Biomimetic Membranes with Transmembrane Proteins: State-of-the-Art in Transmembrane Protein Applications
Abstract
1. Introduction
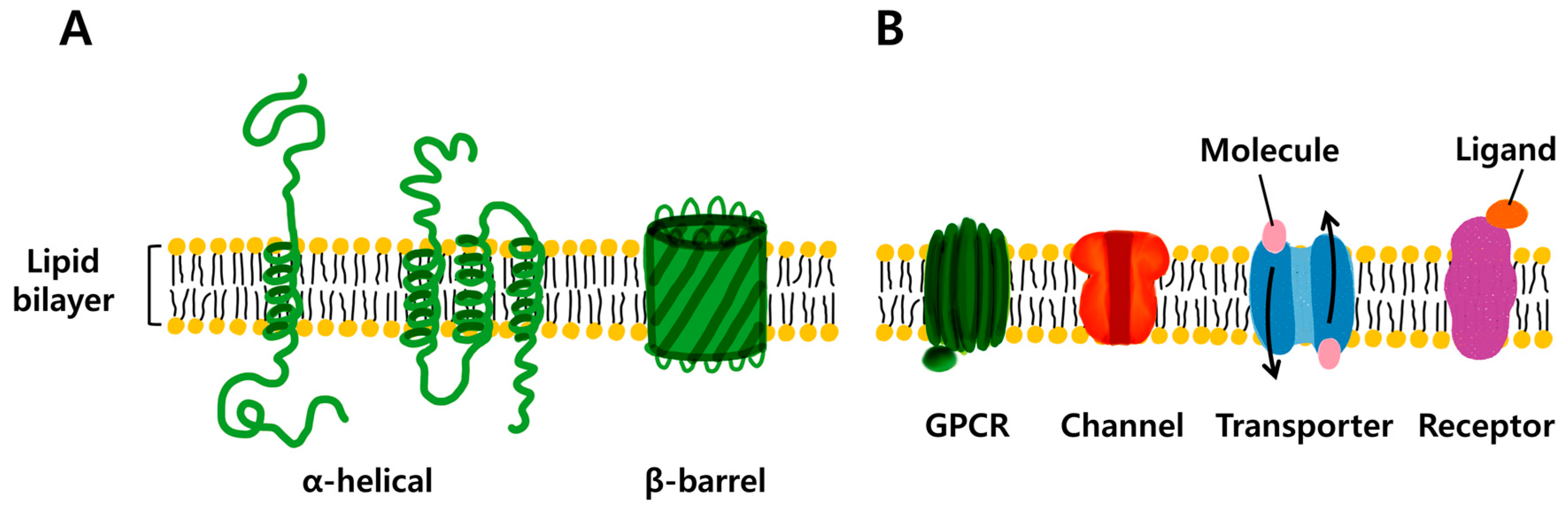
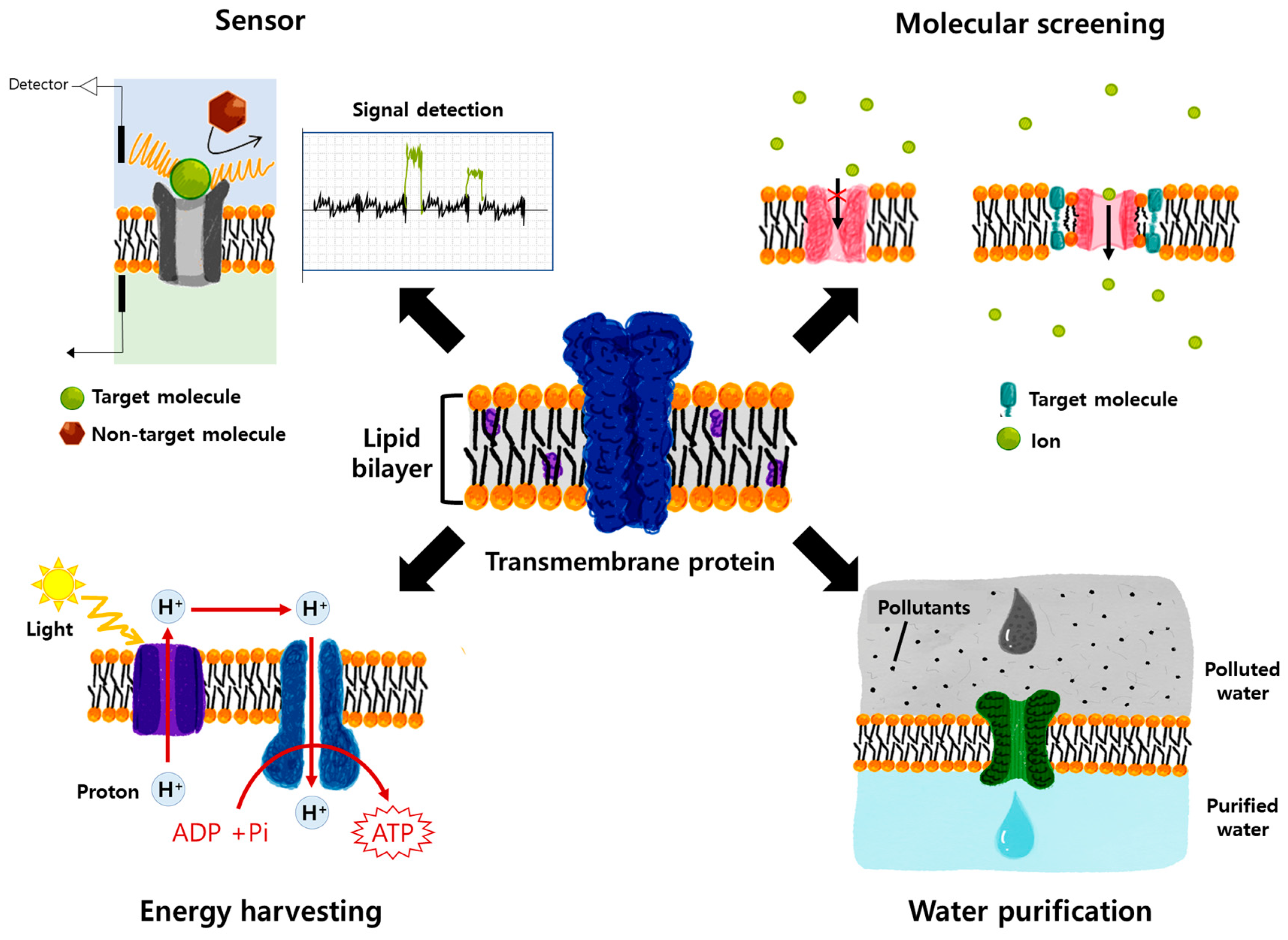
2. Potential Applications of Transmembrane Proteins
2.1. Membrane Protein (MP)-Based Biosensor System
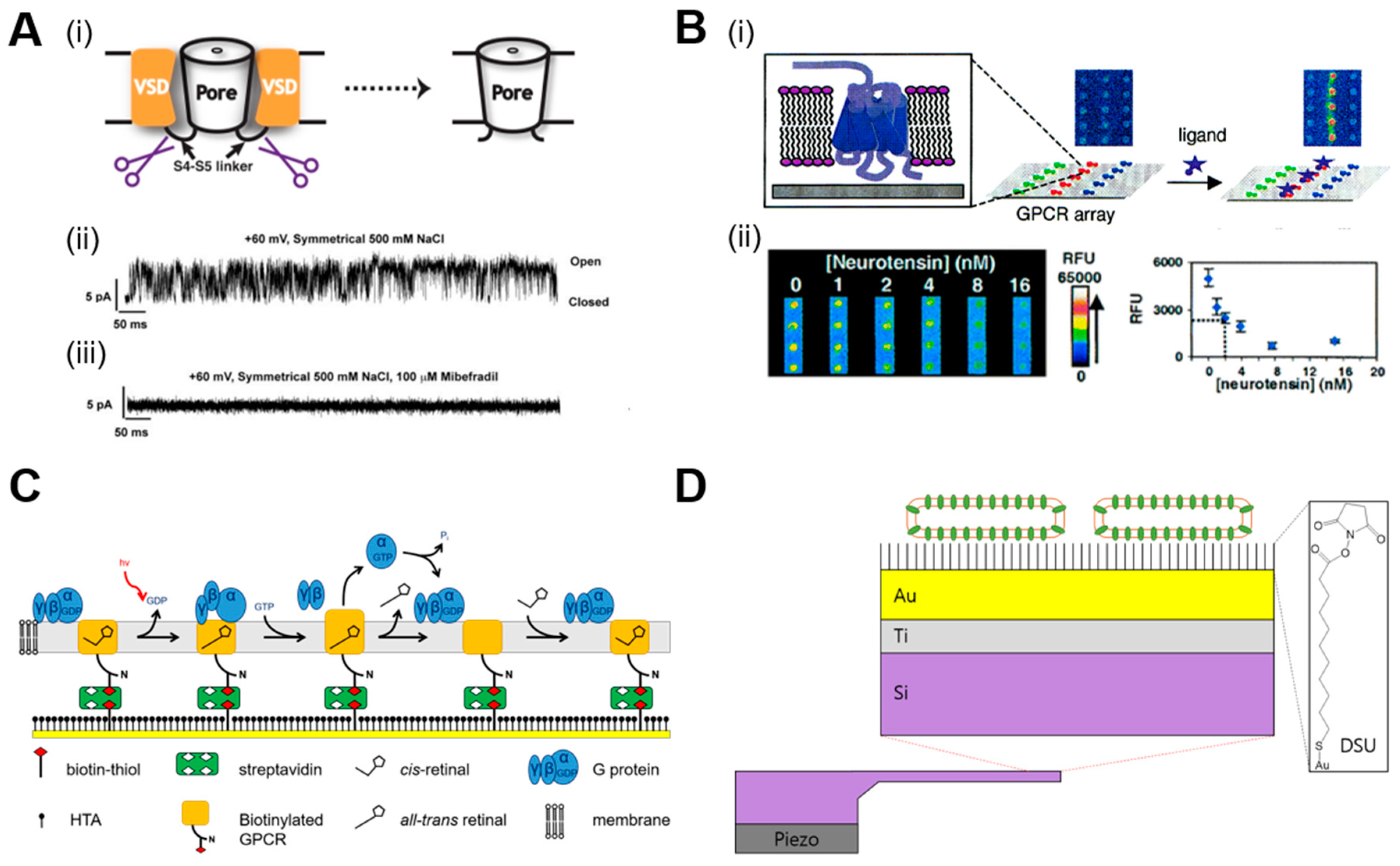
2.1.1. Biological Nanopore Sensors
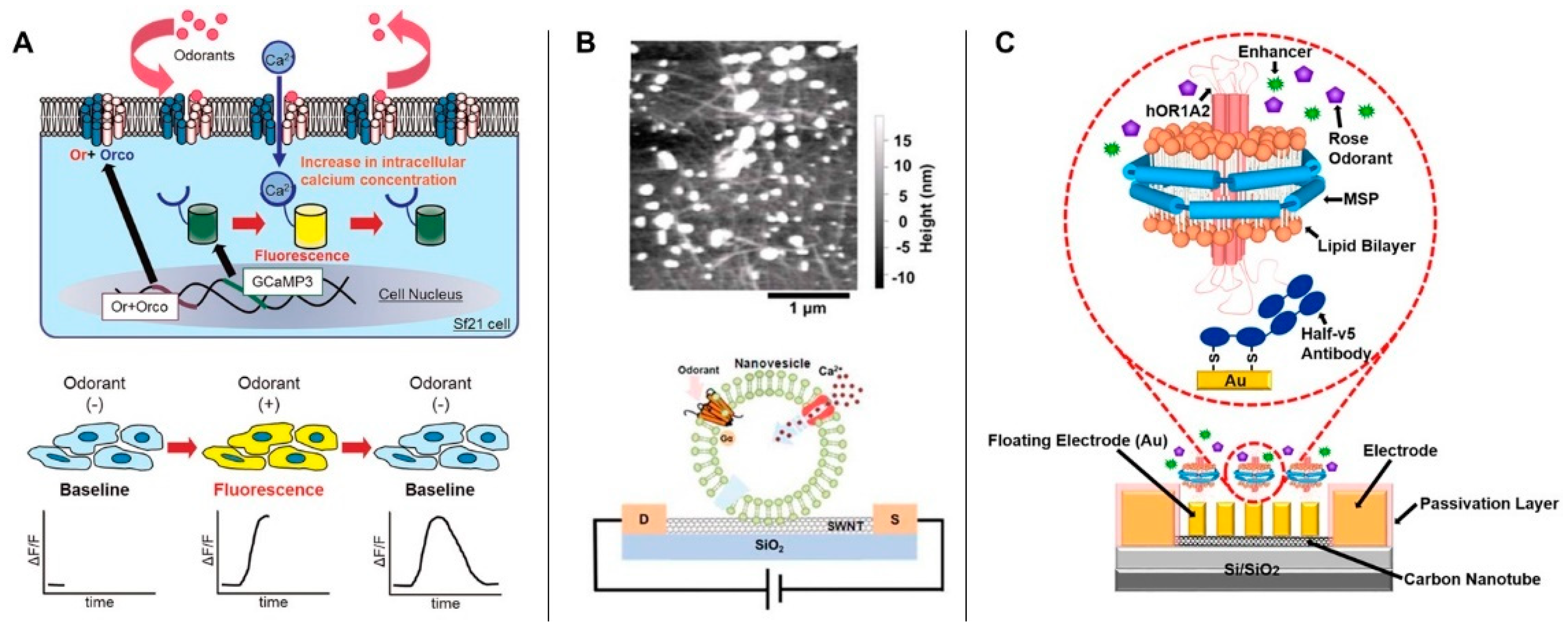
2.1.2. Target-Specific Receptor-Mediated Biosensors
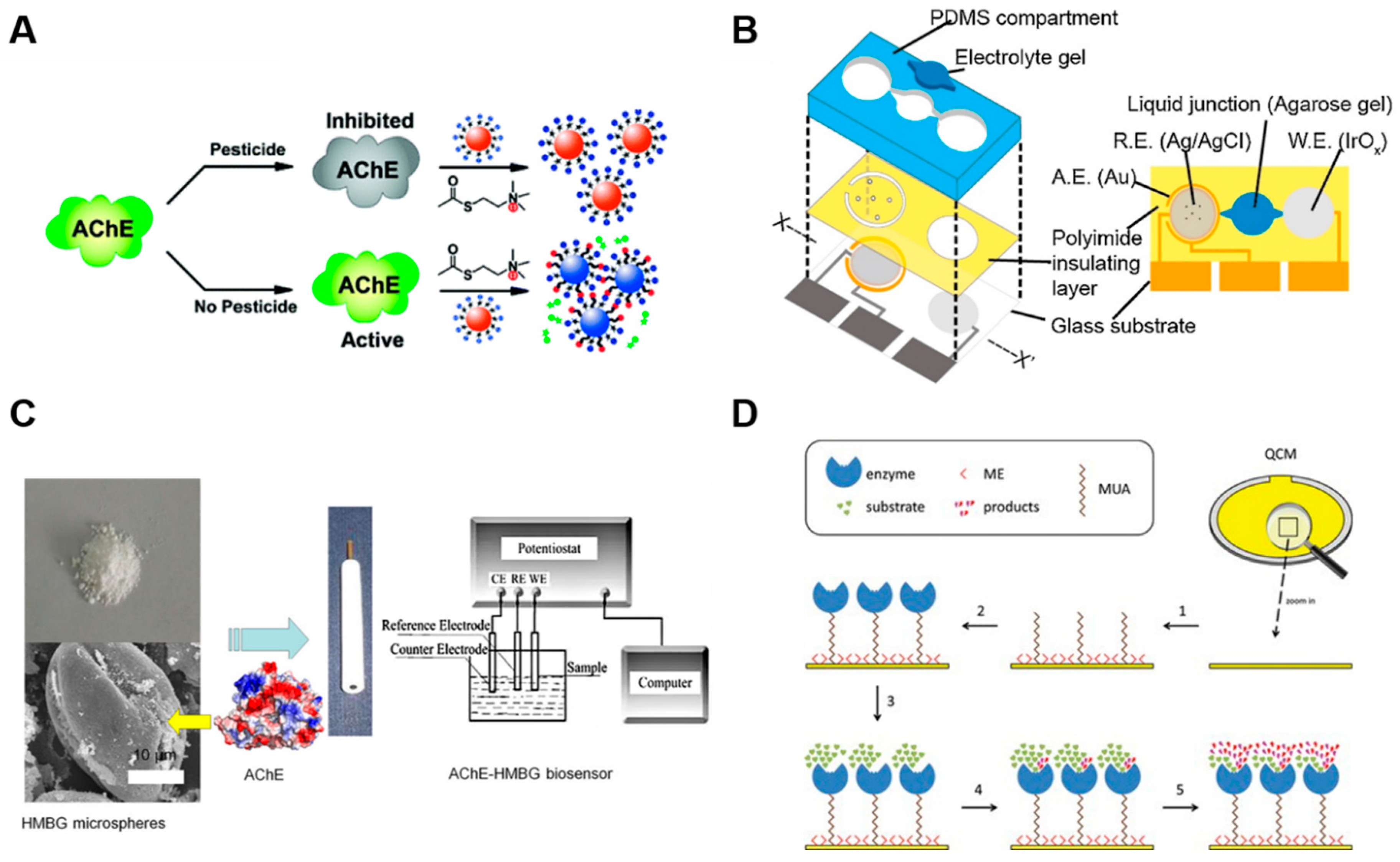
2.1.3. Acetylcholinesterase-Based Sensors
2.2. Drug and Membrane/Membrane Protein Interaction Studies
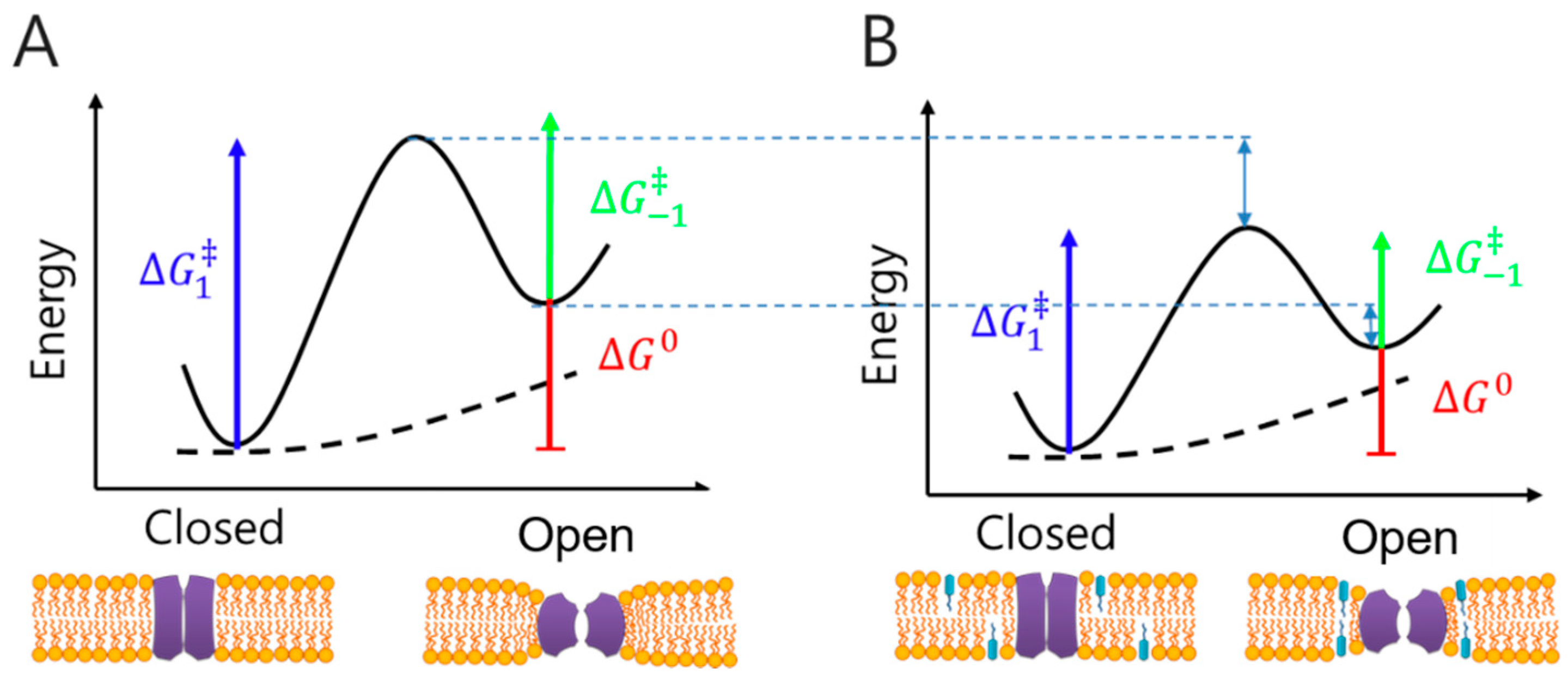
2.2.1. Small Molecule and Transmembrane Protein Interaction Studies
2.2.2. Small Molecule and Membrane/Membrane Protein Interaction Studies
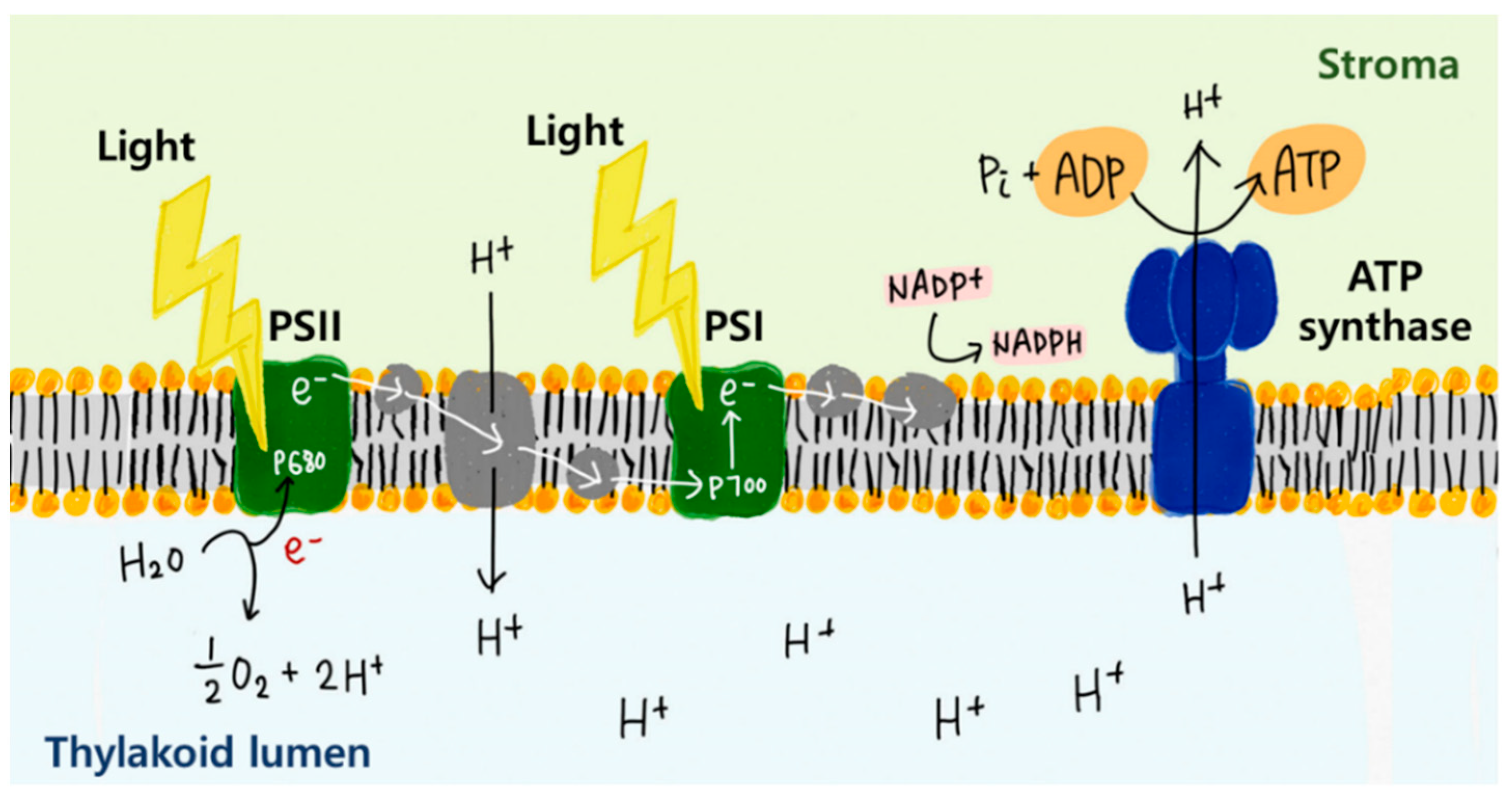
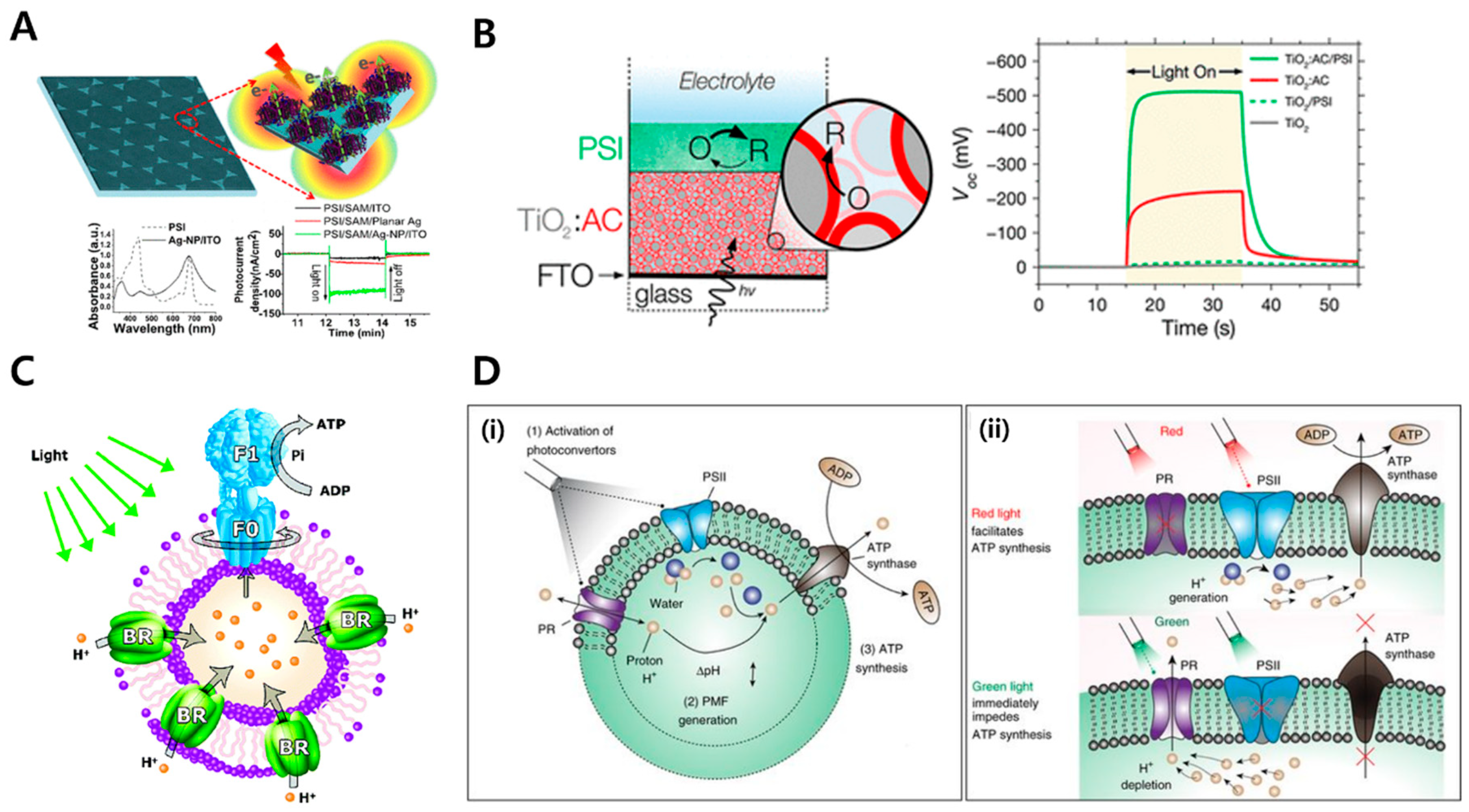
2.3. Energy Harvesting Using Membrane Proteins
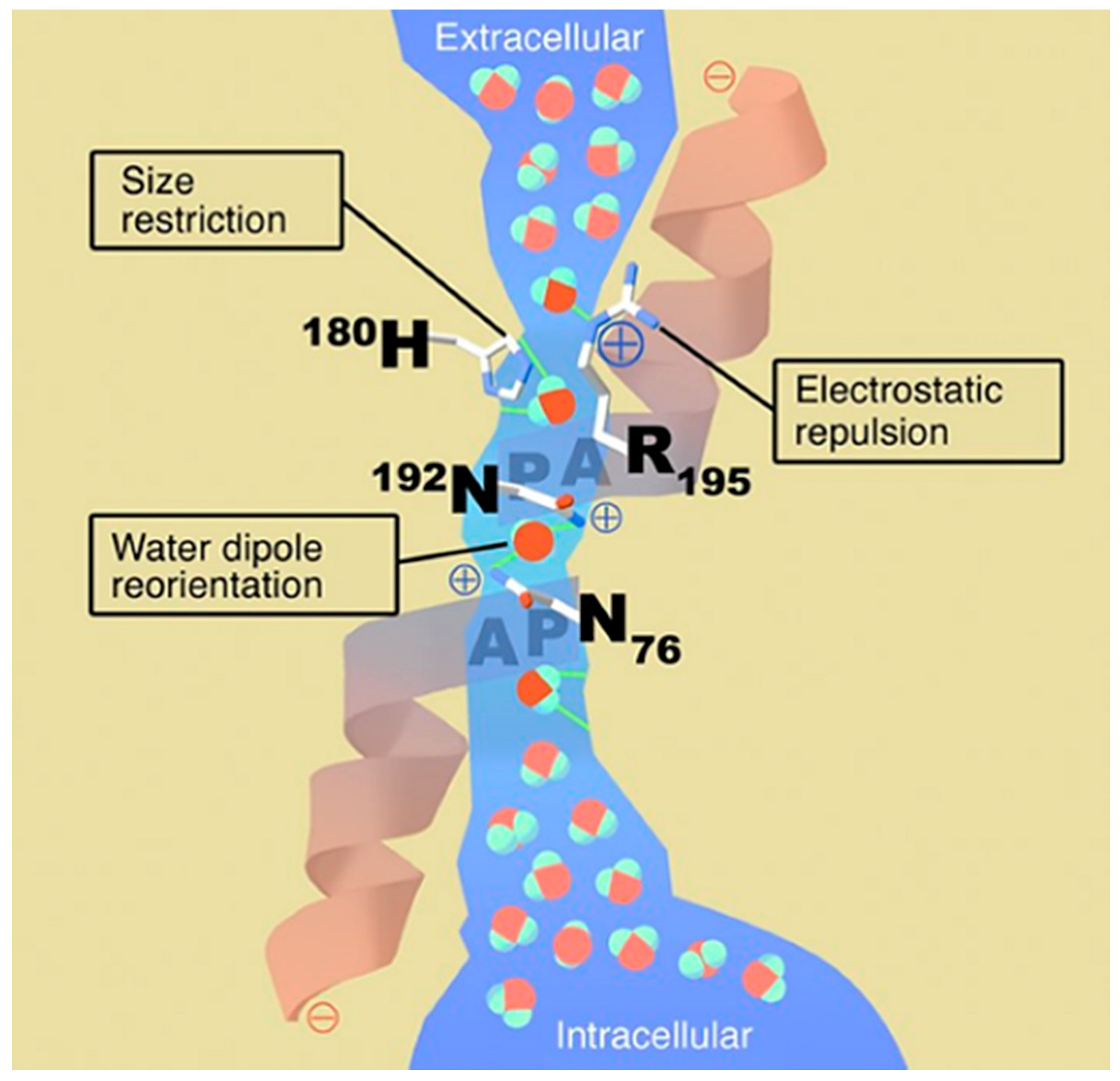
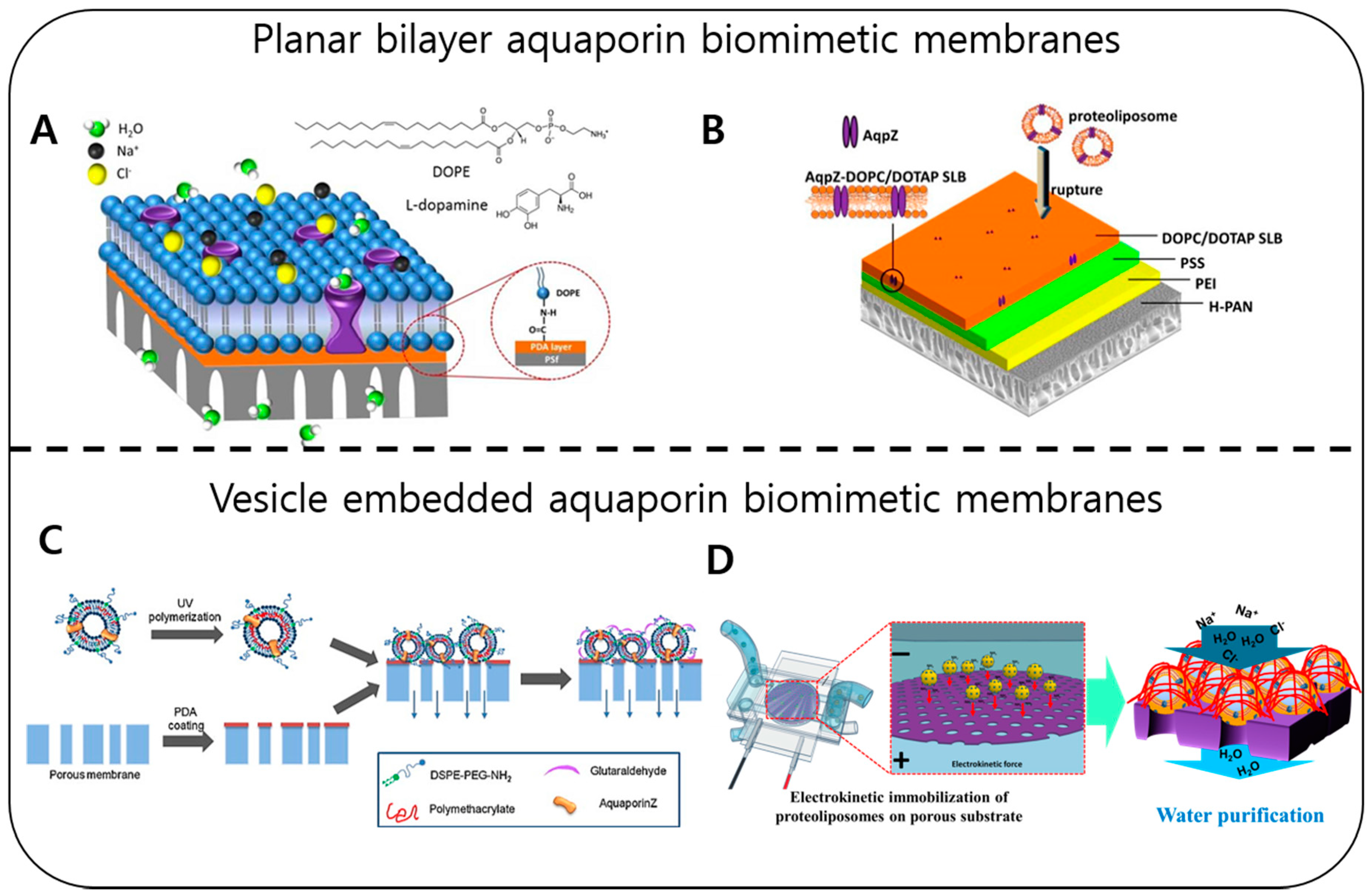
2.4. Water Purification with Membrane Proteins (Water Channels)
3. Conclusions and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Rohloff, J.C.; Gelinas, A.D.; Jarvis, T.C.; Ochsner, U.A.; Schneider, D.J.; Gold, L.; Janjic, N. Nucleic acid ligands with protein-like side chains: Modified aptamers and their use as diagnostic and therapeutic agents. Mol. Ther. Nucleic Acids 2014, 3, e201. [Google Scholar] [CrossRef]
- Nakayama, E.; Yokoyama, A.; Miyamoto, H.; Igarashi, M.; Kishida, N.; Matsuno, K.; Marzi, A.; Feldmann, H.; Ito, K.; Saijo, M.; et al. Enzyme-linked immunosorbent assay for detection of filovirus species-specific antibodies. Clin. Vaccine Immunol. 2010, 17, 1723–1728. [Google Scholar] [CrossRef] [PubMed]
- Isanga, J.; Mukunzi, D.; Chen, Y.; Suryoprabowo, S.; Liu, L.; Kuang, H.; Xu, C. Development of a monoclonal antibody assay and a lateral flow strip test for the detection of paromomycin residues in food matrices. Food Agric. Immunol. 2017, 28, 355–373. [Google Scholar] [CrossRef]
- Fricker, G.; Kromp, T.; Wendel, A.; Blume, A.; Zirkel, J.; Rebmann, H.; Setzer, C.; Quinkert, R.O.; Martin, F.; Müller-Goymann, C. Phospholipids and lipid-based formulations in oral drug delivery. Pharm. Res. 2010, 27, 1469–1486. [Google Scholar] [CrossRef] [PubMed]
- Arai, S.; Nakashima, K.; Tanino, T.; Ogino, C.; Kondo, A.; Fukuda, H. Production of biodiesel fuel from soybean oil catalyzed by fungus whole-cell biocatalysts in ionic liquids. Enzyme Microb. Technol. 2010, 46, 51–55. [Google Scholar] [CrossRef]
- Moraes, I.; Evans, G.; Sanchez-Weatherby, J.; Newstead, S.; Stewart, P.D.S. Membrane protein structure determination—The next generation. Biochim. Biophys. Acta Biomembr. 2014, 1838, 78–87. [Google Scholar] [CrossRef] [PubMed]
- Doerr, A. Membrane protein structures. Nat. Methods 2009, 6, 35. [Google Scholar] [CrossRef]
- Lin, S.H.; Guidotti, G. Chapter 35 Purification of Membrane Proteins. Methods Enzymol. 2009, 463, 619–629. [Google Scholar] [CrossRef]
- Braun, P.; LaBaer, J. High throughput protein production for functional proteomics. Trends Biotechnol. 2003, 21, 383–388. [Google Scholar] [CrossRef]
- Xiong, J. Essential Bioinformatics; Cambridge University Press: Cambridge, UK, 2013; Volume 53, ISBN 9788578110796. [Google Scholar]
- Michalik, M.; Orwick-Rydmark, M.; Habeck, M.; Alva, V.; Arnold, T.; Linke, D. An evolutionarily conserved glycine-tyrosine motif forms a folding core in outer membrane proteins. PLoS ONE 2017, 12, e0182016. [Google Scholar] [CrossRef]
- Mizrachi, D.; Chen, Y.; Liu, J.; Peng, H.M.; Ke, A.; Pollack, L.; Turner, R.J.; Auchus, R.J.; Delisa, M.P. Making water-soluble integral membrane proteins in vivo using an amphipathic protein fusion strategy. Nat. Commun. 2015, 6, 6826. [Google Scholar] [CrossRef] [PubMed]
- Fioroni, M.; Dworeck, T.; Rodriguez-Ropero, F. [Beta]-Barrel Channel Proteins as Tools in Nanotechnology: Biology, Basic Science and Advanced Applications; Springer: Berlin, Germay, 2014; ISBN 940077429X. [Google Scholar]
- Kim, Y.H.; Hang, L.; Cifelli, J.L.; Sept, D.; Mayer, M.; Yang, J. Frequency-Based Analysis of Gramicidin A Nanopores Enabling Detection of Small Molecules with Picomolar Sensitivity. Anal. Chem. 2018, 90, 1635–1642. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Lim, M.C.; Ryu, H.; Shim, J.; Kim, S.M.; Kim, Y.R.; Jeon, T.J. Nanopore based detection of: Bacillus thuringiensis HD-73 spores using aptamers and versatile DNA hairpins. Nanoscale 2018, 10, 11955–11961. [Google Scholar] [CrossRef] [PubMed]
- Fuwad, A.; Ryu, H.; Lee, J.H.; Kim, D.; Yoo, Y.E.; Kim, Y.R.; Kim, S.M.; Jeon, T.J. An electrokinetic approach to fabricating aquaporin biomimetic membranes for water purification. Desalination 2019, 452, 9–16. [Google Scholar] [CrossRef]
- Pamu, R.; Sandireddy, V.P.; Kalyanaraman, R.; Khomami, B.; Mukherjee, D. Plasmon-Enhanced Photocurrent from Photosystem I Assembled on Ag Nanopyramids. J. Phys. Chem. Lett. 2018, 9, 970–977. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.T.; Armbruster, M.E.; Gargye, A.; Cliffel, D.E.; Jennings, G.K. Photosystem I Multilayer Films for Photovoltage Enhancement in Natural Dye-Sensitized Solar Cells. ACS Appl. Energy Mater. 2018, 1, 301–305. [Google Scholar] [CrossRef]
- Chio, H.-J.; Montemagno, C.D. Artificial Organelle: ATP Synthesis from Cellular Mimetic Polymersomes. Nano Lett. 2005, 5, 2538–2542. [Google Scholar] [CrossRef]
- Lee, K.Y.; Park, S.-J.; Lee, K.A.; Kim, S.-H.; Kim, H.; Meroz, Y.; Mahadevan, L.; Jung, K.-H.; Ahn, T.K.; Parker, K.K.; et al. Photosynthetic artificial organelles sustain and control ATP-dependent reactions in a protocellular system. Nat. Biotechnol. 2018, 36, 530–535. [Google Scholar] [CrossRef]
- Misawa, N.; Osaki, T.; Takeuchi, S. Membrane protein-based biosensors. J. R. Soc. Interface 2018, 15, 20170952. [Google Scholar] [CrossRef]
- Dekker, C. Solid-state nanopores. Nat. Nanotechnol. 2007, 2, 209–215. [Google Scholar] [CrossRef]
- Venkatesan, B.M.; Bashir, R. Nanopore sensors for nucleic acid analysis. Nat. Nanotechnol. 2011, 6, 615–624. [Google Scholar] [CrossRef] [PubMed]
- Howorka, S. Building membrane nanopores. Nat. Nanotechnol. 2017, 12, 619–630. [Google Scholar] [CrossRef] [PubMed]
- Ying, Y.-L.; Cao, C.; Long, Y.-T. Single molecule analysis by biological nanopore sensors. Analyst 2014, 139, 3826–3835. [Google Scholar] [CrossRef] [PubMed]
