G-Quadruplex Modulation of SP1 Functional Binding Sites at the KIT Proximal Promoter
Abstract
1. Introduction
2. Results
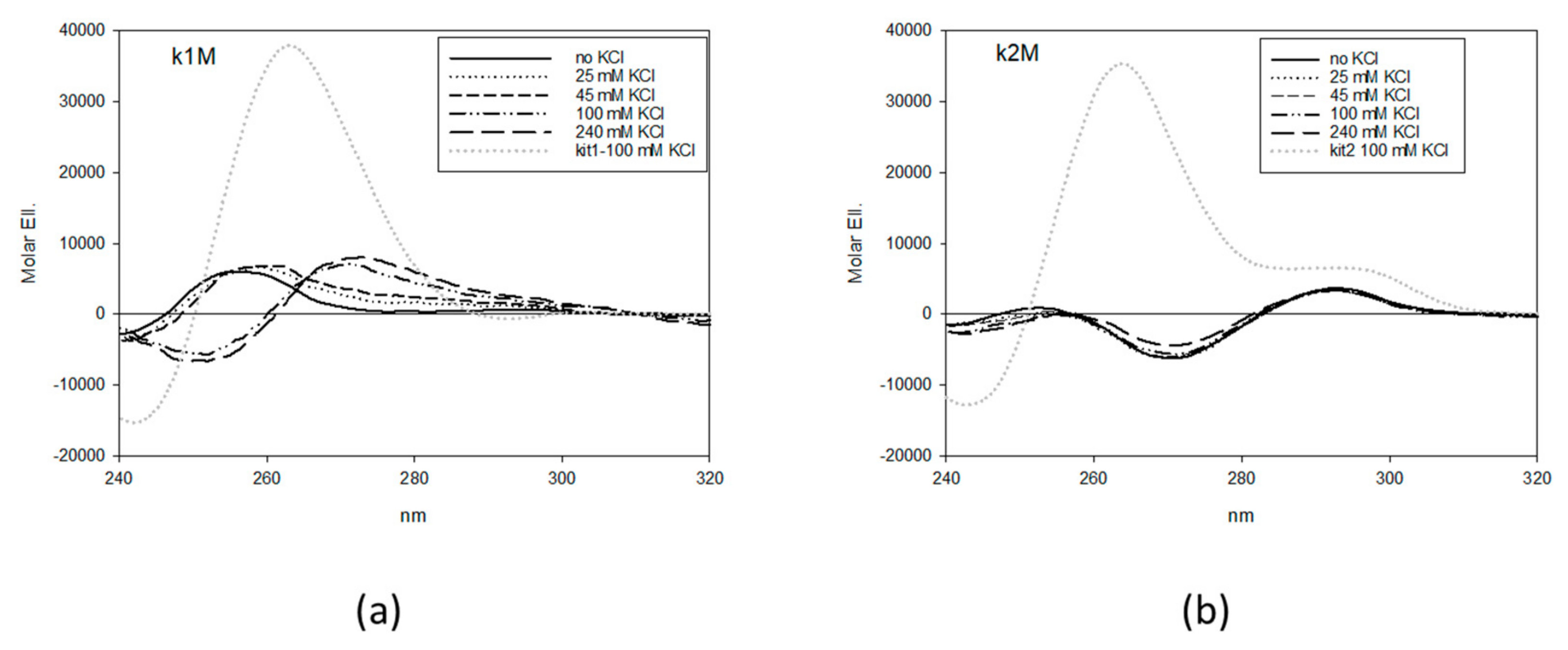
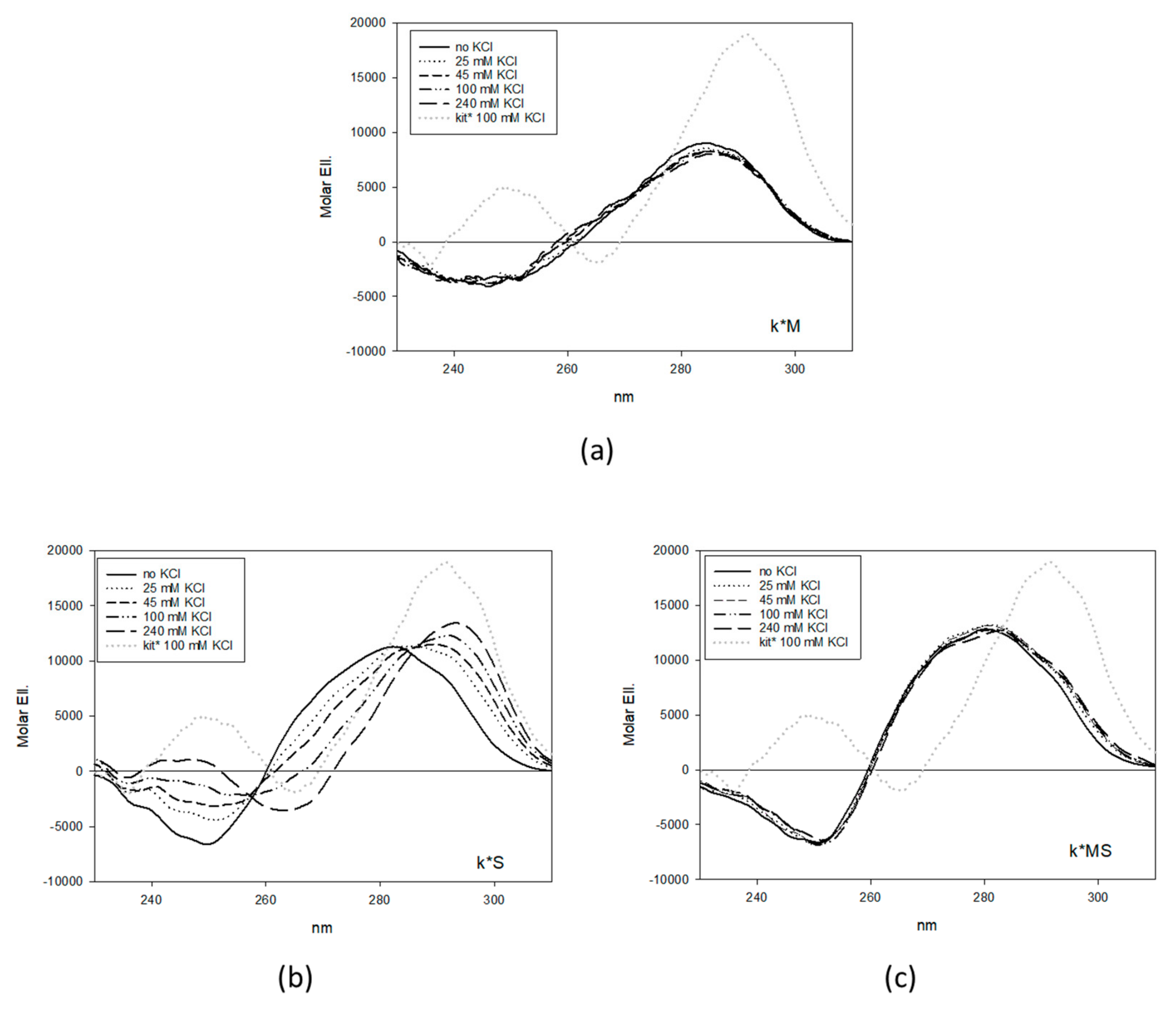
2.1. Design and Selection of Mutated Sequences
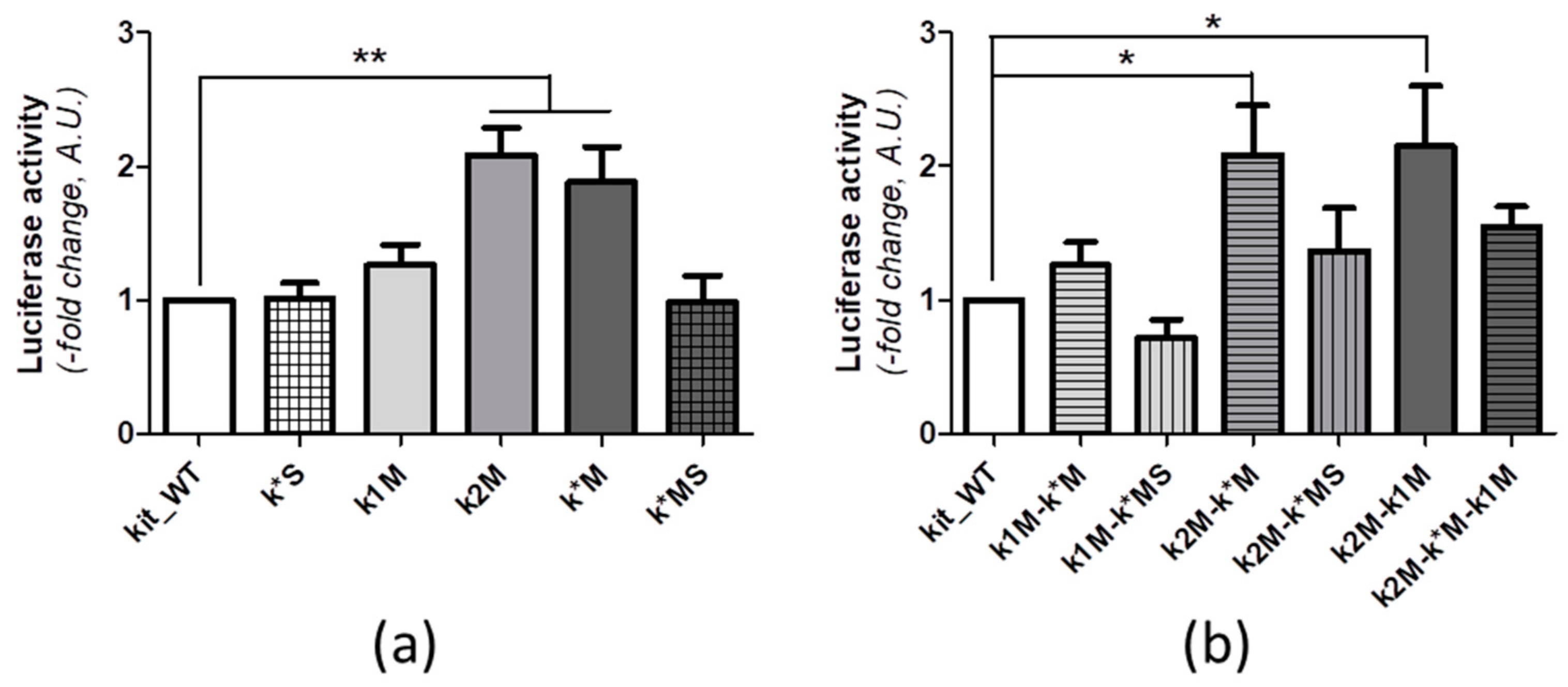
2.2. Site-Directed Mutagenesis and Transfection
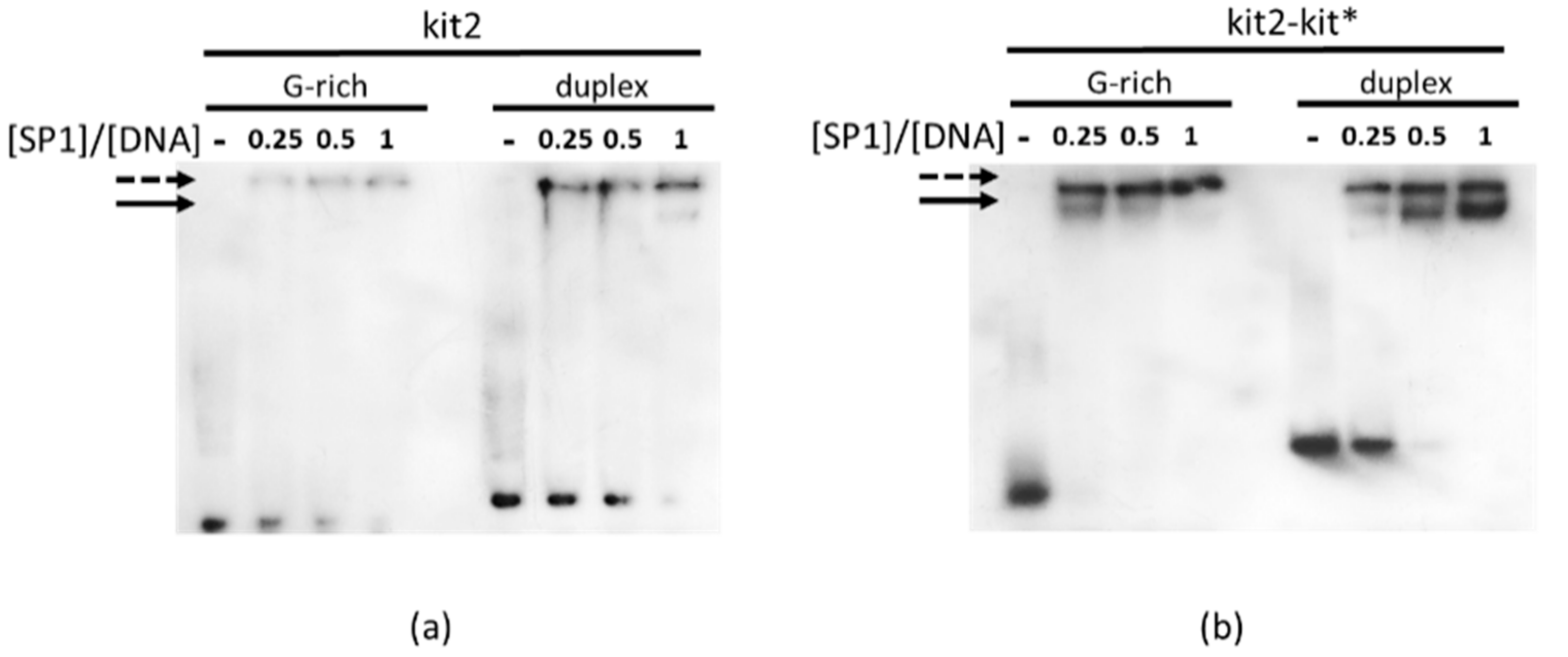
2.3. SP1 Binding at kit2 and kit2-kit*
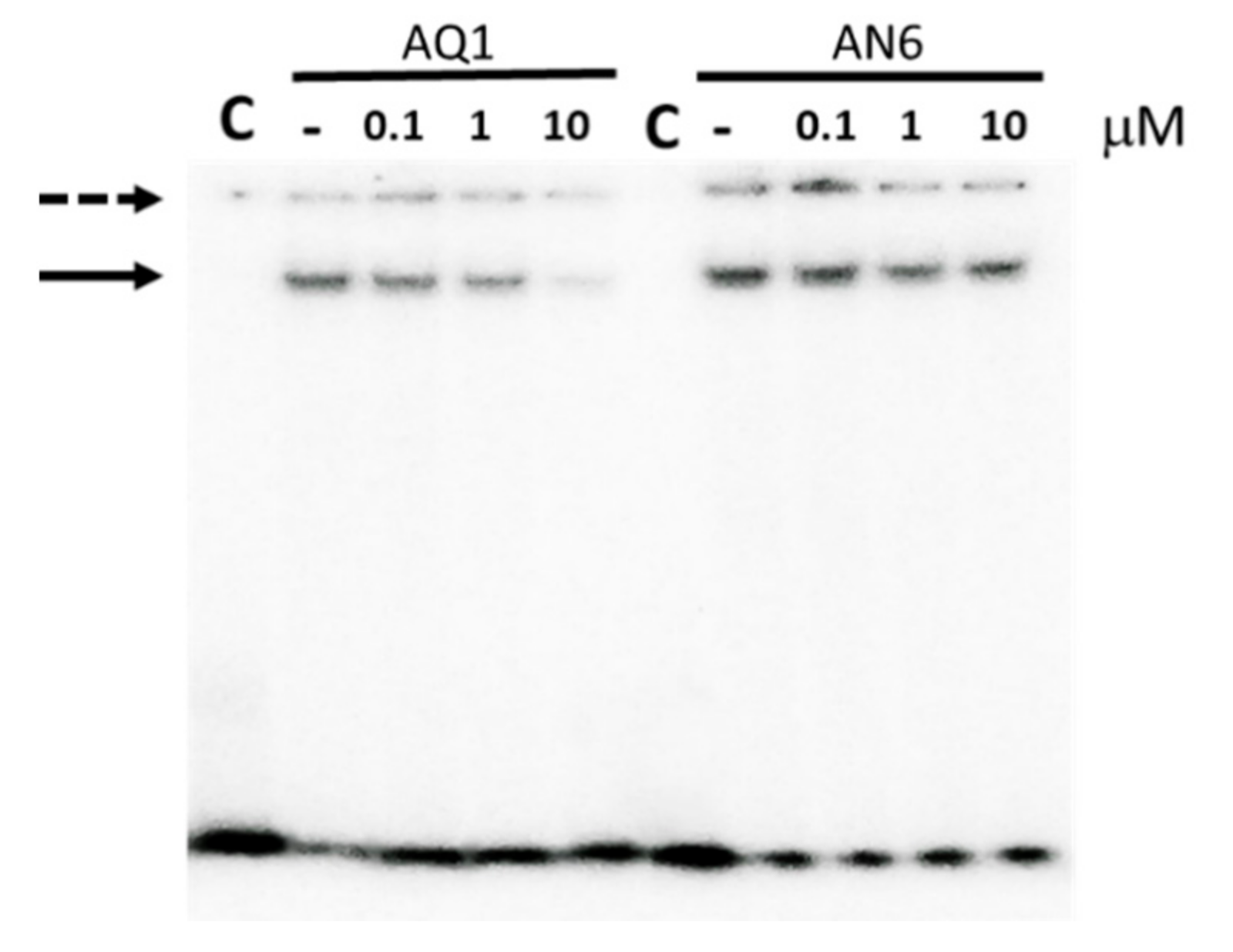
2.4. Binding of Candidate Ligands at kit* and kit2-kit*
3. Discussion
4. Materials and Methods
4.1. CD Analysis
4.2. Electrophoretic Mobility Shift Assay (EMSA)
4.3. Surface Plasmon Resonance (SPR)
4.4. Fluorescence
4.5. Site-Directed Mutagenesis
4.6. Cell Culture
4.7. MCF-7 Transfection
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| G4 | G-quadruplex |
| CD | Circular Dichroism |
| EMSA | Electrophoretic Mobility Shift Assay |
| SPR | Surface Plasmon Resonance |
| AP | 2-aminopurine |
References
- Liang, J.; Wu, Y.L.; Chen, B.J.; Zhang, W.; Tanaka, Y.; Sugiyama, H. The C-Kit Receptor-Mediated Signal Transduction and Tumor-Related Diseases. Int. J. Biol. Sci. 2013, 9, 435–443. [Google Scholar] [CrossRef] [PubMed]
- Pittoni, P.; Piconese, S.; Tripodo, C.; Colombo, M.P. Tumor-intrinsic and -extrinsic roles of c-Kit: Mast cells as the primary off-target of tyrosine kinase inhibitors. Oncogene 2011, 30, 757–769. [Google Scholar] [CrossRef] [PubMed]
