Protein Interactomic Analysis of SAPKs and ABA-Inducible bZIPs Revealed Key Roles of SAPK10 in Rice Flowering
Abstract
1. Introduction
2. Results
2.1. Identification of SAPKs and ABA-inducible bZIPs
2.2. Interactomic Analysis of SAPKs and ABA-inducible bZIPs in Yeast
2.3. SAPK10 Phosphorylates OsbZIP77
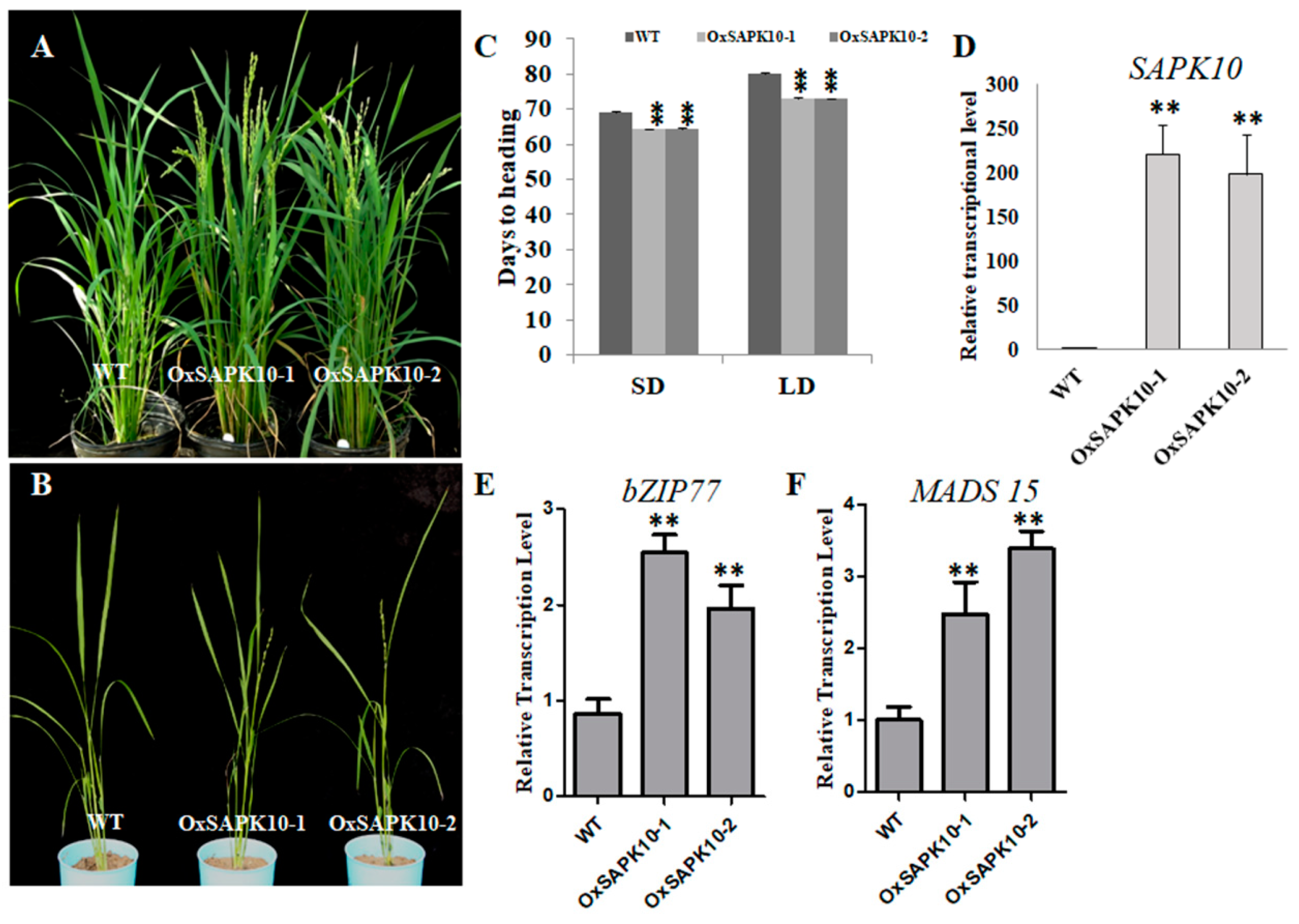
2.4. Over-Expression of SAPK10 Confers early Flowering
3. Materials and Methods
3.1. Plant Materials and ABA Treatment
3.2. Real-Time PCR
3.3. Yeast-two-Hybrid Assay
3.4. In Vitro Pull-Down Assay
3.5. Kinase Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Appendix A. Accession Numbers
References
- Shu, K.; Luo, X.; Meng, Y.; Yang, W. Toward a Molecular Understanding of Abscisic Acid Actions in Floral Transition. Plant Cell Physiol. 2018, 59, 215–221. [Google Scholar] [CrossRef] [PubMed]
- Cutler, S.R.; Rodriguez, P.L.; Finkelstein, R.R.; Abrams, S.R. Abscisic acid: Emergence of a core signaling network. Annu. Rev. Plant Biol. 2010, 61, 651–679. [Google Scholar] [CrossRef] [PubMed]
- Umezawa, T.; Nakashima, K.; Miyakawa, T.; Kuromori, T.; Tanokura, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Molecular basis of the core regulatory network in ABA responses: Sensing, signaling and transport. Plant Cell Physiol. 2010, 51, 1821–1839. [Google Scholar] [CrossRef] [PubMed]
- Fujii, H.; Zhu, J.K.; Jagendorf, A.T. Arabidopsis Mutant Deficient in 3 Abscisic Acid-Activated Protein Kinases Reveals Critical Roles in Growth, Reproduction, and Stress. Proc. Natl. Acad. Sci. USA 2009, 106, 8380–8385. [Google Scholar] [CrossRef] [PubMed]
- Fujita, Y.; Yoshida, T.; Yamaguchishinozaki, K. Pivotal role of the AREB/ABF-SnRK2 pathway in ABRE-mediated transcription in response to osmotic stress in plants. Physiol. Plant. 2013, 147, 15–27. [Google Scholar] [CrossRef] [PubMed]
- Skubacz, A.; Daszkowskagolec, A.; Szarejko, I. The Role and Regulation of ABI5 (ABA-Insensitive 5) in Plant Development, Abiotic Stress Responses and Phytohormone Crosstalk. Front. Plant Sci. 2016, 7, 1884. [Google Scholar] [CrossRef] [PubMed]
- Kulik, A.; Wawer, I.; Krzywińska, E.; Bucholc, M.; Dobrowolska, G. SnRK2 protein kinases—Key regulators of plant response to abiotic stresses. Omics J. Integr. Biol. 2011, 15, 859–872. [Google Scholar] [CrossRef]
- Finkelstein, R.R.; Lynch, T.J. The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 2000, 12, 599–609. [Google Scholar] [CrossRef]
- Lopezmolina, L.; Mongrand, S.; Chua, N.H. A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc. Natl. Acad. Sci. USA 2001, 98, 4782–4787. [Google Scholar] [CrossRef]
- Choi, H.; Hong, J.; Ha, J.; Kang, J.; Kim, S.Y. ABFs, a family of ABA-responsive element binding factors. J. Biol. Chem. 2000, 275, 1723–1730. [Google Scholar] [CrossRef]
- Wang, Y.; Li, L.; Ye, T.; Lu, Y.; Chen, X.; Wu, Y. The inhibitory effect of ABA on floral transition is mediated by ABI5 in Arabidopsis. J. Exp. Bot. 2013, 64, 675–684. [Google Scholar] [CrossRef]