- Kasianowicz, J.J.; Brandin, E.; Branton, D.; Deamer, D.W. Characterization of individual polynucleotide molecules using a membrane channel. Proc. Natl. Acad. Sci. USA 1996, 93, 13770–13773. [Google Scholar] [CrossRef] [PubMed]
- Mathe, J.; Aksimentiev, A.; Nelson, D.R.; Schulten, K.; Meller, A. Orientation discrimination of single-stranded DNA inside the α-hemolysin membrane channel. Proc. Natl. Acad. Sci. USA 2005, 102, 12377–12382. [Google Scholar] [CrossRef]
- Akeson, M.; Branton, D.; Kasianowicz, J.J.; Brandin, E.; Deamer, D.W. Microsecond Time-Scale Discrimination Among Polycytidylic Acid, Polyadenylic Acid, and Polyuridylic Acid as Homopolymers or as Segments Within Single RNA Molecules. Biophys. J. 1999, 77, 3227–3233. [Google Scholar] [CrossRef]
- Rodrigues, C.G.; Machado, D.C.; Chevtchenko, S.F.; Krasilnikov, O.V. Mechanism of KCl enhancement in detection of nonionic polymers by nanopore sensors. Biophys. J. 2008, 95, 5186–5192. [Google Scholar] [CrossRef]
- Ayub, M.; Stoddart, D.; Bayley, H. Nucleobase Recognition by Truncated α-Hemolysin Pores. ACS Nano 2015, 9, 7895–7903. [Google Scholar] [CrossRef]
- Yao, J.; Liu, B.; Qin, F. Rapid temperature jump by infrared diode laser irradiation for patch-clamp studies. Biophys. J. 2009, 96, 3611–3619. [Google Scholar] [CrossRef]
- Angevine, C.E.; Seashols-Williams, S.J.; Reiner, J.E. Infrared Laser Heating Applied to Nanopore Sensing for DNA Duplex Analysis. Anal. Chem. 2016, 88, 2645–2651. [Google Scholar] [CrossRef]
- Johnson, R.P.; Fleming, A.M.; Perera, R.T.; Burrows, C.J.; White, H.S. Dynamics of a DNA Mismatch Site Held in Confinement Discriminate Epigenetic Modifications of Cytosine. J. Am. Chem. Soc. 2017, 139, 2750–2756. [Google Scholar] [CrossRef]
- Johnson, R.P.; Fleming, A.M.; Burrows, C.J.; White, H.S. Effect of an Electrolyte Cation on Detecting DNA Damage with the Latch Constriction of α-Hemolysin. J. Phys. Chem. Lett. 2014, 5, 3781–3786. [Google Scholar] [CrossRef] [PubMed]
- Johnson, R.P.; Fleming, A.M.; Jin, Q.; Burrows, C.J.; White, H.S. Temperature and electrolyte optimization of the α-hemolysin latch sensing zone for detection of base modification in double-stranded DNA. Biophys. J. 2014, 107, 924–931. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Tao, C.; Chien, M.; Hellner, B.; Balijepalli, A.; Robertson, J.W.F.; Li, Z.; Russo, J.J.; Reiner, J.E.; Kasianowicz, J.J.; et al. PEG-Labeled Nucleotides and Nanopore Detection for Single Molecule DNASequencing by Synthesis. Sci. Rep. 2012, 2, 684. [Google Scholar] [CrossRef] [PubMed]
- Fuller, C.W.; Kumar, S.; Porel, M.; Chien, M.; Bibillo, A.; Stranges, P.B.; Dorwart, M.; Tao, C.; Li, Z.; Guo, W.; et al. Real-time single-molecule electronic DNA sequencing by synthesis using polymer-tagged nucleotides on a nanopore array. Proc. Natl. Acad. Sci. USA 2016, 113, 5233–5238. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Han, Y.; Zhou, S.; Guan, X. Real-time label-free measurement of HIV-1 protease activity by nanopore analysis. Biosens. Bioelectron. 2014, 62, 158–162. [Google Scholar] [CrossRef] [PubMed]
- Kukwikila, M.; Howorka, S. Nanopore-Based Electrical and Label-Free Sensing of Enzyme Activity in Blood Serum. Anal. Chem. 2015, 87, 9149–9154. [Google Scholar] [CrossRef]
- Rotem, D.; Jayasinghe, L.; Salichou, M.; Bayley, H. Protein detection by nanopores equipped with aptamers. J. Am. Chem. Soc. 2012, 134, 2781–2787. [Google Scholar] [CrossRef] [PubMed]
- Kawano, R.; Osaki, T.; Sasaki, H.; Takinoue, M.; Yoshizawa, S.; Takeuchi, S. Rapid detection of a cocaine-binding aptamer using biological nanopores on a chip. J. Am. Chem. Soc. 2011, 133, 8474–8477. [Google Scholar] [CrossRef]
- Zhou, S.; Wang, L.; Chen, X.; Guan, X. Label-Free Nanopore Single-Molecule Measurement of Trypsin Activity. ACS Sens. 2016, 1, 607–613. [Google Scholar] [CrossRef]
- Cao, C.; Ying, Y.-L.; Hu, Z.-L.; Liao, D.-F.; Tian, H.; Long, Y.-T. Discrimination of oligonucleotides of different lengths with a wild-type aerolysin nanopore. Nat. Nanotechnol. 2016, 11, 713–718. [Google Scholar] [CrossRef]
- Franceschini, L.; Soskine, M.; Biesemans, A.; Maglia, G. A nanopore machine promotes the vectorial transport of DNA across membranes. Nat. Commun. 2013, 4, 2415. [Google Scholar] [CrossRef]
- Franceschini, L.; Brouns, T.; Willems, K.; Carlon, E.; Maglia, G. DNA Translocation through Nanopores at Physiological Ionic Strengths Requires Precise Nanoscale Engineering. ACS Nano 2016, 10, 8394–8402. [Google Scholar] [CrossRef]
- Laszlo, A.H.; Derrington, I.M.; Brinkerhoff, H.; Langford, K.W.; Nova, I.C.; Samson, J.M.; Bartlett, J.J.; Pavlenok, M.; Gundlach, J.H.; Designed, J.H.G.; et al. Detection and mapping of 5-methylcytosine and 5-hydroxymethylcytosine with nanopore MspA. Proc. Natl. Acad. Sci. USA 2013, 110, 18904–18909. [Google Scholar] [CrossRef]
- Meller, A.; Nivon, L.; Brandin, E.; Golovchenko, J.; Branton, D. Rapid nanopore discrimination between single polynucleotide molecules. Proc. Natl. Acad. Sci. USA 2000, 97, 1079–1084. [Google Scholar] [CrossRef]
- Shi, W.; Friedman, A.K.; Baker, L.A. Nanopore Sensing. Anal. Chem. 2017, 89, 157–188. [Google Scholar] [CrossRef]
- Wang, Y.; Gu, L.Q.; Tian, K. The aerolysin nanopore: from peptidomic to genomic applications. Nanoscale 2018, 10, 13857–13866. [Google Scholar] [CrossRef]
- Deamer, D.; Akeson, M.; Branton, D. Three decades of nanopore sequencing. Nat. Biotechnol. 2016, 34, 518–524. [Google Scholar] [CrossRef]
- Hall, A.R.; Scott, A.; Rotem, D.; Mehta, K.K.; Bayley, H.; Dekker, C. Hybrid pore formation by directed insertion of α-haemolysin into solid-state nanopores. Nat. Nanotechnol. 2010, 5, 874–877. [Google Scholar] [CrossRef]
- Barati Farimani, A.; Dibaeinia, P.; Aluru, N.R. DNA Origami–Graphene Hybrid Nanopore for DNA Detection. ACS Appl. Mater. Interfaces 2017, 9, 92–100. [Google Scholar] [CrossRef]
- Cherf, G.M.; Lieberman, K.R.; Rashid, H.; Lam, C.E.; Karplus, K.; Akeson, M. Automated forward and reverse ratcheting of DNA in a nanopore at 5-Å precision. Nat. Biotechnol. 2012, 30, 344–348. [Google Scholar] [CrossRef]
- Majd, S.; Yusko, E.C.; Billeh, Y.N.; Macrae, M.X.; Yang, J.; Mayer, M. Applications of biological pores in nanomedicine, sensing, and nanoelectronics. Curr. Opin. Biotechnol. 2010, 21, 439–476. [Google Scholar] [CrossRef]
- Ying, Y.-L.; Zhang, J.; Gao, R.; Long, Y.-T. Nanopore-Based Sequencing and Detection of Nucleic Acids. Angew. Chem. Int. Ed. 2013, 52, 13154–13161. [Google Scholar] [CrossRef]