- Ashman, L.K.; Griffith, R. Therapeutic targeting of c-KIT in cancer. Expert Opin. Investig. Drugs 2013, 22, 103–115. [Google Scholar] [CrossRef] [PubMed]
- Francisco, A.P.; Paulo, A. Oncogene Expression Modulation in Cancer Cell Lines by DNA G-Quadruplex-Interactive Small Molecules. Curr. Med. Chem. 2017, 24, 4873–4904. [Google Scholar]
- Rankin, S.; Reszka, A.P.; Huppert, J.; Zloh, M.; Parkinson, G.N.; Todd, A.K.; Ladame, S.; Balasubramanian, S.; Neidle, S. Putative DNA quadruplex formation within the human c-kit oncogene. J. Am. Chem. Soc. 2005, 127, 10584–10589. [Google Scholar] [CrossRef]
- Hsu, S.T.; Varnai, P.; Bugaut, A.; Reszka, A.P.; Neidle, S.; Balasubramanian, S. A G-Rich Sequence within the c-kit Oncogene Promoter Forms a Parallel G-Quadruplex Having Asymmetric G-Tetrad Dynamics. J. Am. Chem. Soc. 2009, 131, 13399–13409. [Google Scholar] [CrossRef]
- Kuryavyi, V.; Phan, A.T.; Patel, D.J. Solution structures of all parallel-stranded monomeric and dimeric G-quadruplex scaffolds of the human c-kit2 promoter. Nucleic Acids Res. 2010, 38, 6757–6773. [Google Scholar] [CrossRef]
- Davis, J.T.; Spada, G.P. Supramolecular architectures generated by self-assembly of guanosine derivatives. Chem. Soc. Rev. 2007, 36, 296–313. [Google Scholar] [CrossRef]
- Rigo, R.; Palumbo, M.; Sissi, C. G-quadruplexes in human promoters: A challenge for therapeutic applications. Biochim. Biophys Acta 2017, 1861, 1399–1413. [Google Scholar] [CrossRef]
