Up-Regulated Expression of Pro-Apoptotic Long Noncoding RNA lincRNA-p21 with Enhanced Cell Apoptosis in Lupus Nephritis
Abstract
1. Introduction
2. Results
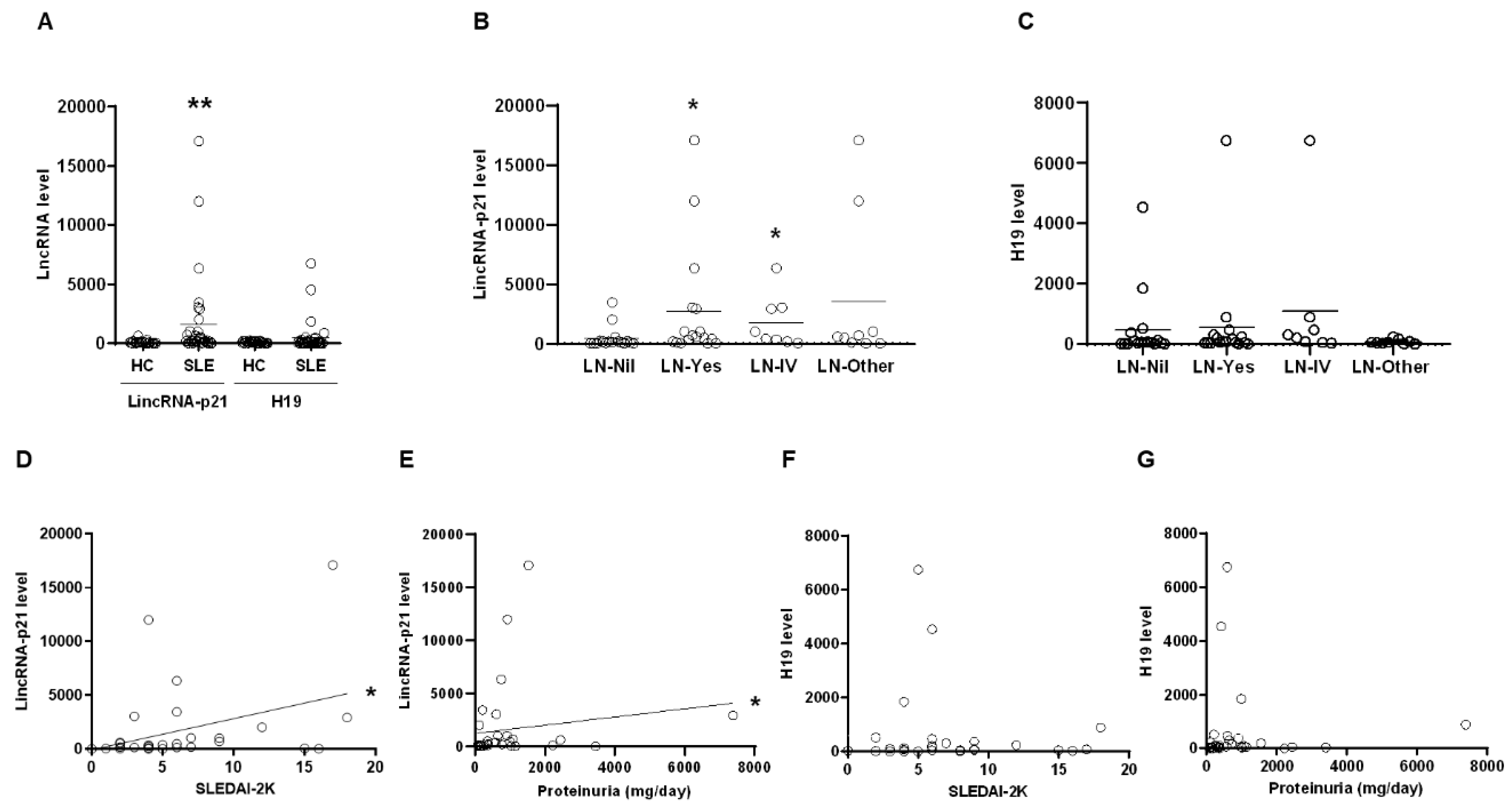
2.1. Up-Regulated Expression of LincRNA-p21 in LN Patient
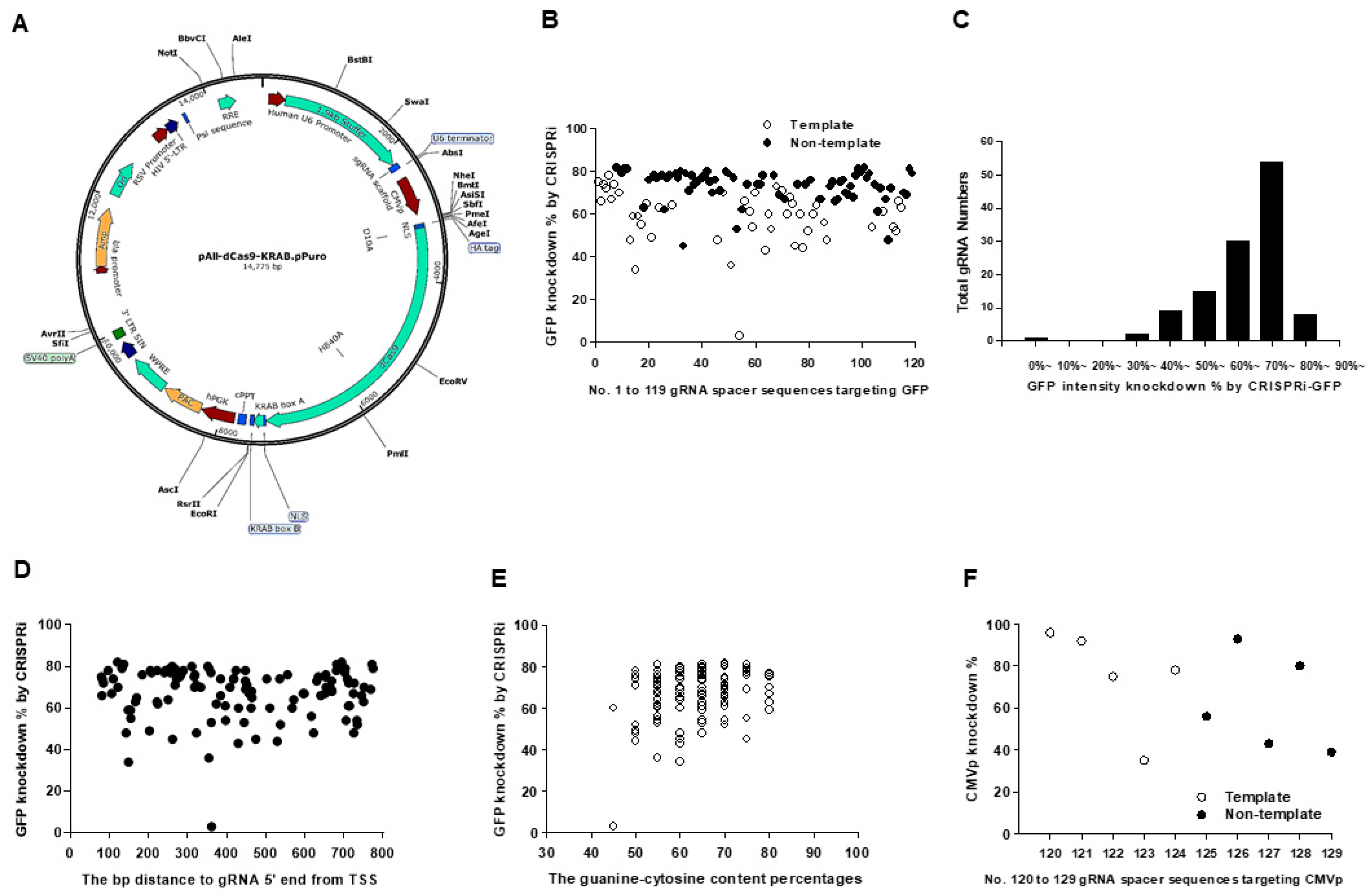
2.2. Transcription Repression Ability in pAll-dCas9-KRAB.pPuro with dCas9/KRAB Domain
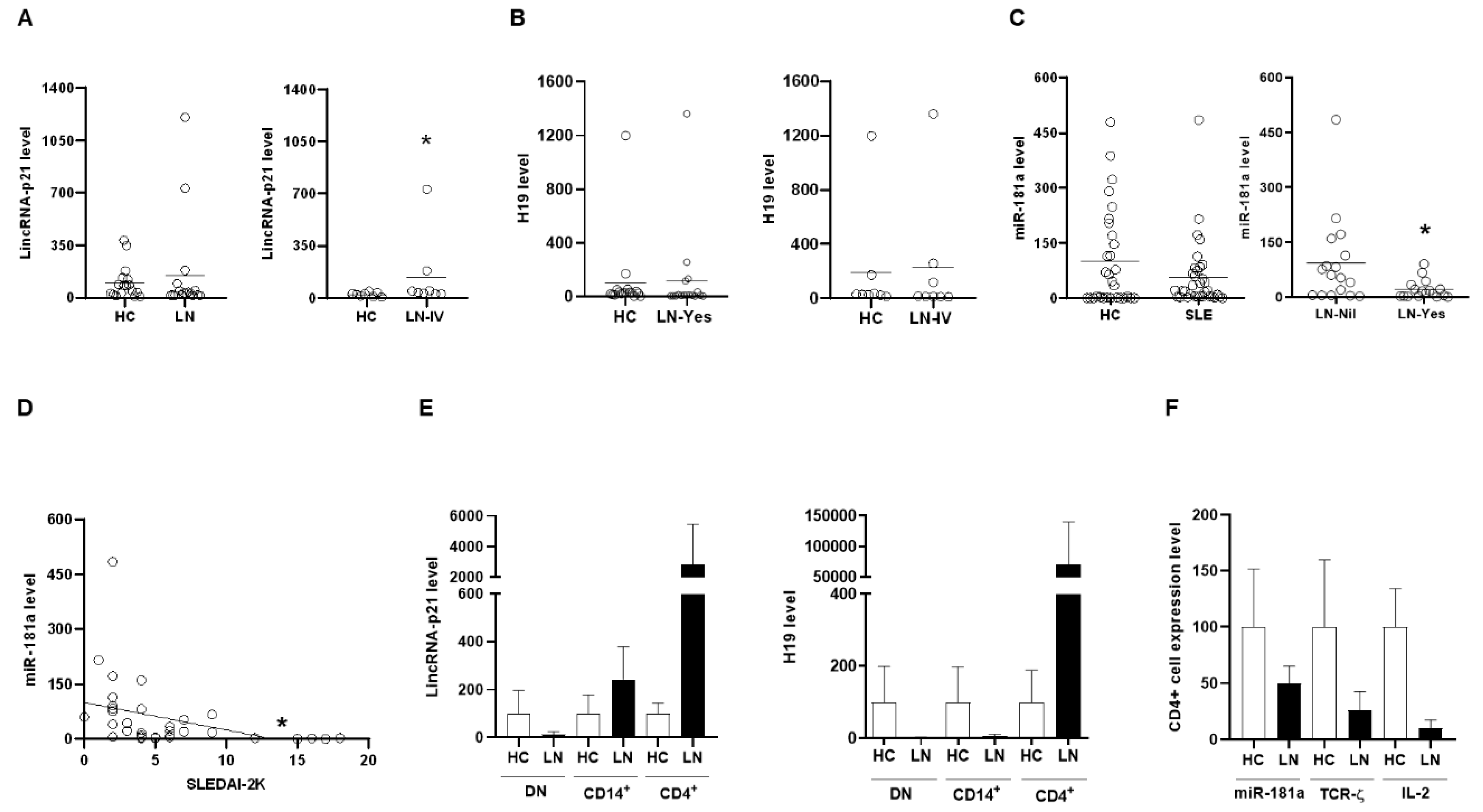
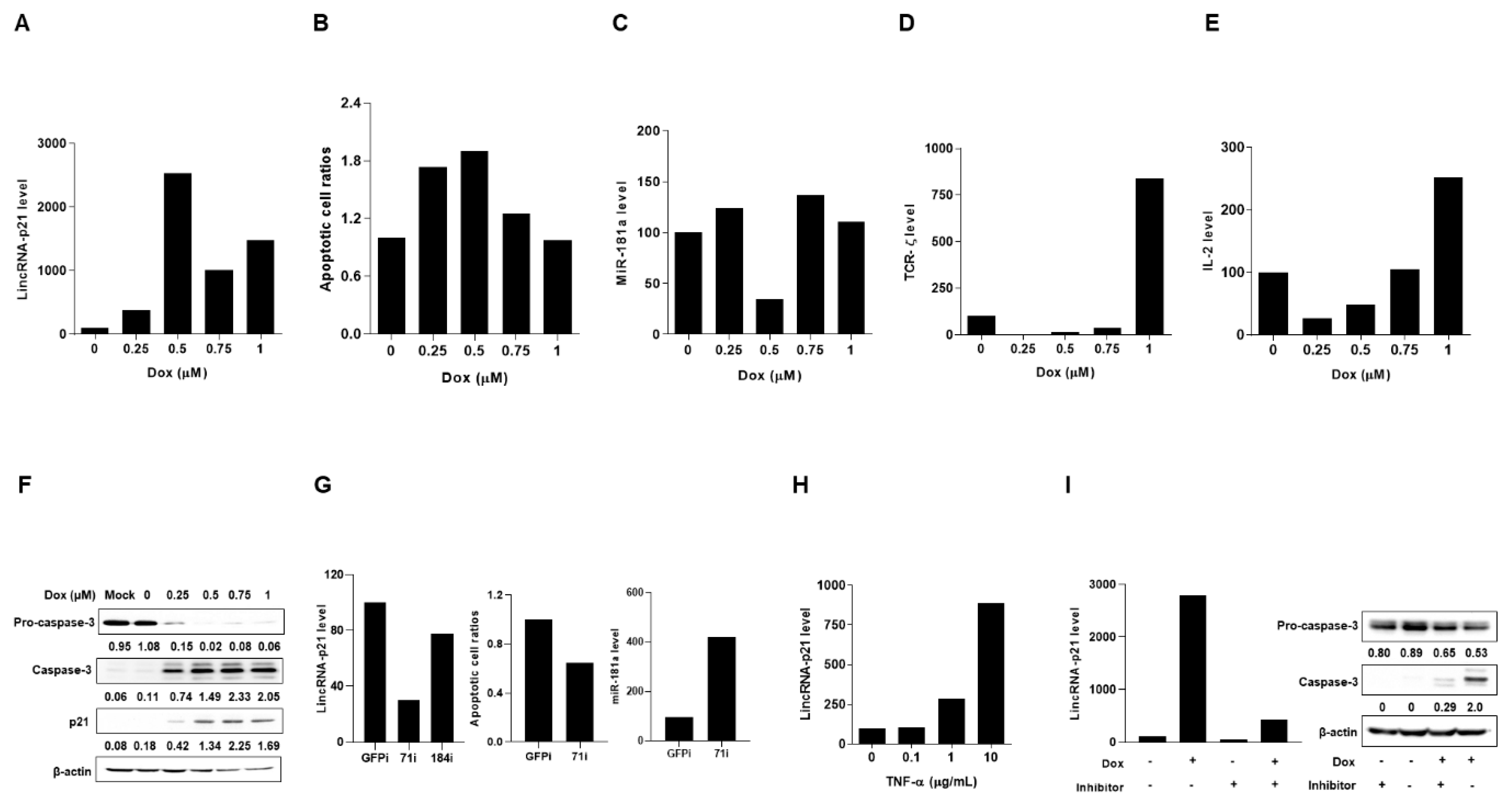
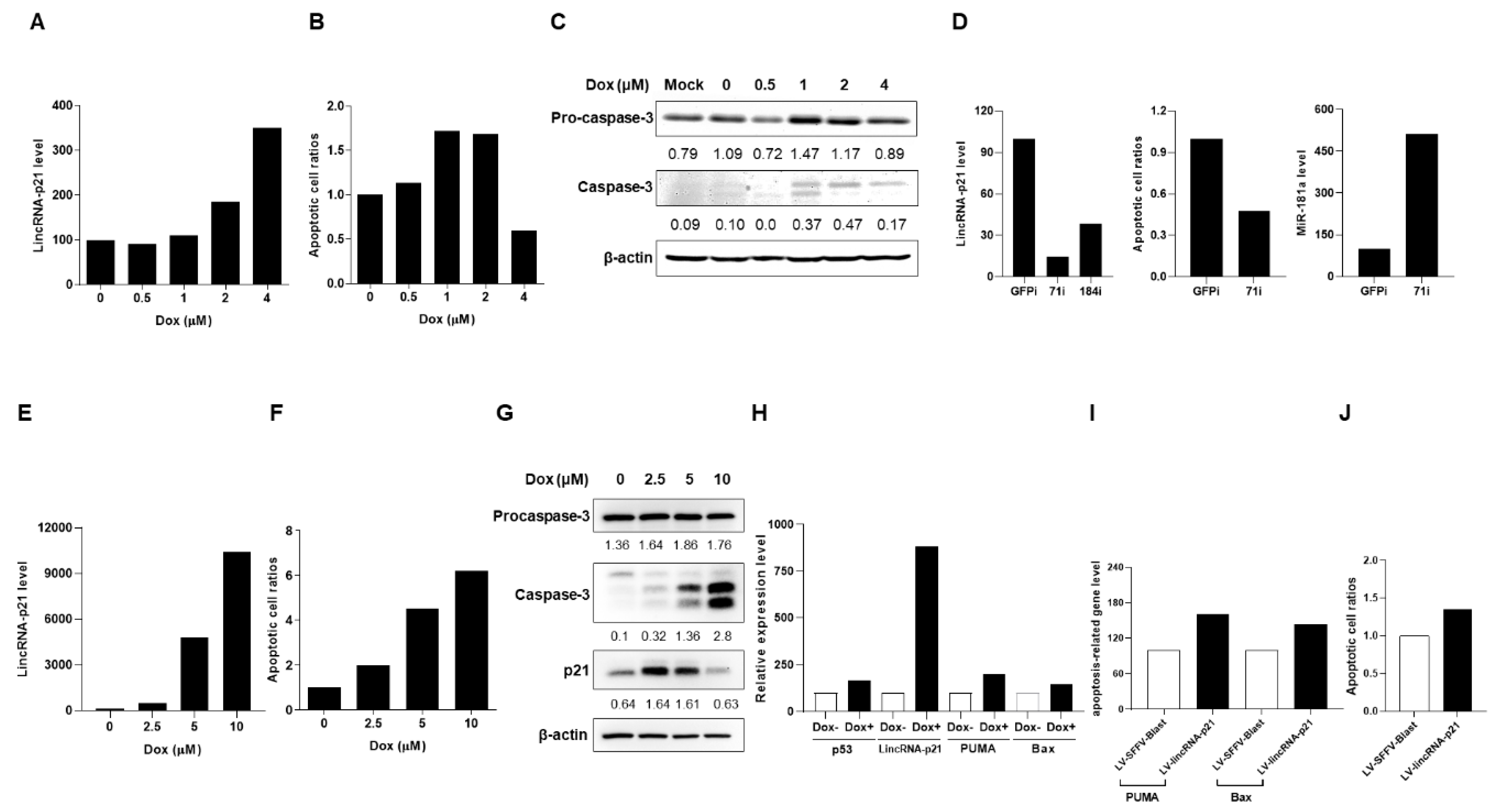
2.3. Up-Regulated Expression of LincRNA-p21 in Apoptotic Human T-Lymphocyte and Kidney Cell Lines
2.4. Up-Regulated Expression of LincRNA-p21 in a LN Mouse Model
3. Discussion
4. Materials and Methods
4.1. LN Patients and Age/Sex-Matched Control Subjects
4.2. Pristane-Induced LN Mouse Model
4.3. Purification of Human and Mouse Cells
4.4. Quantitative Real Time Polymerase Chain Reaction (qRT-PCR)
4.5. pAll-dCas9-KRAB.pPuro/green Fluorescence Protein (GFP) Reporter Construction
4.6. Transcription Repression Assessment
4.7. Construction of LV-Based CRISPRi Targeting and Overexpression of LincRNA-p21
4.8. Production of Stable Transfectants
4.9. Doxorubicin (Dox)-Induced Cell Apoptosis
4.10. Immunoblotting Assessment
4.11. Enzyme-Linked Immunosorbent Assay (ELISA)
4.12. Proteinuria Detection
4.13. Histopathological and Immunofluorescence Analyses
4.14. Statistical Analyses
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ceRNAs | Competing endogenous RNAs |
| CRISPRi | CRISPR interference |
| dsDNA | Double strand DNA |
| Dox | Doxorubicin |
| GN | Glomerulonephritis |
| HC | Healthy control |
| IC | Immune complexes |
| i.p. | intraperitoneally |
| LV | Lentiviral vector |
| lncRNA | Long noncoding RNAs |
| LN | Lupus nephritis |
| miRNA | microRNAs |
| MNCs | mononuclear cells |
| PBS | Phosphate-buffered saline |
| qRT-PCR | quantitative real-time polymerase chain reaction |
| RT | Reverse transcription |
| SEM | standard error of the mean |
| SLE | Systemic lupus erythematosus |
| SLEDAI | SLE disease activity index |
| TCR | T cell receptor |