- Wang, S.; Ji, Z.; Yan, E.; Haque, F.; Guo, P. Three-step channel conformational changes common to DNA packaging motors of bacterial viruses T3, T4, SPP1, and Phi29. Virology 2017, 500, 285–291. [Google Scholar] [CrossRef]
- Haque, F.; Guo, P. Bacteriophage Phi29 DNA-Packaging Motor for Translocation and Sensing of Double-Stranded DNA. In Nanopores; Springer: Boston, MA, USA, 2011; pp. 129–150. ISBN 978-1-4419-8251-3. [Google Scholar]
- Su, C.Y.; Menuz, K.; Carlson, J.R. Olfactory Perception: Receptors, Cells, and Circuits. Cell 2009, 139, 45–59. [Google Scholar] [CrossRef]
- Laska, M.; Galizia, C.G.; Giurfa, M.; Menzel, R. Olfactory Discrimination Ability and Odor Structure–Activity Relationships in Honeybees. Chem. Senses 1999, 24, 429–438. [Google Scholar] [CrossRef]
- Mombaerts, P. Seven-Transmembrane Proteins as Odorant and Chemosensory Receptors. Science 1999, 286, 707–711. [Google Scholar] [CrossRef]
- Du, L.; Wu, C.; Peng, H.; Zhao, L.; Huang, L.; Wang, P. Bioengineered olfactory sensory neuron-based biosensor for specific odorant detection. Biosens. Bioelectron. 2013, 40, 401–406. [Google Scholar] [CrossRef]
- Sato, K.; Takeuchi, S. Chemical Vapor Detection Using a Reconstituted Insect Olfactory Receptor Complex. Angew. Chem. Int. Ed. 2014, 53, 11798–11802. [Google Scholar] [CrossRef] [PubMed]
- Termtanasombat, M.; Mitsuno, H.; Misawa, N.; Yamahira, S.; Sakurai, T.; Yamaguchi, S.; Nagamune, T.; Kanzaki, R. Cell-Based Odorant Sensor Array for Odor Discrimination Based on Insect Odorant Receptors. J. Chem. Ecol. 2016, 42, 716–724. [Google Scholar] [CrossRef]
- Mitsuno, H.; Sakurai, T.; Namiki, S.; Mitsuhashi, H.; Kanzaki, R. Novel cell-based odorant sensor elements based on insect odorant receptors. Biosens. Bioelectron. 2015, 65, 287–294. [Google Scholar] [CrossRef]
- Lee, J.Y.; Ko, H.J.; Lee, S.H.; Park, T.H. Cell-based measurement of odorant molecules using surface plasmon resonance. Enzyme Microb. Technol. 2006, 39, 375–380. [Google Scholar] [CrossRef]
- Ahn, J.H.; Lim, J.H.; Park, J.; Oh, E.H.; Son, M.; Hong, S.; Park, T.H. Screening of target-specific olfactory receptor and development of olfactory biosensor for the assessment of fungal contamination in grain. Sens. Actuators B Chem. 2015, 210, 9–16. [Google Scholar] [CrossRef]
- Lee, M.; Yang, H.; Kim, D.; Yang, M.; Park, T.H.; Hong, S. Human-like smelling of a rose scent using an olfactory receptor nanodisc-based bioelectronic nose. Sci. Rep. 2018, 8, 13945. [Google Scholar] [CrossRef]
- Kim, T.H.; Lee, S.H.; Lee, J.; Song, H.S.; Oh, E.H.; Park, T.H.; Hong, S. Single-carbon-atomic-resolution detection of odorant molecules using a human olfactory receptor-based bioelectronic nose. Adv. Mater. 2009, 21, 91–94. [Google Scholar] [CrossRef]
- Loutfi, A.; Coradeschi, S.; Kumar, G.; Shankar, P.; Bosco, J.; Rayappan, B. Electronic noses for food quality: A review. J. Food Eng. 2015, 144, 103–111. [Google Scholar] [CrossRef]
- Gardner, J.W.; Vincent, T.A. Energy Expenditure. Sensors 2016, 16, 947. [Google Scholar] [CrossRef] [PubMed]
- Capelli, L.; Sironi, S.; Del Rosso, R. Electronic noses for environmental monitoring applications. Sensors 2014, 14, 19979–20007. [Google Scholar] [CrossRef]
- Pohanka, M. Cholinesterases in Biorecognition and Biosensors Construction: A Review. Anal. Lett. 2013, 46, 1849–1868. [Google Scholar] [CrossRef]
- Marrs, T.C.; Maynard, R.L. Neurotranmission systems as targets for toxicants: A review. Cell Biol. Toxicol. 2013, 29, 381–396. [Google Scholar] [CrossRef]
- White, B.J.; Andrew Legako, J.; James Harmon, H. Spectrophotometric detection of cholinesterase inhibitors with an integrated acetyl-/butyrylcholinesterase surface. Sens. Actuators B Chem. 2003, 89, 107–111. [Google Scholar] [CrossRef]
- Chang, J.; Li, H.; Hou, T.; Li, F. Paper-based fluorescent sensor for rapid naked-eye detection of acetylcholinesterase activity and organophosphorus pesticides with high sensitivity and selectivity. Biosens. Bioelectron. 2016, 86, 971–977. [Google Scholar] [CrossRef] [PubMed]
- Kostelnik, A.; Cegan, A.; Pohanka, M. Acetylcholinesterase Inhibitors Assay Using Colorimetric pH Sensitive Strips and Image Analysis by a Smartphone. Int. J. Anal. Chem. 2017, 2017, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Guo, L.; Bao, Y.; Xie, J. A simple, label-free AuNPs-based colorimetric ultrasensitive detection of nerve agents and highly toxic organophosphate pesticide. Biosens. Bioelectron. 2011, 28, 152–157. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Chen, W.; Wei, J.; Li, X.; Wang, Z.; Jiang, X. A Highly Sensitive, Dual-Readout Assay Based on Gold Nanoparticles for Organophosphorus and Carbamate Pesticides. Anal. Chem. 2012, 84, 4185–4191. [Google Scholar] [CrossRef] [PubMed]
- Hai, N.N.; Chinh, V.D.; Chi, T.K.; Thuy, U.T.D.; Nghia, N.X.; Cao, D.T.; Nga, P.T. Optical Detection of the Pesticide by Functionalized Quantum Dots as Fluorescence-Based Biosensor. Key Eng. Mater. 2011, 495, 314–318. [Google Scholar] [CrossRef]
- Zhang, J.; Luo, A.; Liu, P.; Wei, S.; Wang, G.; Wei, S. Detection of Organophosphorus Pesticides Using Potentiometric Enzymatic Membrane Biosensor Based on Methylcellulose Immobilization. Anal. Sci. 2009, 25, 511–515. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yokokawa, M.; Satake, T.; Suzuki, H. A micro IrOx potentiometric sensor for direct determination of organophosphate pesticides. Sens. Actuators B Chem. 2015, 220, 859–863. [Google Scholar] [CrossRef]
- Lv, Z.; Luo, R.; Xi, L.; Chen, Y.; Wang, H. An Amperometric Acetylcholinesterase Sensor Based on the Bio-templated Synthesis of Hierarchical Mesoporous Bioactive Glass Microspheres. J. Electron. Mater. 2017, 46, 6578–6587. [Google Scholar] [CrossRef]
- Wang, K.; Li, H.-N.; Wu, J.; Ju, C.; Yan, J.-J.; Liu, Q.; Qiu, B. TiO2-decorated graphene nanohybrids for fabricating an amperometric acetylcholinesterase biosensor. Analyst 2011, 136, 3349. [Google Scholar] [CrossRef]
- Ivanov, A.N.; Younusov, R.R.; Evtugyn, G.A.; Arduini, F.; Moscone, D.; Palleschi, G. Acetylcholinesterase biosensor based on single-walled carbon nanotubes—Co phtalocyanine for organophosphorus pesticides detection. Talanta 2011, 85, 216–221. [Google Scholar] [CrossRef]