- Varshney, D.; Spiegel, J.; Zyner, K.; Balasubramanian, S. The regulation and functions of DNA and RNA G-quadruplexes. Nat. Rev. Mol. Cell Biol. 2020, 21, 459–474. [Google Scholar] [CrossRef]
- Zorzan, E.; Elgendy, R.; Giantin, M.; Dacasto, M.; Sissi, C. Whole-Transcriptome Profiling of Canine and Human in Vitro Models Exposed to a G-Quadruplex Binding Small Molecule. Sci. Rep. 2018, 8, 17107. [Google Scholar] [CrossRef] [PubMed]
- Ceschi, S.; Sissi, C. ARMC-Quadruplex Nucleic Acid as Target. for Medicinal Chemistry; Neidle, S., Ed.; Elsevier: Amsterdam, The Netherlands, 2020; Volume 54, pp. 409–439. [Google Scholar]
- Kotar, A.; Rigo, R.; Sissi, C.; Plavec, J. Two-quartet kit* G-quadruplex is formed via double-stranded pre-folded structure. Nucleic Acids Res. 2019, 47, 2641–2653. [Google Scholar] [CrossRef] [PubMed]
- Phan, A.T.; Kuryavyi, V.; Burge, S.; Neidle, S.; Patel, D.J. Two-Repeat Human Telomeric d(TAGGGTTAGGGT) Sequence Forms Interconverting Parallel and Antiparallel G-Quadruplexes in Solution: Distinct Topologies, Thermodynamic Properties, and Folding/Unfolding Kinetics. J. Am. Chem. Soc. 2007, 129, 4386–4392. [Google Scholar] [CrossRef] [PubMed]
- Wei, D.; Husby, J.; Neidle, S. Flexibility and structural conservation in a c-KIT G-quadruplex. Nucleic Acids Res. 2015, 43, 629–644. [Google Scholar] [CrossRef]
