] [PubMed]
- Jerabek-Willemsen, M.; Wienken, C.J.; Braun, D.; Baaske, P.; Duhr, S. Molecular interaction studies using microscale thermophoresis. Assay Drug Dev. Technol. 2011, 9, 342–353. [Google Scholar] [CrossRef] [PubMed]
- Mrozowich, T.; MeierStephenson, V.; Patel, T.R. Microscale thermophoresis: Warming up to a new biomolecular interaction technique. Biochemist 2019, 41, 8–12. [Google Scholar] [CrossRef]
- Liu, F.; Putnam, A.; Jankowsky, E. ATP hydrolysis is required for DEAD-box protein recycling but not for duplex unwinding. Proc. Natl. Acad. Sci. USA 2008, 105, 20209–20214. [Google Scholar] [CrossRef]
- Xing, Z.; Wang, S.; Tran, E.J. Characterization of the mammalian DEAD-box protein DDX5 reveals functional conservation with S. cerevisiae ortholog Dbp2 in transcriptional control and glucose metabolism. RNA 2017, 23, 1125–1138. [Google Scholar] [CrossRef]
- Wu, G.; Xing, Z.; Tran, E.J.; Yang, D. DDX5 helicase resolves G-quadruplex and is involved in MYC gene transcriptional activation. Proc. Natl. Acad. Sci. USA 2019, 116, 20453–20461. [Google Scholar] [CrossRef]
- Floor, S.N.; Condon, K.J.; Sharma, D.; Jankowsky, E.; Doudna, J.A. Autoinhibitory interdomain interactions and subfamily-specific extensions redefine the catalytic core of the human DEAD-box protein DDX3. J. Biol. Chem. 2016, 291, 2412–2421. [Google Scholar] [CrossRef]
- Ding, Y.; Chan, C.Y.; Lawrence, C.E. S fold web server for statistical folding and rational design of nucleic acids. Nucleic Acids Res. 2004, 32 (Suppl. 2), W135–W141. [Google Scholar] [CrossRef]
- User Manual for the Monolith NT.115; NanoTemper Technologies: Munich, Germany, 2018.
- Seidel, S.A.; Dijkman, P.M.; Lea, W.A.; van den Bogaart, G.; Jerabek-Willemsen, M.; Lazic, A.; Joseph, J.S.; Srinivasan, P.; Baaske, P.; Simeonov, A.; et al. Microscale thermophoresis quantifies biomolecular interactions under previously challenging conditions. Methods 2013, 59, 301–315. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nelson, C.; Mrozowich, T.; Gemmill, D.L.; Park, S.M.; Patel, T.R. Human DDX3X Unwinds Japanese Encephalitis and Zika Viral 5′ Terminal Regions. Int. J. Mol. Sci. 2021, 22, 413. https://doi.org/10.3390/ijms22010413
Nelson C, Mrozowich T, Gemmill DL, Park SM, Patel TR. Human DDX3X Unwinds Japanese Encephalitis and Zika Viral 5′ Terminal Regions. International Journal of Molecular Sciences. 2021; 22(1):413. https://doi.org/10.3390/ijms22010413
Chicago/Turabian StyleNelson, Corey, Tyler Mrozowich, Darren L. Gemmill, Sean M. Park, and Trushar R. Patel. 2021. "Human DDX3X Unwinds Japanese Encephalitis and Zika Viral 5′ Terminal Regions" International Journal of Molecular Sciences 22, no. 1: 413. https://doi.org/10.3390/ijms22010413
APA StyleNelson, C., Mrozowich, T., Gemmill, D. L., Park, S. M., & Patel, T. R. (2021). Human DDX3X Unwinds Japanese Encephalitis and Zika Viral 5′ Terminal Regions. International Journal of Molecular Sciences, 22(1), 413. https://doi.org/10.3390/ijms22010413