| TUNEL | Terminal deoxynucleotidyl transferase dUTP nick end labeling |
| UPC | Urine protein concentration |
References
- Yu, F.; Haas, M.; Glassock, R.; Zhao, M.H. Redefining lupus nephritis: Clinical implications of pathophysiologic subtypes. Nat. Rev. Nephrol. 2017, 13, 483–495. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Anders, H.J. Lupus nephritis: From pathogenesis to targets for biologic treatment. Nephron. Clin. Pract. 2014, 128, 224–231. [Google Scholar] [CrossRef] [PubMed]
- Mistry, P.; Kaplan, M.J. Cell death in the pathogenesis of systemic lupus erythematosus and lupus nephritis. Clin. Immunol. 2017, 185, 59–73. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Tong, Y.; Liu, Y.; Hu, H. Self-dsDNA in the pathogenesis of systemic lupus erythematosus. Clin. Exp. Immunol. 2018, 191, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Dhir, V.; Singh, A.P.; Aggarwal, A.; Naik, S.; Misra, R. Increased T-lymphocyte apoptosis in lupus correlates with disease activity and may be responsible for reduced T-cell frequency: A cross-sectional and longitudinal study. Lupus 2009, 18, 785–791. [Google Scholar] [CrossRef] [PubMed]
- Ransohoff, J.D.; Wei, Y.; Khavari, P.A. The functions and unique features of long intergenic non-coding RNA. Nat. Rev. Mol. Cell. Biol. 2018, 19, 143–157. [Google Scholar] [CrossRef]
- Hur, K.; Kim, S.H.; Kim, J.M. Potential implications of long noncoding RNAs in autoimmune diseases. Immune Netw. 2019, 19, e4. [Google Scholar] [CrossRef]
- Taheri, M.; Eghtedarian, R.; Dinger, M.E.; Ghafouri-Fard, S. Exploring the role of non-coding RNAs in the pathophysiology of systemic lupus erythematosus. Biomolecules 2020, 10, 937. [Google Scholar] [CrossRef] [PubMed]
- Miret, C.; Molina, R.; Filella, X.; García-Carrasco, M.; Claver, G.; Ingelmo, M.; Ballesta, A.; Font, J. Relationship of p53 with other oncogenes, cytokines and systemic lupus erythematosus activity. Tumor Biol. 2003, 24, 185–188. [Google Scholar] [CrossRef] [PubMed]