- Wei, D.; Parkinson, G.N.; Reszka, A.P.; Neidle, S. Crystal structure of a c-kit promoter quadruplex reveals the structural role of metal ions and water molecules in maintaining loop conformation. Nucleic Acids Res. 2012, 40, 4691–4700. [Google Scholar] [CrossRef]
- Rigo, R.; Dean, W.L.; Gray, R.D.; Chaires, J.B.; Sissi, C. Conformational profiling of a G-rich sequence within the c-KIT promoter. Nucleic Acids Res. 2017, 45, 13056–13067. [Google Scholar] [CrossRef]
- Rigo, R.; Sissi, C. Characterization of G4-G4 Crosstalk in the c-KIT Promoter Region. Biochemistry 2017, 56, 4309–4312. [Google Scholar] [CrossRef]
- Ducani, C.; Bernardinelli, G.; Hogberg, B.; Keppler, B.K.; Terenzi, A. Interplay of Three G-Quadruplex Units in the KIT Promoter. J. Am. Chem. Soc. 2019, 141, 10205–10213. [Google Scholar] [CrossRef]
- Vandenbank, G.R.; Chen, Y.; Friday, E.; Pavlik, K.; Anthony, B.; Decastro, C.; Kaufman, R.E. Complex regulation of human c-kit transcription by promoter repressors, activators, and specific myb elements. Cell Growth Differ. 1996, 7, 1383. [Google Scholar]
- Maeda, K.; Nishiyama, C.; Ogawa, H.; Okumura, K. GATA2 and Sp1 Positively Regulate the c-kit Promoter in Mast Cells. J. Immunol. 2010, 185, 4252–4260. [Google Scholar] [CrossRef]
- Park, G.H.; Plummer, H.K., III; Krystal, G.W. Selective Sp1 binding is critical for maximal activity of the human c-kit promoter. Blood 1998, 92, 4138–4149. [Google Scholar] [CrossRef] [PubMed]