- Cooper, M.A.; Singleton, V.T. A survey of the 2001 to 2005 quartz crystal microbalance biosensor literature: Applications of acoustic physics to the analysis of biomolecular interactions. J. Mol. Recognit. 2007, 20, 154–184. [Google Scholar] [CrossRef]
- Bueno, P.R.; Gonçalves, L.M.; dos Santos, F.C.; dos Santos, M.L.; Barros, A.A.; Faria, R.C. Electrogravimetric Analysis by Quartz-Crystal Microbalance on the Consumption of the Neurotransmitter Acetylcholine by Acetylcholinesterase. Anal. Lett. 2013, 46, 258–265. [Google Scholar] [CrossRef]
- Hossain, S.M.Z.; Luckham, R.E.; Smith, A.M.; Lebert, J.M.; Davies, L.M.; Pelton, R.H.; Filipe, C.D.M.; Brennan, J.D. Development of a Bioactive Paper Sensor for Detection of Neurotoxins Using Piezoelectric Inkjet Printing of Sol−Gel-Derived Bioinks. Anal. Chem. 2009, 81, 5474–5483. [Google Scholar] [CrossRef]
- Arduini, F.; Forchielli, M.; Amine, A.; Neagu, D.; Cacciotti, I.; Nanni, F.; Moscone, D.; Palleschi, G. Screen-printed biosensor modified with carbon black nanoparticles for the determination of paraoxon based on the inhibition of butyrylcholinesterase. Microchim. Acta 2015, 182, 643–651. [Google Scholar] [CrossRef]
- Cho, Y.A.; Lee, H.S.; Cha, G.S.; Lee, Y.T. Fabrication of butyrylcholinesterase sensor using polyurethane-based ion-selective membranes. Biosens. Bioelectron. 1999, 14, 435–438. [Google Scholar] [CrossRef]
- Pohanka, M. Butyrylcholinesterase as a biochemical marker. Bratisl. Med. J. 2013, 114, 726–734. [Google Scholar] [CrossRef]
- Andreescu, S.; Marty, J.-L. Twenty years research in cholinesterase biosensors: From basic research to practical applications. Biomol. Eng. 2006, 23, 1–15. [Google Scholar] [CrossRef]
- Periasamy, A.P.; Umasankar, Y.; Chen, S.-M.; Periasamy, A.P.; Umasankar, Y.; Chen, S.-M. Nanomaterials-Acetylcholinesterase Enzyme Matrices for Organophosphorus Pesticides Electrochemical Sensors: A Review. Sensors 2009, 9, 4034–4055. [Google Scholar] [CrossRef]
- Badawy, M.E.I.; El-Aswad, A.F. Bioactive paper sensor based on the acetylcholinesterase for the rapid detection of organophosphate and carbamate pesticides. Int. J. Anal. Chem. 2014, 2014, 536823. [Google Scholar] [CrossRef]
- Timur, S.; Telefoncu, A. Acetylcholinesterase (AChE) Electrodes Based on Gelatin and Chitosan Matrices for the Pesticide Detection. Artif. Cells Blood Substit. Biotechnol. 2004, 32, 427–442. [Google Scholar] [CrossRef]
- Cui, H.-F.; Wu, W.-W.; Li, M.-M.; Song, X.; Lv, Y.; Zhang, T.-T. A highly stable acetylcholinesterase biosensor based on chitosan-TiO2-graphene nanocomposites for detection of organophosphate pesticides. Biosens. Bioelectron. 2018, 99, 223–229. [Google Scholar] [CrossRef] [PubMed]
- Bull, S.C.; Doig, A.J. Properties of protein drug target classes. PLoS ONE 2015, 10, e0117955. [Google Scholar] [CrossRef]
- Bleicher, K.H.; Böhm, H.-J.; Müller, K.; Alanine, A.I. Hit and lead generation: Beyond high-throughput screening. Nat. Rev. Drug Discov. 2003, 2, 369–378. [Google Scholar] [CrossRef] [PubMed]
- Brink, C.B.; Harvey, B.H.; Bodenstein, J.; Venter, D.P.; Oliver, D.W. Recent advances in drug action and therapeutics: Relevance of novel concepts in G-protein-coupled receptor and signal transduction pharmacology. Br. J. Clin. Pharmacol. 2004, 57, 373–387. [Google Scholar] [CrossRef] [PubMed]
- Sriram, K.; Insel, P.A. GPCRs as targets for approved drugs: How many targets and how many drugs? Mol. Pharmacol. 2018, 93. [Google Scholar] [CrossRef] [PubMed]
- Terstappen, G.C.; Reggiani, A. In silico research in drug discovery. Trends Pharmacol. Sci. 2001, 22, 23–26. [Google Scholar] [CrossRef]
- Sethi, R.S. Transducer aspects of biosensors. Biosens. Bioelectron. 1994, 9, 243–264. [Google Scholar] [CrossRef]
- Hamill, O.P.; Marty, A.; Neher, E.; Sakmann, B.; Sigworth, F.J. Improved patch-clamp techniques for high-resolution current recording from cells and cell-free membrane patches. Pflüg. Arch. Eur. J. Physiol. 1981, 391, 85–100. [Google Scholar] [CrossRef]
- Herold, K.F.; Sanford, R.L.; Lee, W.; Schultz, M.F.; Ingólfsson, H.I.; Andersen, O.S.; Hemmings, H.C. Volatile anesthetics inhibit sodium channels without altering bulk lipid bilayer properties. J. Gen. Physiol. 2014, 144, 545–560. [Google Scholar] [CrossRef]
- Moyer, B.D.; Loffing, J.; Schwiebert, E.M.; Loffing-Cueni, D.; Halpin, P.A.; Karlson, K.H.; Ismailov, I.I.; Guggino, W.B.; Langford, G.M.; Stanton, B.A. Membrane trafficking of the cystic fibrosis gene product, cystic fibrosis transmembrane conductance regulator, tagged with green fluorescent protein in madin-darby canine kidney cells. J. Biol. Chem. 1998, 273, 21759–21768. [Google Scholar] [CrossRef] [PubMed]
- Shaya, D.; Kreir, M.; Robbins, R.A.; Wong, S.; Hammon, J.; Bruggemann, A.; Minor, D.L. Voltage-gated sodium channel (NaV) protein dissection creates a set of functional pore-only proteins. Proc. Natl. Acad. Sci. USA 2011, 108, 12313–12318. [Google Scholar] [CrossRef] [PubMed]
- Hirano-Iwata, A.; Ishinari, Y.; Yoshida, M.; Araki, S.; Tadaki, D.; Miyata, R.; Ishibashi, K.; Yamamoto, H.; Kimura, Y.; Niwano, M. Reconstitution of Human Ion Channels into Solvent-free Lipid Bilayers Enhanced by Centrifugal Forces. Biophys. J. 2016, 110, 2207–2215. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Frutos, A.G.; Lahiri, J. Membrane protein microarrays. J. Am. Chem. Soc. 2002, 124, 2394–2395. [Google Scholar] [CrossRef] [PubMed]
- Dockendorff, C.; Gandhi, D.M.; Kimball, I.H.; Eum, K.S.; Rusinova, R.; Ingólfsson, H.I.; Kapoor, R.; Peyear, T.; Dodge, M.W.; Martin, S.F.; et al. Synthetic Analogues of the Snail Toxin 6-Bromo-2-mercaptotryptamine Dimer (BrMT) Reveal That Lipid Bilayer Perturbation Does Not Underlie Its Modulation of Voltage-Gated Potassium Channels. Biochemistry 2018, 57, 2733–2743. [Google Scholar] [CrossRef]
- Bieri, C.; Ernst, O.P.; Heyse, S.; Hofmann, K.P.; Vogel, H. Micropatterned immobilization of a G protein-coupled receptor and direct detection of G protein activation. Nat. Biotechnol. 1999, 17, 1105–1108. [Google Scholar] [CrossRef]
- Patching, S.G. Surface plasmon resonance spectroscopy for characterisation of membrane protein–ligand interactions and its potential for drug discovery. Biochim. Biophys. Acta Biomembr. 2014, 1838, 43–55. [Google Scholar] [CrossRef] [PubMed]
- Braun, T.; Ghatkesar, M.K.; Backmann, N.; Grange, W.; Boulanger, P.; Letellier, L.; Lang, H.-P.; Bietsch, A.; Gerber, C.; Hegner, M. Quantitative time-resolved measurement of membrane protein–ligand interactions using microcantilever array sensors. Nat. Nanotechnol. 2009, 4, 179–185. [Google Scholar] [CrossRef]