- Amirinejad, R.; Rezaei, M.; Shirvani-Farsani, Z. An update on long intergenic noncoding RNA p21: A regulatory molecule with various significant functions in cancer. Cell. Biosci. 2020, 10, 82. [Google Scholar] [CrossRef] [PubMed]
- Huarte, M.; Guttman, M.; Feldser, D.; Garber, M.; Koziol, M.J.; Kenzelmann-Broz, D.; Khalil, A.M.; Zuk, O.; Amit, I.; Rabani, M.; et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 2010, 142, 409–419. [Google Scholar] [CrossRef] [PubMed]
- Spurlock, C.F., 3rd; Tossberg, J.T.; Matlock, B.K.; Olsen, N.J.; Aune, T.M. Methotrexate inhibits NF-κB activity via long intergenic (noncoding) RNA-p21 induction. Arthritis Rheumatol. 2014, 66, 2947–2957. [Google Scholar] [CrossRef] [PubMed]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Amado, T.; Schmolka, N.; Metwally, H.; Silva-Santos, B.; Gomes, A.Q. Cross-regulation between cytokine and microRNA pathways in T cells. Eur. J. Immunol. 2015, 45, 1584–1595. [Google Scholar] [CrossRef] [PubMed]
- Lashine, Y.A.; Seoudi, A.M.; Salah, S.; Abdelaziz, A.I. Expression signature of microRNA-181-a reveals its crucial role in the pathogenesis of paediatric systemic lupus erythematosus. Clin. Exp. Rheumatol. 2011, 29, 351–357. [Google Scholar]
- Stypińska, B.; Paradowska-Gorycka, A. Cytokines and microRNAs as candidate biomarkers for systemic lupus erythematosus. Int. J. Mol. Sci. 2015, 16, 24194–24218. [Google Scholar] [CrossRef]
- Kampmann, M. CRISPRi and CRISPRa screens in mammalian cells for precision biology and medicine. ACS Chem. Biol. 2018, 13, 406–416. [Google Scholar] [CrossRef]
- Urrutia, R. KRAB-containing zinc-finger repressor proteins. Genome Biol. 2003, 4, 231. [Google Scholar] [CrossRef]
- Li, Q.J.; Chau, J.; Ebert, P.J.; Sylvester, G.; Min, H.; Liu, G.; Braich, R.; Manoharan, M.; Soutschek, J.; Skare, P.; et al. MiR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell 2007, 129, 147–161. [Google Scholar] [CrossRef]
- Moulton, V.R.; Tsokos, G.C. Abnormalities of T cell signaling in systemic lupus erythematosus. Arthritis Res. Ther. 2011, 13, 207. [Google Scholar] [CrossRef]