- Raiber, E.A.; Kranaster, R.; Lam, E.; Nikan, M.; Balasubramanian, S. A non-canonical DNA structure is a binding motif for the transcription factor SP1 in vitro. Nucleic Acids Res. 2012, 40, 1499–1508. [Google Scholar] [CrossRef] [PubMed]
- Zorzan, E.; Da Ros, S.; Musetti, C.; Shahidian, L.Z.; Coelho, N.F.; Bonsembiante, F.; Letard, S.; Gelain, M.E.; Palumbo, M.; Dubreuil, P.; et al. Screening of candidate G-quadruplex ligands for the human c-KIT promotorial region and their effects in multiple in-vitro models. Oncotarget 2016, 7, 21658–21675. [Google Scholar] [CrossRef] [PubMed]
- Fleming, A.M.; Zhou, J.; Wallace, S.S.; Burrows, C.J. A role for the fifth G-track in G-quadruplex forming oncogene promoter sequences during oxidative stress: Do these “spare tires” have an evolved function? ACS Cent. Sci. 2015, 1, 226–233. [Google Scholar] [CrossRef] [PubMed]
- Xin-min Li, X.; Zheng, K.; Zhang, J.; Liu, H.; He, Y.; Yuan, B.; Hao, Y.; Tan, Z. Guanine-vacancy–bearing G-quadruplexes responsive to guanine derivatives. Proc. Natl. Acad. Sci USA 2015, 112, 14581–14586. [Google Scholar]
- Yamamoto, K.; Tojo, A.; Aoki, N.; Shibuya, M. Characterization of the promoter region of the human c-kit proto-oncogene. Jpn J. Cancer Res. 1993, 84, 1136–1144. [Google Scholar] [CrossRef]