- Pantoliano, M.W.; Petrella, E.C.; Kwasnoski, J.D.; Lobanov, V.S.; Myslik, J.; Graf, E.; Carver, T.; Asel, E.; Springer, B.A.; Lane, P.; et al. High-Density Miniaturized Thermal Shift Assays as a General Strategy for Drug Discovery. J. Biomol. Screen. 2001, 6, 429–440. [Google Scholar] [CrossRef]
- Maynard, J.A.; Lindquist, N.C.; Sutherland, J.N.; Lesuffleur, A.; Warrington, A.E.; Rodriguez, M.; Oh, S.-H. Surface plasmon resonance for high-throughput ligand screening of membrane-bound proteins. Biotechnol. J. 2009, 4, 1542–1558. [Google Scholar] [CrossRef] [PubMed]
- Naumann, R.L.C.; Nowak, C.; Knoll, W. Proteins in biomimetic membranes: Promises and facts. Soft Matter 2011, 7, 9535. [Google Scholar] [CrossRef]
- Makowski, G.S. Advances in Clinical Chemistry; Elsevier/Academic Press: Cambridge, MA, USA, 2009; Volume 49, ISBN 9780123747983. [Google Scholar]
- McCabe, I.P.; Forstner, M.B. Polymer Supported Lipid Bilayers. Open J. Biophys. 2013, 3, 59–69. [Google Scholar] [CrossRef]
- Studer, A.; Demarche, S.; Langenegger, D.; Tiefenauer, L. Integration and recording of a reconstituted voltage-gated sodium channel in planar lipid bilayers. Biosens. Bioelectron. 2011, 26, 1924–1928. [Google Scholar] [CrossRef]
- Ryu, H.; Choi, S.; Park, J.; Yoo, Y.-E.; Yoon, J.S.; Seo, Y.H.; Kim, Y.-R.; Kim, S.M.; Jeon, T.-J. Automated Lipid Membrane Formation Using a Polydimethylsiloxane Film for Ion Channel Measurements. Anal. Chem. 2014, 86, 8910–8915. [Google Scholar] [CrossRef] [PubMed]
- May, S.; Andreasson-Ochsner, M.; Fu, Z.; Low, Y.X.; Tan, D.; de Hoog, H.-P.M.; Ritz, S.; Nallani, M.; Sinner, E.-K. In Vitro Expressed GPCR Inserted in Polymersome Membranes for Ligand-Binding Studies. Angew. Chem. Int. Ed. 2013, 52, 749–753. [Google Scholar] [CrossRef] [PubMed]
- Fox, C.B.; Harris, J.M. Confocal Raman microscopy for simultaneous monitoring of partitioning and disordering of tricyclic antidepressants in phospholipid vesicle membranes. J. Raman Spectrosc. 2010, 41, 498–507. [Google Scholar] [CrossRef]
- Go, M.-L.; Ngiam, T.-L.; Rogers, J.A. Thermodynamics of the Partitioning of 7-Chloro-4-(4’-methoxy)anilinoquinoline and Its Cyclized Analog in Octanol-Buffer and Liposome Systems. Chem. Pharm. Bull. (Tokyo) 1995, 43, 289–294. [Google Scholar] [CrossRef]
- Lundbæk, J.A.; Collingwood, S.A.; Ingólfsson, H.I.; Kapoor, R.; Andersen, O.S. Lipid bilayer regulation of membrane protein function: Gramicidin channels as molecular force probes. J. R. Soc. Interface 2010, 7, 373–395. [Google Scholar] [CrossRef]
- Ingólfsson, H.I.; Andersen, O.S. Screening for Small Molecules’ Bilayer-Modifying Potential Using a Gramicidin-Based Fluorescence Assay. Assay Drug Dev. Technol. 2010, 8, 427–436. [Google Scholar] [CrossRef]
- Zhang, M.; Peyear, T.; Patmanidis, I.; Greathouse, D.V.; Marrink, S.J.; Andersen, O.S.; Ingólfsson, H.I. Fluorinated Alcohols’ Effects on Lipid Bilayer Properties. Biophys. J. 2018, 115, 679–689. [Google Scholar] [CrossRef]
- Ryu, H.; Lee, H.; Iwata, S.; Choi, S.; Kim, M.K.; Kim, Y.R.; Maruta, S.; Kim, S.M.; Jeon, T.J. Investigation of Ion Channel Activities of Gramicidin A in the Presence of Ionic Liquids Using Model Cell Membranes. Sci. Rep. 2015, 5, 11935. [Google Scholar] [CrossRef] [PubMed]
- Ingólfsson, H.I.; Thakur, P.; Herold, K.F.; Hobart, E.A.; Ramsey, N.B.; Periole, X.; de Jong, D.H.; Zwama, M.; Yilmaz, D.; Hall, K.; et al. Phytochemicals Perturb Membranes and Promiscuously Alter Protein Function. ACS Chem. Biol. 2014, 9, 1788–1798. [Google Scholar] [CrossRef] [PubMed]
- Kundu, S.K.; Chakraborty, C.; Yagihara, S.; Teoh, S.L.; Das, S. Anesthetic Molecule Interaction of Noble Gases with Proteins and Lipids and their Effect: A Review. Curr. Drug Deliv. 2018, 15, 1381–1392. [Google Scholar] [CrossRef] [PubMed]
- Luo, K.; Jeong, K.-B.; Oh, J.-M.; Choi, S.-J.; Jeon, T.-J.; Kim, Y.-R. Investigation of membrane condensation induced by CaCO3 nanoparticles and its effect on membrane protein function. RSC Adv. 2017, 7, 49858–49862. [Google Scholar] [CrossRef]
- Ramsey, N.B.; Andersen, O.S. Bilayer Effects of Antimalarial Compounds. PLoS ONE 2015, 10, e0142401. [Google Scholar] [CrossRef]
- Hoffert, M.I.; Caldeira, K.; Jain, A.K.; Haites, E.F.; Harvey, L.D.D.; Potter, S.D.; Schlesinger, M.E.; Schneider, S.H.; Watts, R.G.; Wigley, T.M.L.; et al. Energy implications of future stabilization of atmospheric CO2 content. Nature 1998, 395, 881–884. [Google Scholar] [CrossRef]
- Kerr, R.A. Peak Oil Production May Already Be Here. Science 2011, 331, 1510–1511. [Google Scholar] [CrossRef] [PubMed]
- Shockley, W.; Queisser, H.J. Detailed Balance Limit of Efficiency of p-n Junction Solar Cells. J. Appl. Phys. 1961, 32, 510–519. [Google Scholar] [CrossRef]
- Deisenhofer, J.; Epp, O.; Miki, K.; Huber, R.; Michel, H. Structure of the protein subunits in the photosynthetic reaction centre of Rhodopseudomonas viridis at 3 Å resolution. Nature 1985, 318, 618–624. [Google Scholar] [CrossRef]
- Eaton-Rye, J.J.; Tripathy, B.C.; Sharkey, T.D. Photosynthesis: Plastid Biology, Energy Conversion and Carbon Assimilation; Springer: Berlin, Germany, 2012; ISBN 940071579X. [Google Scholar]
- Anderson, J.M.; Boardman, N.K. Fractionation of the photochemical systems of photosynthesis I. Chlorophyll contents and photochemical activities of particles isolated from spinach chloroplasts. Biochim. Biophys. Acta Biophys. Incl. Photosynth. 1966, 112, 403–421. [Google Scholar] [CrossRef]
- Bengis, C.; Nelson, N.A. Purification and properties of the photosystem I reaction center from chloroplasts. J. Biol. Chem. 1975, 250, 2783–2788. [Google Scholar]
- Chitnis, P.R. PHOTOSYSTEM I: Function and Physiology. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001, 52, 593–626. [Google Scholar] [CrossRef] [PubMed]
- Armond, P.A.; Arntzen, C.J. Localization and Characterization of Photosystem II in Grana and Stroma Lamellae. Plant Physiol. 1977, 59, 398–404. [Google Scholar] [CrossRef] [PubMed]
- Barber, J. Photosystem II: A multisubunit membrane protein that oxidises water. Curr. Opin. Struct. Biol. 2002, 12, 523–530. [Google Scholar] [CrossRef]
- Nathans, J. Rhodopsin: Structure, Function, and Genetics. Biochemistry 1992, 31, 4923–4931. [Google Scholar] [CrossRef] [PubMed]
- Lozier, R.H.; Bogomolni, R.A.; Stoeckenius, W. Bacteriorhodopsin: A light-driven proton pump in Halobacterium Halobium. Biophys. J. 1975, 15, 955–962. [Google Scholar] [CrossRef]