- Idborg, H.; Eketjäll, S.; Pettersson, S.; Gustafsson, J.T.; Zickert, A.; Kvarnström, M.; Oke, V.; Jakobsson, P.J.; Gunnarsson, I.; Svenungsson, E.; et al. TNF-α and plasma albumin as biomarkers of disease activity in systemic lupus erythematosus. Lupus Sci. Med. 2018, 5, e000260. [Google Scholar] [CrossRef] [PubMed]
- Hadjicharalambous, M.R.; Lindsay, M.A. Long non-coding RNAs and the innate immune response. Noncoding RNA 2019, 5, 34. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, H.; Han, S.; Lee, P.Y.; Khaybullin, R.; Shumyak, S.; Lu, L.; Chatha, A.; Afaneh, A.; Zhang, Y.; Xie, C.; et al. Pathogenesis of diffuse alveolar hemorrhage in murine lupus. Arthritis Rheumatol. 2017, 69, 1280–1293. [Google Scholar] [CrossRef] [PubMed]
- Emlen, W.; Niebur, J.; Kadera, R. Accelerated in vitro apoptosis of lymphocytes from patients with systemic lupus erythematosus. J. Immunol. 1994, 152, 3685–3692. [Google Scholar]
- Denny, M.F.; Chandaroy, P.; Killen, P.D.; Caricchio, R.; Lewis, E.E.; Richardson, B.C.; Lee, K.D.; Gavalchin, J.; Kaplan, M.J. Accelerated macrophage apoptosis induces autoantibody formation and organ damage in systemic lupus erythematosus. J. Immunol. 2006, 176, 2095–2104. [Google Scholar] [CrossRef]
- Cui, J.H.; Qiao, Q.; Guo, Y.; Zhang, Y.Q.; Cheng, H.; He, F.R.; Zhang, J. Increased apoptosis and expression of FasL, Bax and caspase-3 in human lupus nephritis class II and IV. J. Nephrol. 2012, 25, 255–261. [Google Scholar] [CrossRef]
- Rapicavoli, N.A.; Qu, K.; Zhang, J.; Mikhail, M.; Laberge, R.M.; Chang, H.Y. A mammalian pseudogene lncRNA at the interface of inflammation and anti-inflammatory therapeutics. Elife 2013, 2, e00762. [Google Scholar] [CrossRef]
- Li, W.; Titov, A.; Morel, L. An update on lupus animal models. Curr. Opin. Rheumatol. 2017, 29, 434–441. [Google Scholar] [CrossRef]
- Reeves, W.H.; Lee, P.Y.; Weinstein, J.S.; Satoh, M.; Lu, L. Induction of autoimmunity by pristane and other naturally occurring hydrocarbons. Trends Immunol. 2009, 30, 455–464. [Google Scholar] [CrossRef]