- Fleming, A.M.; Zhu, J.; Ding, Y.; Burrows, C.J. Location dependence of the transcriptional response of a potential G-quadruplex in gene promoters under oxidative stress. Nucleic Acids Res. 2019, 47, 5049–5060. [Google Scholar] [CrossRef]
- Salsbury, A.M.; Dean, T.J.; Lemkul, J.A. Polarizable Molecular Dynamics Simulations of Two c-kit Oncogene Promoter G-Quadruplexes: Effect of Primary and Secondary Structure on Loop and Ion Sampling. J. Chem. Theory Comput. 2020, 16, 3430–3444. [Google Scholar] [CrossRef]
- Zagotto, G.; Ricci, A.; Vasquez, E.; Sandoli, A.; Benedetti, S.; Palumbo, M.; Sissi, C. Tuning G-quadruplex vs double-stranded DNA recognition in regioisomeric lysyl-peptidyl-anthraquinone conjugates. Bioconjugate Chem. 2011, 22, 2126–2135. [Google Scholar] [CrossRef]
- Folini, M.; Pivetta, C.; Zagotto, G.; De Marco, C.; Palumbo, M.; Zaffaroni, N.; Sissi, C. Remarkable interference with telomeric function by a G-quadruplex selective bisantrene regioisomer. Biochem. Pharmacol. 2010, 79, 1781–1790. [Google Scholar] [CrossRef]


| Name | Sequence 1 |
|---|---|
| kit1 | 5′-AGGGAGGGCGCTGGGAGGAGGG-3′ |
| k1M | 5′-AGGGAGTTCGCTGGGAGGAGGG-3′ |
| kit2 | 5′-CGGGCGGGCGCGAGGGAGGGG-3′ |
| k2M | 5′-CGTGCGTGCGCGAGGGAGGGG-3′ |
| kit* | 5′-GGCGAGGAGGGGCGTGGCCGGC-3′ |
| k*M | 5′-GTCGATGAGGGGCGTGGCCGGC-3′ |
| k*S | 5′-GGCGAGGATTGGCGTGGCCGGC-3′ |
| k*MS | 5′-GGCGAGGAGTTGCGTGGCCGGC-3′ |
| kit2-kit* | 5′-CGGGCGGGCGCGAGGGAGGGGAGGCGAGGAGGGGCGTGGCCGGC-3′ |