- Kobayashi, Y.; Yamamoto, S.; Minami, H.; Kagaya, Y.; Hattori, T. Differential activation of the rice sucrose nonfermenting1-related protein kinase2 family by hyperosmotic stress and abscisic acid. Plant Cell 2004, 16, 1163–1177. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, Y.; Murata, M.; Minami, H.; Yamamoto, S.; Kagaya, Y.; Hobo, T.; Yamamoto, A.; Hattori, T. Abscisic acid-activated SNRK2 protein kinases function in the gene-regulation pathway of ABA signal transduction by phosphorylating ABA response element-binding factors. Plant J. 2005, 44, 939–949. [Google Scholar] [CrossRef] [PubMed]
- Chae, M.J.; Lee, J.S.; Nam, M.H.; Cho, K.; Hong, J.Y.; Yi, S.A.; Suh, S.C.; Yoon, I.S. A rice dehydration-inducible SNF1-related protein kinase 2 phosphorylates an abscisic acid responsive element-binding factor and associates with ABA signaling. Plant Mol. Biol. 2007, 63, 151–169. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.; Moon, S.J.; Min, M.K.; Choi, E.H.; Kim, J.A.; Koh, E.Y.; Yoon, I.; Byun, M.O.; Yoo, S.D.; Kim, B.G. Functional characterization and reconstitution of ABA signaling components using transient gene expression in rice protoplasts. Front. Plant Sci. 2015, 6, 614. [Google Scholar] [CrossRef] [PubMed]
- Tang, N.; Zhang, H.; Li, X.; Xiao, J.; Xiong, L. Constitutive activation of transcription factor OsbZIP46 improves drought tolerance in rice. Plant Physiol. 2012, 158, 1755–1768. [Google Scholar] [CrossRef]
- Zong, W.; Tang, N.; Yang, J.; Peng, L.; Ma, S.; Xu, Y.; Li, G.; Xiong, L. Feedback regulation of ABA signaling and biosynthesis by a bZIP transcription factor targets drought resistance related genes. Plant Physiol. 2016, 171, 2810–2825. [Google Scholar] [CrossRef] [PubMed]
- Fujii, H.; Chinnusamy, V.; Rodrigues, A.; Rubio, S.; Antoni, R.; Park, S.Y.; Cutler, S.R.; Sheen, J.; Rodriguez, P.L.; Zhu, J.K. In vitro reconstitution of an abscisic acid signalling pathway. Nature 2009, 462, 660–664. [Google Scholar] [CrossRef]
- Yang, M.; Wu, Z.; Fields, S. Protein-peptide interactions analyzed with the yeast two-hybrid system. Nucleic Acids Res. 1995, 23, 1152–1156. [Google Scholar] [CrossRef] [PubMed]
- Qiu, J.; Hou, Y.; Tong, X.; Wang, Y.; Lin, H.; Liu, Q.; Zhang, W.; Li, Z.; Nallamilli, B.R.; Zhang, J. Quantitative phosphoproteomic analysis of early seed development in rice (Oryza sativa L.). Plant Mol. Biol. 2016, 90, 249–265. [Google Scholar] [CrossRef]
- Ahmad, Z.; Huang, K.P. Dephosphorylation of rabbit skeletal muscle glycogen synthase (phosphorylated by cyclic AMP-independent synthase kinase 1) by phosphatases. J. Biol. Chem. 1981, 256, 757–760. [Google Scholar] [PubMed]
- Taoka, K.-I.; Ohki, I.; Tsuji, H.; Furuita, K.; Hayashi, K.; Yanase, T.; Yamaguchi, M.; Nakashima, C.; Purwestri, Y.A.; Tamaki, S. 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen. Nature 2011, 476, 332–335. [Google Scholar] [CrossRef] [PubMed]