- Gilbert, L.A.; Horlbeck, M.A.; Adamson, B.; Villalta, J.E.; Chen, Y.; Whitehead, E.H.; Guimaraes, C.; Panning, B.; Ploegh, H.L.; Bassik, M.C.; et al. Genome-scale CRISPR-mediated control of gene repression and activation. Cell 2014, 159, 647–661. [Google Scholar] [CrossRef]
- Yeo, N.C.; Chavez, A.; Lance-Byrne, A.; Chan, Y.; Menn, D.; Milanova, D.; Kuo, C.C.; Guo, X.; Sharma, S.; Tung, A.; et al. An enhanced CRISPR repressor for targeted mammalian gene regulation. Nat. Methods 2018, 15, 611–616. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Wu, D.; Gao, L.; Liu, X.; Jin, Y.; Wang, D.; Wang, T.; Li, X. Competing endogenous RNA networks in human cancer: Hypothesis, validation, and perspectives. Oncotarget 2016, 7, 13479–13490. [Google Scholar] [CrossRef] [PubMed]
- Ballesteros-Tato, A.; Papillion, A. Mechanisms of action of low-dose IL-2 restoration therapies in SLE. Curr. Opin. Immunol. 2019, 61, 39–45. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Zhang, R.; Shao, M.; Zhao, X.; Miao, M.; Chen, J.; Liu, J.; Zhang, X.; Zhang, X.; Jin, Y.; et al. Efficacy and safety of low-dose IL-2 in the treatment of systemic lupus erythematosus: A randomised, double-blind, placebo-controlled trial. Ann. Rheum. Dis. 2020, 79, 141–149. [Google Scholar] [CrossRef]
- Hochberg, M.C. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1997, 40, 1725. [Google Scholar] [CrossRef]
- Bonomini, F.; Dos Santos, M.; Veronese, F.V.; Rezzani, R. NLRP3 Inflammasome modulation by melatonin supplementation in chronic pristane-induced lupus nephritis. Int. J. Mol. Sci. 2019, 20, 3466. [Google Scholar] [CrossRef]
- Zhou, T.; Benda, C.; Dunzinger, S.; Huang, Y.; Ho, J.C.; Yang, J.; Wang, Y.; Zhang, Y.; Zhuang, Q.; Li, Y.; et al. Generation of human induced pluripotent stem cells from urine samples. Nat. Protoc. 2012, 7, 2080–2089. [Google Scholar] [CrossRef]
- Bell, C.L.; Tenenhouse, H.S.; Scriver, C.R. Initiation and characterization of primary mouse kidney epithelial cultures. In Vitro Cell Dev. Biol. 1988, 24, 683–695. [Google Scholar] [CrossRef]