| kit2-kit*-kit1 2 | 5′-CGGGCGGGCGCGAGGGAGGGGAGGCGAGGAGGGGCGTGGCCGGCG CGCAGAGGGAGGGCGCTGGGAGGAGGG-3′ |
| Ligand | kit* 1 | kit*5AP 2 | kit*8AP 2 |
|---|---|---|---|
| AN6 | 6.71 ± 1.26 | 2.24 ± 0.17 | 1.78 ± 0.15 |
| AQ1 | 0.51 ± 0.09 | 0.32 ± 0.02 | 0.31 ± 0.02 |
| Mutations | Primer Pair 1 | Oligonucleotide Sequence 5′-3′ | Primer Design |
|---|---|---|---|
| k2M | F | ACCCGTGCGTGCGCGAGGGAGGGGAGGCGAGGA | ex novo |
| R | CGCGCACGCACGGGTCTCGCTTCTTCCCGGCGG | ex novo | |
| k1M | F | CGGCGCGCAGAGGGAGTTCGCTGGGAGGAGGGGCTGCT | ex novo |
| R | GCGAACTCCCTCTGCGCGCCGGCCACGCCCCTCCTCGC | ex novo | |
| k*M | F | GAGGGAGGGGAGTCGATGAGGGGCGTGGCCGGCG | ex novo |
| R | ATCGACTCCCCTCCCTCGCGCCCGCCCGGGTCTC | ex novo | |
| k*S | F | GGATTGGCGTGGCCGGCGCGCAGAGGGAG | ex novo |
| R | CCACGCCAATCCTCGCCTCCCCTCCCTCGC | ex novo | |
| k*MS | F | GGAGTTGCGTGGCCGGCGCGCAGAGGGAG | ex novo |
| R | CCACGCAACTCCTCGCCTCCCCTCCCTCGC | ex novo | |
| k2M-k*M | F | ACCCGTGCGTGCGCGAGGGAGGGGAGTCGATGA | ex novo |
| R | CGCGCACGCACGGGTCTCGCTTCTTCCCGGCGG | k2M-R | |
| k2M-k*MS | F | GGAGTTGCGTGGCCGGCGCGCAGAGGGAG | k*MS-F |
| R | CCACGCAACTCCTCGCCTCCCCTCCCTCGC | k*MS-R | |
| k1M-k*M | F | GAGGGAGGGGAGTCGATGAGGGGCGTGGCCGGCG | k*M-F |
| R | ATCGACTCCCCTCCCTCGCGCCCGCCCGGGTCTC | k*M-R | |
| k1M-k*MS | F | GGAGTTGCGTGGCCGGCGCGCAGAGGGAG | k*MS-F |
| R | CCACGCAACTCCTCGCCTCCCCTCCCTCGC | k*MS-R | |
| k2M-k1M | F | ACCCGTGCGTGCGCGAGGGAGGGGAGGCGAGGA | k2M-F |