- Lin, Q.; Wu, F.; Sheng, P.; Zhang, Z.; Zhang, X.; Guo, X.; Wang, J.; Cheng, Z.; Wang, J.; Wang, H. The SnRK2-APC/CTE regulatory module mediates the antagonistic action of gibberellic acid and abscisic acid pathways. Nat. Commun. 2015, 6, 7981. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Li, C.; Wu, Z.; Jia, Y.; Wang, H.; Sun, S.; Mao, C.; Wang, X. Abscisic Acid Regulates Auxin Homeostasis in Rice Root Tips to Promote Root Hair Elongation. Front. Plant Sci. 2017, 8, 1121. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Nallamilli, B.R.; Mujahid, H.; Peng, Z. OsMADS6 plays an essential role in endosperm nutrient accumulation and is subject to epigenetic regulation in rice (Oryza sativa). Plant J. 2010, 64, 604–617. [Google Scholar] [CrossRef] [PubMed]
- Hiei, Y.; Komari, T.; Kubo, T. Transformation of rice mediated by Agrobacterium tumefaciens. Plant. Mol. Biol. 1997, 35, 205–218. [Google Scholar] [CrossRef] [PubMed]
- Bello, B.K.; Hou, Y.; Zhao, J.; Jiao, G.; Wu, Y.; Li, Z.; Wang, Y.; Tong, X.; Wang, W.; Yuan, W.; et al. NF-YB1-YC12-bHLH144 complex directly activates Wx to regulate grain quality in rice (Oryza sativa L.). Plant. Biotechnol. J. 2018. [Google Scholar] [CrossRef]



| SAPK1 | SAPK2 | SAPK3 | SAPK4 | SAPK5 | SAPK6 | SAPK7 | SAPK8 | SAPK9 | SAPK10 | |
|---|---|---|---|---|---|---|---|---|---|---|
| bZIP10 | Y | |||||||||
| bZIP11 | Y | |||||||||
| bZIP35 | Y | Y | Y | |||||||
| bZIP52 | Auto | Auto | Auto | Auto | Auto | Auto | Auto | Auto | Auto | Auto |
| bZIP55 | Y | Y | ||||||||
| bZIP71 | Y | |||||||||
| bZIP75 | Y | |||||||||
| bZIP77 | Y | Y | Y | |||||||
| bZIP83 | Y | Y |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, X.; Li, Z.; Hou, Y.; Wang, Y.; Wang, H.; Tong, X.; Ao, H.; Zhang, J. Protein Interactomic Analysis of SAPKs and ABA-Inducible bZIPs Revealed Key Roles of SAPK10 in Rice Flowering. Int. J. Mol. Sci. 2019, 20, 1427. https://doi.org/10.3390/ijms20061427
Liu X, Li Z, Hou Y, Wang Y, Wang H, Tong X, Ao H, Zhang J. Protein Interactomic Analysis of SAPKs and ABA-Inducible bZIPs Revealed Key Roles of SAPK10 in Rice Flowering. International Journal of Molecular Sciences. 2019; 20(6):1427. https://doi.org/10.3390/ijms20061427
Chicago/Turabian StyleLiu, Xixi, Zhiyong Li, Yuxuan Hou, Yifeng Wang, Huimei Wang, Xiaohong Tong, Hejun Ao, and Jian Zhang. 2019. "Protein Interactomic Analysis of SAPKs and ABA-Inducible bZIPs Revealed Key Roles of SAPK10 in Rice Flowering" International Journal of Molecular Sciences 20, no. 6: 1427. https://doi.org/10.3390/ijms20061427
APA StyleLiu, X., Li, Z., Hou, Y., Wang, Y., Wang, H., Tong, X., Ao, H., & Zhang, J. (2019). Protein Interactomic Analysis of SAPKs and ABA-Inducible bZIPs Revealed Key Roles of SAPK10 in Rice Flowering. International Journal of Molecular Sciences, 20(6), 1427. https://doi.org/10.3390/ijms20061427