- Kühlbrandt, W. Bacteriorhodopsin—The movie. Nature 2000, 406, 569–570. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, Z.; Okafor, F.; Azim, S.; Laughlin, T.F. ATP Synthase: A Molecular Therapeutic Drug Target for Antimicrobial and Antitumor Peptides. Curr. Med. Chem. 2013, 20, 1956–1973. [Google Scholar] [CrossRef]
- Ahmad, Z.; Cox, J.L. ATP synthase: The right size base model for nanomotors in nanomedicine. Sci. World J. 2014, 2014. [Google Scholar] [CrossRef] [PubMed]
- Boyer, P.D. The ATP synthase—A splendid molecular machine. Annu. Rev. Biochem. 1997, 66, 717–749. [Google Scholar] [CrossRef]
- Weber, J.; Senior, A.E. ATP synthesis driven by proton transport in F1F0-ATP synthase. FEBS Lett. 2003, 545, 61–70. [Google Scholar] [CrossRef]
- Stock, D.; Gibbons, C.; Arechaga, I.; Leslie, A.G.W.; Walker, J.E. The rotary mechanism of ATP-synthase. Curr. Opin. Struct. Biol. 2000, 10, 672–679. [Google Scholar] [CrossRef]
- Mershin, A.; Matsumoto, K.; Kaiser, L.; Yu, D.; Vaughn, M.; Nazeeruddin, M.K.; Bruce, B.D.; Graetzel, M.; Zhang, S. Self-assembled photosystem-I biophotovoltaics on nanostructured TiO2 and ZnO. Sci. Rep. 2012, 2, 234. [Google Scholar] [CrossRef] [PubMed]
- LeBlanc, G.; Chen, G.; Gizzie, E.A.; Jennings, G.K.; Cliffel, D.E. Enhanced Photocurrents of Photosystem I Films on p-Doped Silicon. Adv. Mater. 2012, 24, 5959–5962. [Google Scholar] [CrossRef]
- Badura, A.; Guschin, D.; Kothe, T.; Kopczak, M.J.; Schuhmann, W.; Rögner, M. Photocurrent generation by photosystem 1 integrated in crosslinked redox hydrogels. Energy Environ. Sci. 2011, 4, 2435. [Google Scholar] [CrossRef]
- Kato, M.; Cardona, T.; Rutherford, A.W.; Reisner, E. Photoelectrochemical Water Oxidation with Photosystem II Integrated in a Mesoporous Indium–Tin Oxide Electrode. J. Am. Chem. Soc. 2012, 134, 8332–8335. [Google Scholar] [CrossRef] [PubMed]
- Mersch, D.; Lee, C.-Y.; Zhang, J.Z.; Brinkert, K.; Fontecilla-Camps, J.C.; Rutherford, A.W.; Reisner, E. Wiring of Photosystem II to Hydrogenase for Photoelectrochemical Water Splitting. J. Am. Chem. Soc. 2015, 137, 8541–8549. [Google Scholar] [CrossRef] [PubMed]
- Robertson, B.; Lukashev, E.P. Rapid pH change due to bacteriorhodopsin measured with a tin-oxide electrode. Biophys. J. 1995, 68, 1507–1517. [Google Scholar] [CrossRef]
- Miyasaka, T.; Atake, T.; Watanabe, T. Generation of Photoinduced Steady Current by Purple Membrane Langmuir-Blodgett Films at Electrode-Electrolyte Interface. Chem. Lett. 2003, 32, 144–145. [Google Scholar] [CrossRef]
- Saga, Y.; Watanabe, T.; Koyama, K.; Miyasaka, T. Mechanism of Photocurrent Generation from Bacteriorhodopsin on Gold Electrodes. J. Phys. Chem. B 1999, 103, 234–238. [Google Scholar] [CrossRef]
- Horn, C.; Steinem, C. Photocurrents Generated by Bacteriorhodopsin Adsorbed on Nano-Black Lipid Membranes. Biophys. J. 2005, 89, 1046–1054. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Liang, D.; Rao, S.; Xiang, Y. Heterogeneous bacteriorhodopsin/gold nanoparticle stacks as a photovoltaic system. Nano Energy 2015, 11, 654–661. [Google Scholar] [CrossRef]
- Allam, N.K.; Yen, C.-W.; Near, R.D.; El-Sayed, M.A. Bacteriorhodopsin/TiO2 nanotube arrays hybrid system for enhanced photoelectrochemical water splitting. Energy Environ. Sci. 2011, 4, 2909–2914. [Google Scholar] [CrossRef]
- Feng, X.; Jia, Y.; Cai, P.; Fei, J.; Li, J. Coassembly of Photosystem II and ATPase as Artificial Chloroplast for Light-Driven ATP Synthesis. ACS Nano 2016, 10, 556–561. [Google Scholar] [CrossRef] [PubMed]
- Kuruma, Y.; Ueda, T. The PURE system for the cell-free synthesis of membrane proteins. Nat. Protoc. 2015, 10, 1328–1344. [Google Scholar] [CrossRef] [PubMed]
- Anderson, S.; Constable, E.C.; Dare-Edwards, M.P.; Goodenough, J.B.; Hamnett, A.; Seddon, K.R.; Wright, R.D. Chemical modification of a titanium (IV) oxide electrode to give stable dye sensitisation without a supersensitiser. Nature 1979, 280, 571–573. [Google Scholar] [CrossRef]
- Boghossian, A.A.; Ham, M.-H.; Choi, J.H.; Strano, M.S. Biomimetic strategies for solar energy conversion: A technical perspective. Energy Environ. Sci. 2011, 4, 3834–3843. [Google Scholar] [CrossRef]
- Badura, A.; Kothe, T.; Schuhmann, W.; Rögner, M. Wiring photosynthetic enzymes to electrodes. Energy Environ. Sci. 2011, 4, 3263–3274. [Google Scholar] [CrossRef]
- Ravi, S.K.; Udayagiri, V.S.; Suresh, L.; Tan, S.C. Emerging Role of the Band-Structure Approach in Biohybrid Photovoltaics: A Path Beyond Bioelectrochemistry. Adv. Funct. Mater. 2018, 28, 1705305. [Google Scholar] [CrossRef]
- Ravi, S.K.; Yu, Z.; Swainsbury, D.J.K.; Ouyang, J.; Jones, M.R.; Tan, S.C. Enhanced Output from Biohybrid Photoelectrochemical Transparent Tandem Cells Integrating Photosynthetic Proteins Genetically Modified for Expanded Solar Energy Harvesting. Adv. Energy Mater. 2017, 7, 1601821. [Google Scholar] [CrossRef]
- Singh, V.K.; Ravi, S.K.; Ho, J.W.; Wong, J.K.C.; Jones, M.R.; Tan, S.C. Biohybrid Photoprotein-Semiconductor Cells with Deep-Lying Redox Shuttles Achieve a 0.7 V Photovoltage. Adv. Funct. Mater. 2018, 28, 1703689. [Google Scholar] [CrossRef]
- Xie, G.; Wen, L.; Jiang, L. Biomimetic smart nanochannels for power harvesting. Nano Res. 2016, 9, 59–71. [Google Scholar] [CrossRef]
- Hellingwerf, K.J.; Arents, J.C.; Scholte, B.J.; Westerhoff, H.V. Bacteriorhodopsin in liposomes. II. Experimental evidence in support of a theoretical model. Biochim. Biophys. Acta 1979, 547, 561–582. [Google Scholar] [CrossRef]
- Saeedi, P.; Moosaabadi, J.M.; Sebtahmadi, S.S.; Behmanesh, M.; Mehrabadi, J.F. Site-directed mutagenesis in bacteriorhodopsin mutants and their characterization for bioelectrical and biotechnological equipment. Biotechnol. Lett. 2012, 34, 455–462. [Google Scholar] [CrossRef]
- Birge, R.R.; Gillespie, N.B.; Izaguirre, E.W.; Kusnetzow, A.; Lawrence, A.F.; Singh, D.; Song, Q.W.; Schmidt, E.; Stuart, J.A.; Seetharaman, S.; et al. Biomolecular Electronics: Protein-Based Associative Processors and Volumetric Memories. J. Phys. Chem. B 1999, 103, 10746–10766. [Google Scholar] [CrossRef]
- Armstrong, R.E.; Warner, J.B. Biology and the Battlefield. Def. Horizons 2003, 24–25, S1. [Google Scholar]
- DaSilva, E.J. Biological warfare, bioterrorism, biodefence and the biological and toxin weapons convention. Electron. J. Biotechnol. 1999, 2, 3–4. [Google Scholar] [CrossRef]