| R | CGCGCACGCACGGGTCTCGCTTCTTCCCGGCGG | k2M-R | |
| k2M-k*M-k1M | F | ACCCGTGCGTGCGCGAGGGAGGGGAGTCGATGA | ex novo |
| R | CGCGCACGCACGGGTCTCGCTTCTTCCCGGCGG | k2M-R |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Da Ros, S.; Nicoletto, G.; Rigo, R.; Ceschi, S.; Zorzan, E.; Dacasto, M.; Giantin, M.; Sissi, C. G-Quadruplex Modulation of SP1 Functional Binding Sites at the KIT Proximal Promoter. Int. J. Mol. Sci. 2021, 22, 329. https://doi.org/10.3390/ijms22010329
Da Ros S, Nicoletto G, Rigo R, Ceschi S, Zorzan E, Dacasto M, Giantin M, Sissi C. G-Quadruplex Modulation of SP1 Functional Binding Sites at the KIT Proximal Promoter. International Journal of Molecular Sciences. 2021; 22(1):329. https://doi.org/10.3390/ijms22010329
Chicago/Turabian StyleDa Ros, Silvia, Giulia Nicoletto, Riccardo Rigo, Silvia Ceschi, Eleonora Zorzan, Mauro Dacasto, Mery Giantin, and Claudia Sissi. 2021. "G-Quadruplex Modulation of SP1 Functional Binding Sites at the KIT Proximal Promoter" International Journal of Molecular Sciences 22, no. 1: 329. https://doi.org/10.3390/ijms22010329
APA StyleDa Ros, S., Nicoletto, G., Rigo, R., Ceschi, S., Zorzan, E., Dacasto, M., Giantin, M., & Sissi, C. (2021). G-Quadruplex Modulation of SP1 Functional Binding Sites at the KIT Proximal Promoter. International Journal of Molecular Sciences, 22(1), 329. https://doi.org/10.3390/ijms22010329