- Peng, J.S.; Chen, S.Y.; Wu, C.L.; Chong, H.E.; Ding, Y.C.; Shiau, A.L.; Wang, C.R. Amelioration of experimental autoimmune arthritis through targeting synovial fibroblasts by the intra-articular delivery of microRNA-140-3p and -5p. Arthritis Rheumatol. 2016, 68, 370–381. [Google Scholar] [CrossRef]
- Chen, S.Y.; Shiau, A.L.; Wu, C.L.; Wang, C.R. P53-derived hybrid peptides induce apoptosis of synovial fibroblasts in the rheumatoid joint. Oncotarget 2017, 8, 115413–115419. [Google Scholar] [CrossRef][Green Version]

| No. | gRNA Spacer Sequence 5′–3′ | Strand | No. | gRNA Spacer Sequence 5′–3′ | Strand |
|---|---|---|---|---|---|
| 1 | AAGGGCGAGGAGCTGTTCAC | T | 66 | GCTGAAGGGCATCGACTTCA | T |
| 2 | AGGGCGAGGAGCTGTTCACC | T | 67 | CAGCTCGATGCGGTTCACCA | NT |
| 3 | GGGCGAGGAGCTGTTCACCG | T | 68 | GAAGGGCATCGACTTCAAGG | T |
| 4 | CGAGGAGCTGTTCACCGGGG | T | 69 | TCAGCTCGATGCGGTTCACC | NT |
| 5 | CACCGGGGTGGTGCCCATCC | T | 70 | GGCATCGACTTCAAGGAGGA | T |
| 6 | GGTGCCCATCCTGGTCGAGC | T | 71 | CGATGCCCTTCAGCTCGATG | NT |
| 7 | CCCATCCTGGTCGAGCTGGA | T | 72 | CAAGGAGGACGGCAACATCC | T |
| 8 | GACCAGGATGGGCACCACCC | NT | 73 | AAGGAGGACGGCAACATCCT | T |
| 9 | GAGCTGGACGGCGACGTAAA | T | 74 | AGGAGGACGGCAACATCCTG | T |
| 10 | CCGTCCAGCTCGACCAGGAT | NT | 75 | CAACATCCTGGGGCACAAGC | T |
| 11 | GCCGTCCAGCTCGACCAGGA | NT | 76 | TGTACTCCAGCTTGTGCCCC | NT |
| 12 | CGTCGCCGTCCAGCTCGACC | NT | 77 | CAGCCACAACGTCTATATCA | T |
| 13 | GGCCACAAGTTCAGCGTGTC | T | 78 | ATGGCCGACAAGCAGAAGAA | T |
| 14 | CAAGTTCAGCGTGTCCGGCG | T | 79 | CGGCCATGATATAGACGTTG | NT |
| 15 | AAGTTCAGCGTGTCCGGCGA | T | 80 | CAAGCAGAAGAACGGCATCA | T |
| 16 | CAGCGTGTCCGGCGAGGGCG | T | 81 | GATGCCGTTCTTCTGCTTGT | NT |
| 17 | AGCGTGTCCGGCGAGGGCGA | T | 82 | CAAGATCCGCCACAACATCG | T |
| 18 | CGCCGGACACGCTGAACTTG | NT | 83 | ATCCGCCACAACATCGAGGA | T |
| 19 | GGCGAGGGCGATGCCACCTA | T | 84 | TGCCGTCCTCGATGTTGTGG | NT |
| 20 | GGCATCGCCCTCGCCCTCGC | NT | 85 | CGCTGCCGTCCTCGATGTTG | NT |
| 21 | CTGAAGTTCATCTGCACCAC | T | 86 | TACCAGCAGAACACCCCCAT | T |
| 22 | CAGGGTCAGCTTGCCGTAGG | NT | 87 | CAGAACACCCCCATCGGCGA | T |
| 23 | CTTCAGGGTCAGCTTGCCGT | NT | 88 | GGTGTTCTGCTGGTAGTGGT | NT |
| 24 | CCGGCAAGCTGCCCGTGCCC | T | 89 | TGGGGGTGTTCTGCTGGTAG | NT |