- Saeedi, P.; Moosaabadi, J.M.; Sebtahmadi, S.S.; Mehrabadi, J.F.; Behmanesh, M.; Mekhilef, S. Potential applications of bacteriorhodopsin mutants. Bioengineered 2012, 3, 326–328. [Google Scholar] [CrossRef] [PubMed]
- Ikonen, M.; Peltonen, J.; Vuorimaa, E.; Lemmetyinen, H. Study of photocycle and spectral properties of bacteriorhodopsin in Langmuir-Blodgett films. Thin Solid Films 1992, 213, 277–284. [Google Scholar] [CrossRef]
- Flanagan, M.T. The deposition of Langmuir-Blodgett films containing purple membrane on lipid- and paraffin-impregnated filters. Thin Solid Films 1983, 99, 133–138. [Google Scholar] [CrossRef]
- Uehara, K.; Kawai, K.; Kouyama, T. Photoelectric response of oriented purple membrane electrodeposited onto poly(vinyl alcohol) film. Thin Solid Films 1993, 232, 271–277. [Google Scholar] [CrossRef]
- Al-Aribe, K.M.; Knopf, G.K.; Bassi, A.S. Photoelectric Monolayers Based on Self-Assembled and Oriented Purple Membrane Patches. J. Microelectromech. Syst. 2011, 20, 800–810. [Google Scholar] [CrossRef]
- He, J.A.; Samuelso, L.; Li, L.; Kumar, J.; Tripathy, S.K. Oriented Bacteriorhodopsin/Polycation Multilayers by Electrostatic Layer-by-Layer Assembly. Langmuir 1998, 14, 1674–1679. [Google Scholar] [CrossRef]
- Bayburt, T.H.; Grinkova, Y.V.; Sligar, S.G. Self-Assembly of Discoidal Phospholipid Bilayer Nanoparticles with Membrane Scaffold Proteins. Nano Lett. 2002, 2, 853–856. [Google Scholar] [CrossRef]
- Boghossian, A.A.; Choi, J.H.; Ham, M.H.; Strano, M.S. Dynamic and reversible self-assembly of photoelectrochemical complexes based on lipid bilayer disks, photosynthetic reaction centers, and single-walled carbon nanotubes. Langmuir 2011, 27, 1599–1609. [Google Scholar] [CrossRef] [PubMed]
- Ham, M.H.; Choi, J.H.; Boghossian, A.A.; Jeng, E.S.; Graff, R.A.; Heller, D.A.; Chang, A.C.; Mattis, A.; Bayburt, T.H.; Grinkova, Y.V.; et al. Photoelectrochemical complexes for solar energy conversion that chemically and autonomously regenerate. Nat. Chem. 2010, 2, 929–936. [Google Scholar] [CrossRef] [PubMed]
- Kozono, D.; Yasui, M.; King, L.S.; Agre, P. Aquaporin water channels: Atomic structure molecular dynamics meet clinical medicine. J. Clin. Investig. 2002, 109, 1395–1399. [Google Scholar] [CrossRef] [PubMed]
- Murata, K.; Mitsuoka, K.; Hirai, T.; Walz, T.; Agre, P.; Heymann, J.B.; Engel, A.; Fujiyoshi, Y. Structural determinants of water permeation through aquaporin-1. Nature 2000, 407, 599–605. [Google Scholar] [CrossRef] [PubMed]
- Agre, P. The aquaporin water channels. Proc. Am. Thorac. Soc. 2006, 3, 5–13. [Google Scholar] [CrossRef]
- Borgnia, M.; Nielsen, S.; Engel, A.; Agre, P. Cellular and molecular biology of the aquaporin water channels. Annu. Rev. Biochem. 1999, 68, 425–458. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, R.; Bhattacharjee, H.; Rosen, B.P. Aquaglyceroporins: Generalized metalloid channels. Biochim. Biophys. Acta (BBA)-Gen. Subj. 2014, 1840, 1583–1591. [Google Scholar] [CrossRef] [PubMed]
- Holm, L.M.; Jahn, T.P.; Møller, A.L.B.; Schjoerring, J.K.; Ferri, D.; Klaerke, D.A.; Zeuthen, T. NH3 and NH 4+ permeability in aquaporin-expressing Xenopus oocytes. Pflüg. Arch. 2005, 450, 415–428. [Google Scholar] [CrossRef] [PubMed]
- Litman, T.; Søgaard, R.; Zeuthen, T. Ammonia and urea permeability of mammalian aquaporins. In Aquaporins; Springer: Berlin, Germany, 2009; pp. 327–358. ISBN 3540798846. [Google Scholar]
- Kruse, E.; Uehlein, N.; Kaldenhoff, R. The aquaporins. Genome Biol. 2006, 7, 206. [Google Scholar] [CrossRef] [PubMed]
- Ishibashi, K. Aquaporin superfamily with unusual npa boxes: S-aquaporins (superfamily, sip-like and subcellular-aquaporins). Cell. Mol. Biol. 2006, 52, 20–27. [Google Scholar]
- Yakata, K.; Tani, K.; Fujiyoshi, Y. Water permeability and characterization of aquaporin-11. J. Struct. Biol. 2011, 174, 315–320. [Google Scholar] [CrossRef] [PubMed]
- Madeira, A.; Fernández-Veledo, S.; Camps, M.; Zorzano, A.; Moura, T.F.; Ceperuelo-Mallafré, V.; Vendrell, J.; Soveral, G. Human aquaporin-11 is a water and glycerol channel and localizes in the vicinity of lipid droplets in human adipocytes. Obesity 2014, 22, 2010–2017. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Grzelakowski, M.; Zilles, J.; Clark, M.; Meier, W. Highly permeable polymeric membranes based on the incorporation of the functional water channel protein Aquaporin Z. Proc. Natl. Acad. Sci. USA 2007, 104, 20719–20724. [Google Scholar] [CrossRef] [PubMed]
- Fuwad, A.; Ryu, H.; Malmstadt, N.; Kim, S.M.; Jeon, T.J. Biomimetic membranes as potential tools for water purification: Preceding and future avenues. Desalination 2019, 458, 97–115. [Google Scholar] [CrossRef]
- Ding, W.; Cai, J.; Yu, Z.; Wang, Q.; Xu, Z.; Wang, Z.; Gao, C. Fabrication of an aquaporin-based forward osmosis membrane through covalent bonding of a lipid bilayer to a microporous support. J. Mater. Chem. A 2015, 3, 20118–20126. [Google Scholar] [CrossRef]
- Wang, M.; Wang, Z.; Wang, X.; Wang, S.; Ding, W.; Gao, C. Layer-by-layer assembly of aquaporin z-incorporated biomimetic membranes for water purification. Environ. Sci. Technol. 2015, 49, 3761–3768. [Google Scholar] [CrossRef] [PubMed]
- Sun, G.; Chung, T.-S.; Jeyaseelan, K.; Armugam, A. Stabilization and immobilization of aquaporin reconstituted lipid vesicles for water purification. Colloids Surf. B Biointerfaces 2013, 102, 466–471. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Qiu, C.; Li, X.; Vararattanavech, A.; Shen, W.; Torres, J.; Helix-Nielsen, C.; Wang, R.; Hu, X.; Fane, A.G. Synthesis of robust and high-performance aquaporin-based biomimetic membranes by interfacial polymerization-membrane preparation and RO performance characterization. J. Membar. Sci. 2012, 423, 422–428. [Google Scholar] [CrossRef]
- Li, X.; Wang, R.; Tang, C.; Vararattanavech, A.; Zhao, Y.; Torres, J.; Fane, T. Preparation of supported lipid membranes for aquaporin Z incorporation. Colloids Surf. B Biointerfaces 2012, 94, 333–340. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Xie, M.; Song, X.; Guo, W.; Ngo, H.H.; Zhou, J.L.; Nghiem, L.D. Biomimetic aquaporin membranes for osmotic membrane bioreactors: Membrane performance and contaminant removal. Bioresour. Technol. 2018, 249, 62–68. [Google Scholar] [CrossRef]
- Li, Z.; Valladares Linares, R.; Bucs, S.; Fortunato, L.; Hélix-Nielsen, C.; Vrouwenvelder, J.S.; Ghaffour, N.; Leiknes, T.O.; Amy, G. Aquaporin based biomimetic membrane in forward osmosis: Chemical cleaning resistance and practical operation. Desalination 2017, 420, 208–215. [Google Scholar] [CrossRef]
- Li, X.; Chou, S.; Wang, R.; Shi, L.; Fang, W.; Chaitra, G.; Tang, C.Y.; Torres, J.; Hu, X.; Fane, A.G. Nature gives the best solution for desalination: Aquaporin-based hollow fiber composite membrane with superior performance. J. Membr. Sci. 2015, 494, 68–77. [Google Scholar] [CrossRef]
- Sun, G.; Chung, T.; Chen, N.; Lu, X.; Zhao, Q. Highly permeable aquaporin-embedded biomimetic membranes featuring a magnetic-aided approach. RSC Adv. 2013, 3, 9178–9184. [Google Scholar] [CrossRef]
- Wang, H.L.; Chung, T.-S.; Tong, Y.W.; Jeyaseelan, K.; Armugam, A.; Duong, H.H.P.; Fu, F.; Seah, H.; Yang, J.; Hong, M. Mechanically robust and highly permeable AquaporinZ biomimetic membranes. J. Membr. Sci. 2013, 434, 130–136. [Google Scholar] [CrossRef]
- Xie, W.; He, F.; Wang, B.; Chung, T.-S.; Jeyaseelan, K.; Armugam, A.; Tong, Y.W. An aquaporin-based vesicle-embedded polymeric membrane for low energy water filtration. J. Mater. Chem. A 2013, 1, 7592–7600. [Google Scholar] [CrossRef]
- Wang, H.; Chung, T.-S.; Tong, Y.W.; Jeyaseelan, K.; Armugam, A.; Chen, Z.; Hong, M.; Meier, W. Highly Permeable and Selective Pore-Spanning Biomimetic Membrane Embedded with Aquaporin Z. Small 2012, 8, 1185–1190. [Google Scholar] [CrossRef] [PubMed]
- Duong, P.H.H.; Chung, T.-S.; Jeyaseelan, K.; Armugam, A.; Chen, Z.; Yang, J.; Hong, M. Planar biomimetic aquaporin-incorporated triblock copolymer membranes on porous alumina supports for nanofiltration. J. Membr. Sci. 2012, 409, 34–43. [Google Scholar] [CrossRef]
- NoNATURAL PURIFICATION OF YOUR WATER WITH AQUAPORIN INSIDE®. Available online: http://www.aquaporin.dk/86/biomimetic-membranes.aspx (accessed on 23 September 2018).

| Nanopore | Pore Size (nm) | Channel Length | Applications | Pore Assembly | Reference |
|---|---|---|---|---|---|
| α-HL | 1.4 | Positive charge (pH 7.0) | RNA, dsDNA, ssDNA, proteins, small molecules | Heptameric | [26,53] |
| MspA | 1.2 | Negatively charged lumen | ssDNA, dsDNA | Octameric | [53,54] |
| ClyA | 3.3 | Negatively charged lumen | dsDNA, ClysA | Dodecameric | [45,55] |
| Ael | 1.0 | Positively charged lumen | ssDNA, proteins | Heptameric | [54] |
| Phi29 | 3.6 | Positively charged lumen | ssDNA, dsDNA, large molecules and proteins | Dodecameric | [56,57] |
| Protein | Detection Method | Transducing Material | Analyte | Limit of Detection | Reference |
|---|---|---|---|---|---|
| AChE | Colorimetric | CS/DTNB | Methomyl, Profenofos | 6.16 × 10−4 mM 0.27mM | [93] |
| AChE | Colorimetric | AuNP | Carbaryl Diazinon Malathion Phorate | 0.1 μg/L 0.1 μg/L 0.3 μg/L 1 μg/L | [78] |
| AChE | Potentiometric | IrOx pH electrode | Potentiometric | 1 μM | [81] |
| AChE | Potentiometric | CS/pH electrode | Malathion, Parathion-methyl, Methamidophos | 0.6–600mM 0.1–100mM 0.1–100mM | [94] |
| AChE | Amperometric | AChE/CS@TiO2-CS/rGO/GC | Dichlorvos | 0.036μM−22.6μM | [95] |
| AChE | Amperometric | Glass microsphere | Malathion | 0.0135 ppb | [82] |
| AChE | Piezoelectric | ME/MUA/QCM | Physostigmine | 1 mg/mL | [86] |
| BChE | Amperometric | CBNPs | Paraoxon | 5 μg/L | [88] |
| BChE | Potentiometric | PU/HPU | paraoxon | 10 nM | [89] |
| Protein | Function | Application | Novelty of Application | Limitation of Application | Reference | |
|---|---|---|---|---|---|---|
| In Common | Each | |||||
| PSI | Light absorption (700 nm) and Electron transfer | Solar energy harvesting (e.g., Biohybrid photovoltaic system) | Clean energy source, the highest theoretical efficiency | Short lifespan, difficulty of achieve maximum theoretical efficiency | Hard to recover when photodamage occurs (almost degraded) | [17,18,148,149,150] |
| PSII | Light absorption (680 nm) and Electron transfer | Photoinhibition of PSII under strong light reduces the activity. | [151,152] | |||
| bR | Proton pumping | Difficulty in overexpression | [153,154,155,156,157,158] | |||
| ATP synthase | ATP synthesis | ATP production | Artificial cell research | Low activity at low temperature | [19,20,159,160] | |
| Membrane Design | Protein Housing | Fabrication Techniques | Membrane Performance | Limitations | References |
|---|---|---|---|---|---|
| Vesicles embedding | Synthetic lipids | Electrokinetic force, pressure force, magnetic adsorption | 3.6–55.2 LMH * 20%–97.8 ± 0.7% salt rejection | Small membrane size, harsh coating conditions, defects in large area membrane | [16,199,202,203] |
| Synthetic polymers | Adsorption chemical bonding, pressure assisted coating | 16.4–22.6 LMH 61%–90% salt rejection | No uniform surface coating, low water desalination property shows high defects in coating | [204,205,206] | |
| Planar bilayer | Synthetic lipids | Chemical bonding, vesicles fusion via electrostatic interactions | 5.5–6.31 LMH 61%–90% salt rejection | Incomplete surface coating, unstable under high pressure, low performance | [195,196] |
| Synthetic polymers | Vesicles rupturing with disulfide-gold conjugation | 8.2 LMH/bar 45.1% salt rejection | Low water purification, incomplete surface coating | [207] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ryu, H.; Fuwad, A.; Yoon, S.; Jang, H.; Lee, J.C.; Kim, S.M.; Jeon, T.-J. Biomimetic Membranes with Transmembrane Proteins: State-of-the-Art in Transmembrane Protein Applications. Int. J. Mol. Sci. 2019, 20, 1437. https://doi.org/10.3390/ijms20061437
Ryu H, Fuwad A, Yoon S, Jang H, Lee JC, Kim SM, Jeon T-J. Biomimetic Membranes with Transmembrane Proteins: State-of-the-Art in Transmembrane Protein Applications. International Journal of Molecular Sciences. 2019; 20(6):1437. https://doi.org/10.3390/ijms20061437
Chicago/Turabian StyleRyu, Hyunil, Ahmed Fuwad, Sunhee Yoon, Huisoo Jang, Jong Chan Lee, Sun Min Kim, and Tae-Joon Jeon. 2019. "Biomimetic Membranes with Transmembrane Proteins: State-of-the-Art in Transmembrane Protein Applications" International Journal of Molecular Sciences 20, no. 6: 1437. https://doi.org/10.3390/ijms20061437
APA StyleRyu, H., Fuwad, A., Yoon, S., Jang, H., Lee, J. C., Kim, S. M., & Jeon, T.-J. (2019). Biomimetic Membranes with Transmembrane Proteins: State-of-the-Art in Transmembrane Protein Applications. International Journal of Molecular Sciences, 20(6), 1437. https://doi.org/10.3390/ijms20061437