| 25 | GGTGGTGCAGATGAACTTCA | NT | 90 | CGCCGATGGGGGTGTTCTGC | NT |
| 26 | CGGTGGTGCAGATGAACTTC | NT | 91 | CACGGGGCCGTCGCCGATGG | NT |
| 27 | GGGCACGGGCAGCTTGCCGG | NT | 92 | GCACGGGGCCGTCGCCGATG | NT |
| 28 | CCAGGGCACGGGCAGCTTGC | NT | 93 | AGCACGGGGCCGTCGCCGAT | NT |
| 29 | CTCGTGACCACCCTGACCTA | T | 94 | CAGCACGGGGCCGTCGCCGA | NT |
| 30 | ACGAGGGTGGGCCAGGGCAC | NT | 95 | GGTTGTCGGGCAGCAGCACG | NT |
| 31 | CACGAGGGTGGGCCAGGGCA | NT | 96 | TGGTTGTCGGGCAGCAGCAC | NT |
| 32 | GTGGTCACGAGGGTGGGCCA | NT | 97 | GTGGTTGTCGGGCAGCAGCA | NT |
| 33 | GGTGGTCACGAGGGTGGGCC | NT | 98 | GTGCTCAGGTAGTGGTTGTC | NT |
| 34 | GTCAGGGTGGTCACGAGGGT | NT | 99 | GGTGCTCAGGTAGTGGTTGT | NT |
| 35 | GGTCAGGGTGGTCACGAGGG | NT | 100 | CGGACTGGGTGCTCAGGTAG | NT |
| 36 | GTAGGTCAGGGTGGTCACGA | NT | 101 | TCAGGGCGGACTGGGTGCTC | NT |
| 37 | CGTAGGTCAGGGTGGTCACG | NT | 102 | GTCTTTGCTCAGGGCGGACT | NT |
| 38 | CTGCACGCCGTAGGTCAGGG | NT | 103 | GGTCTTTGCTCAGGGCGGAC | NT |
| 39 | GCACTGCACGCCGTAGGTCA | NT | 104 | CAACGAGAAGCGCGATCACA | T |
| 40 | AGCACTGCACGCCGTAGGTC | NT | 105 | GTTGGGGTCTTTGCTCAGGG | NT |
| 41 | GCTGAAGCACTGCACGCCGT | NT | 106 | CTCGTTGGGGTCTTTGCTCA | NT |
| 42 | GCTTCATGTGGTCGGGGTAG | NT | 107 | TCTCGTTGGGGTCTTTGCTC | NT |
| 43 | CGTGCTGCTTCATGTGGTCG | NT | 108 | GCGCGATCACATGGTCCTGC | T |
| 44 | TCGTGCTGCTTCATGTGGTC | NT | 109 | TGTGATCGCGCTTCTCGTTG | NT |
| 45 | GTCGTGCTGCTTCATGTGGT | NT | 110 | ATGTGATCGCGCTTCTCGTT | NT |
| 46 | TTCAAGTCCGCCATGCCCGA | T | 111 | CATGTGATCGCGCTTCTCGT | NT |
| 47 | AGAAGTCGTGCTGCTTCATG | NT | 112 | CTGGAGTTCGTGACCGCCGC | T |
| 48 | CATGCCCGAAGGCTACGTCC | T | 113 | TGGAGTTCGTGACCGCCGCC | T |
| 49 | GACGTAGCCTTCGGGCATGG | NT | 114 | ACCGCCGCCGGGATCACTCT | T |
| 50 | CTGGACGTAGCCTTCGGGCA | NT | 115 | CGCCGGGATCACTCTCGGCA | T |
| 51 | GGAGCGCACCATCTTCTTCA | T | 116 | CGGCGGTCACGAACTCCAGC | NT |
| 52 | CGCTCCTGGACGTAGCCTTC | NT | 117 | GCCGAGAGTGATCCCGGCGG | NT |
| 53 | GCGCTCCTGGACGTAGCCTT | NT | 118 | CATGCCGAGAGTGATCCCGG | NT |
| 54 | ACCATCTTCTTCAAGGACGA | T | 119 | GTCCATGCCGAGAGTGATCC | NT |
| 55 | TGAAGAAGATGGTGCGCTCC | NT | 120 | CGGTAGGCGTGTACGGTGGG | T |
| 56 | CAACTACAAGACCCGCGCCG | T | 121 | AAATGGGCGGTAGGCGTGTA | T |
| 57 | GCCGTCGTCCTTGAAGAAGA | NT | 122 | CATTGACGCAAATGGGCGGT | T |
| 58 | CCGCGCCGAGGTGAAGTTCG | T | 123 | CTCCGCCCCATTGACGCAAA | T |
| 59 | CGCGCCGAGGTGAAGTTCGA | T | 124 | GTTTTGGCACCAAAATCAAC | T |
| 60 | GAAGTTCGAGGGCGACACCC | T | 125 | CTACCGCCCATTTGCGTCAA | NT |
| 61 | CTCGAACTTCACCTCGGCGC | NT | 126 | ACCGCCCATTTGCGTCAATG | NT |
| 62 | CCTCGAACTTCACCTCGGCG | NT | 127 | GCCCATTTGCGTCAATGGGG | NT |
| 63 | GTCGCCCTCGAACTTCACCT | NT | 128 | CAAACTCCCATTGACGTCAA | NT |
| 64 | GGTGAACCGCATCGAGCTGA | T | 129 | TCCCATTGACGTCAATGGGG | NT |
| 65 | GTGAACCGCATCGAGCTGAA | T |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Y.-C.; Kuo, P.-Y.; Chou, Y.-C.; Chong, H.-E.; Hsieh, Y.-T.; Yang, M.-L.; Wu, C.-L.; Shiau, A.-L.; Wang, C.-R. Up-Regulated Expression of Pro-Apoptotic Long Noncoding RNA lincRNA-p21 with Enhanced Cell Apoptosis in Lupus Nephritis. Int. J. Mol. Sci. 2021, 22, 301. https://doi.org/10.3390/ijms22010301
Chen Y-C, Kuo P-Y, Chou Y-C, Chong H-E, Hsieh Y-T, Yang M-L, Wu C-L, Shiau A-L, Wang C-R. Up-Regulated Expression of Pro-Apoptotic Long Noncoding RNA lincRNA-p21 with Enhanced Cell Apoptosis in Lupus Nephritis. International Journal of Molecular Sciences. 2021; 22(1):301. https://doi.org/10.3390/ijms22010301
Chicago/Turabian StyleChen, Yi-Cheng, Pin-Yu Kuo, Yu-Chi Chou, Hao-Earn Chong, Yu-Tung Hsieh, Mei-Lin Yang, Chao-Liang Wu, Ai-Li Shiau, and Chrong-Reen Wang. 2021. "Up-Regulated Expression of Pro-Apoptotic Long Noncoding RNA lincRNA-p21 with Enhanced Cell Apoptosis in Lupus Nephritis" International Journal of Molecular Sciences 22, no. 1: 301. https://doi.org/10.3390/ijms22010301
APA StyleChen, Y.-C., Kuo, P.-Y., Chou, Y.-C., Chong, H.-E., Hsieh, Y.-T., Yang, M.-L., Wu, C.-L., Shiau, A.-L., & Wang, C.-R. (2021). Up-Regulated Expression of Pro-Apoptotic Long Noncoding RNA lincRNA-p21 with Enhanced Cell Apoptosis in Lupus Nephritis. International Journal of Molecular Sciences, 22(1), 301. https://doi.org/10.3390/ijms22010301






