Distribution of Gadolinium in Rat Heart Studied by Fast Field Cycling Relaxometry and Imaging SIMS
Abstract
1. Introduction
2. Results and Discussion
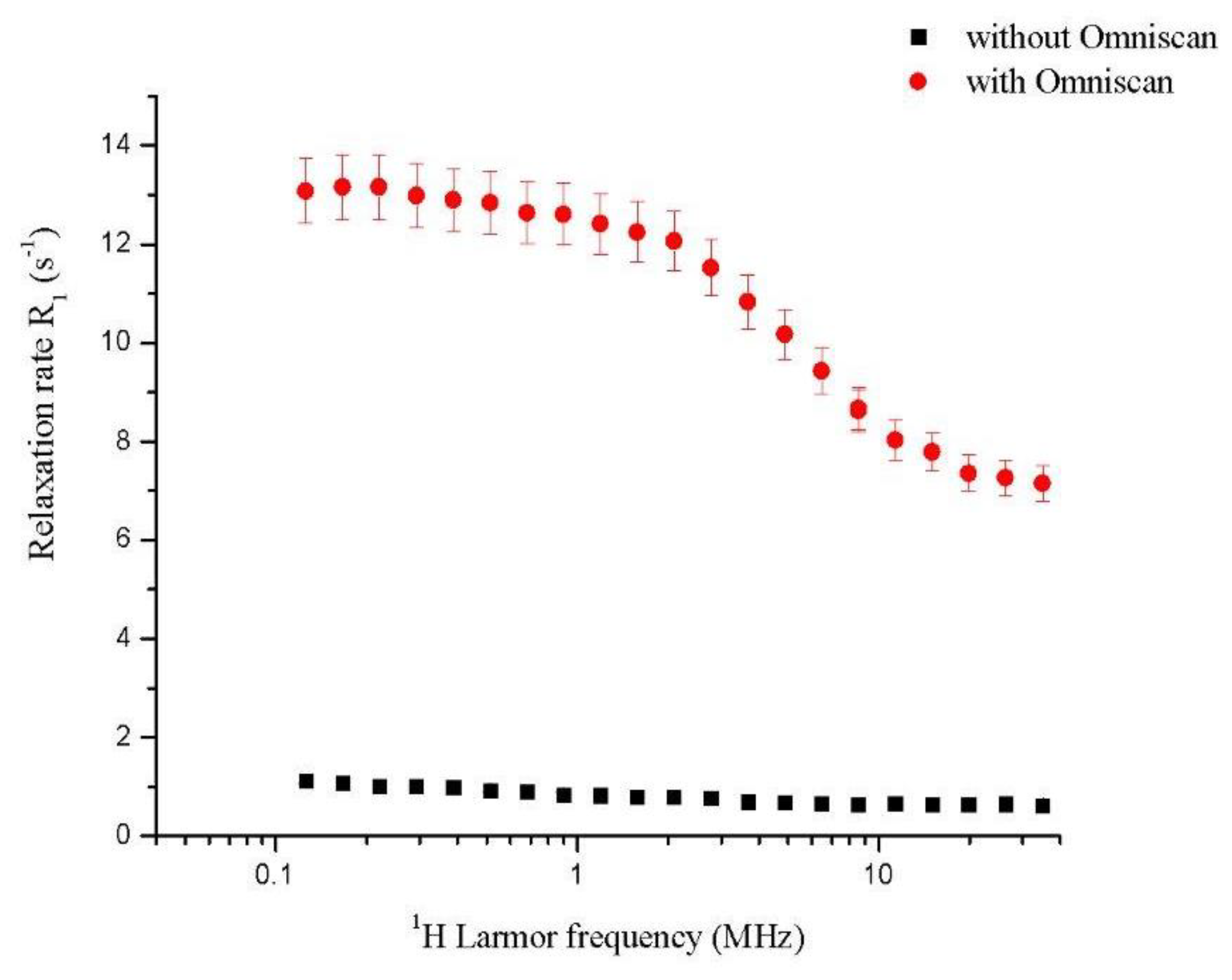
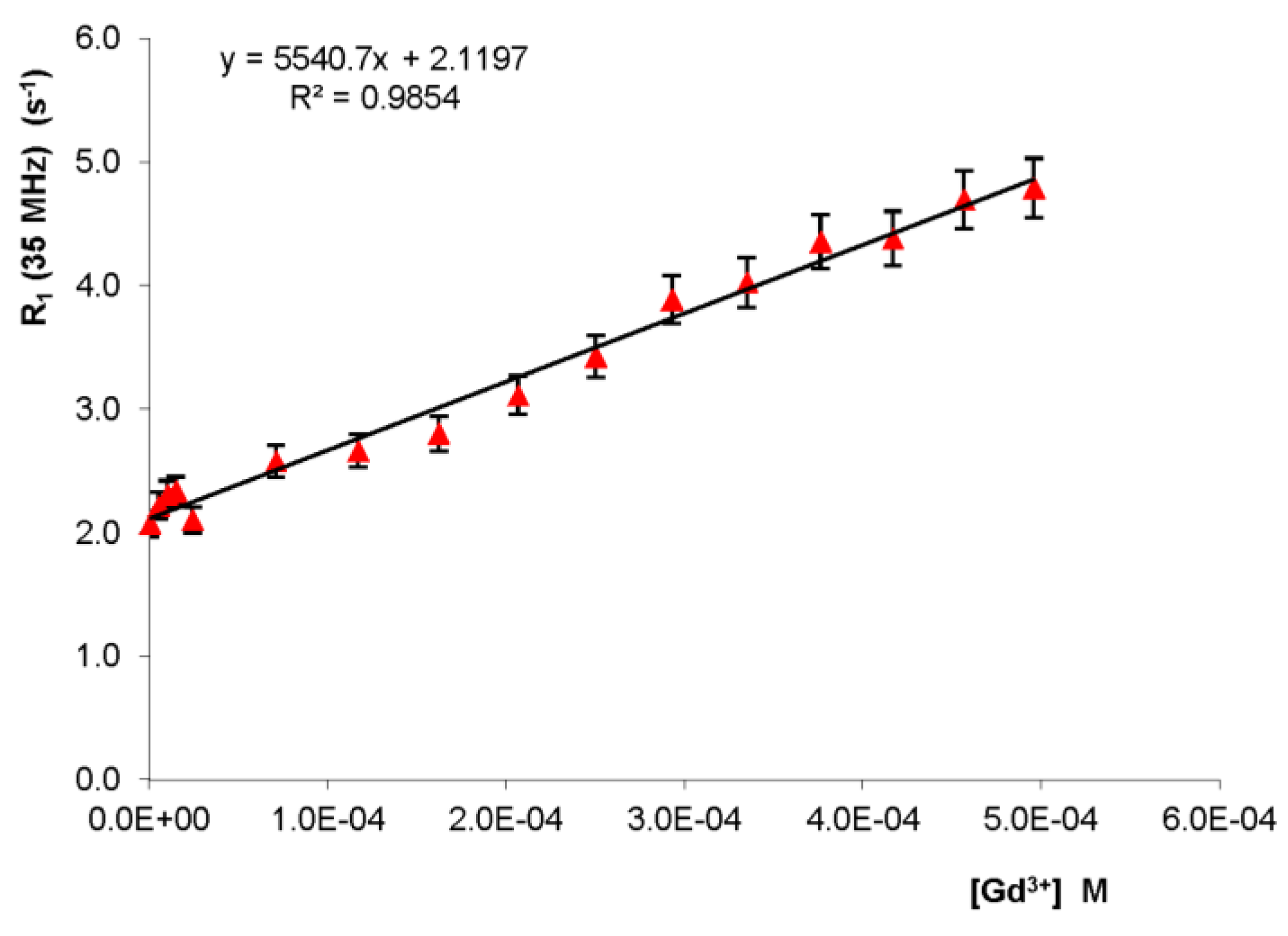
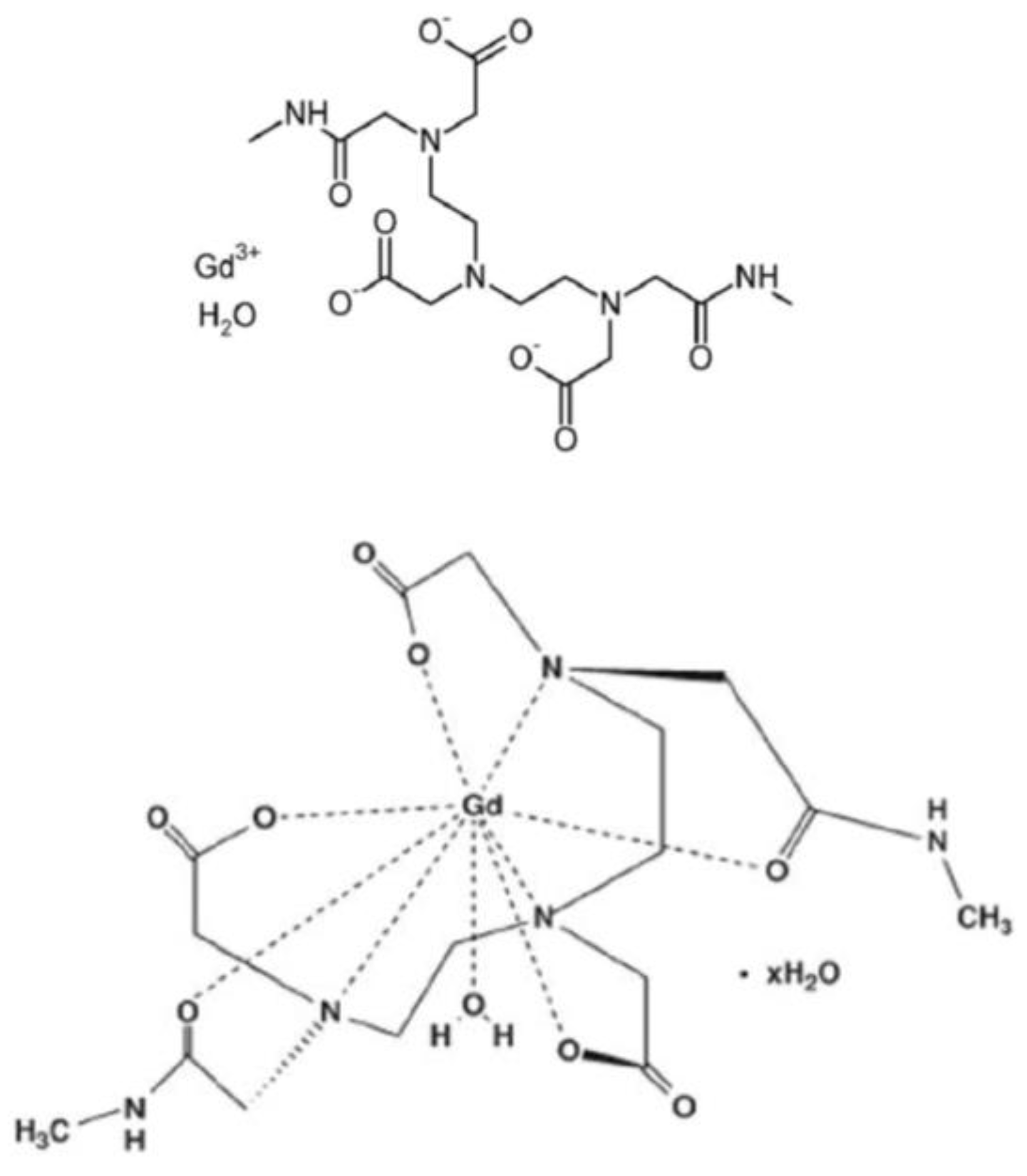
2.1. Fast Field Cycling Relaxometry Fast field cycling relaxometry
2.2. Analysis of Heart Tissue Sections
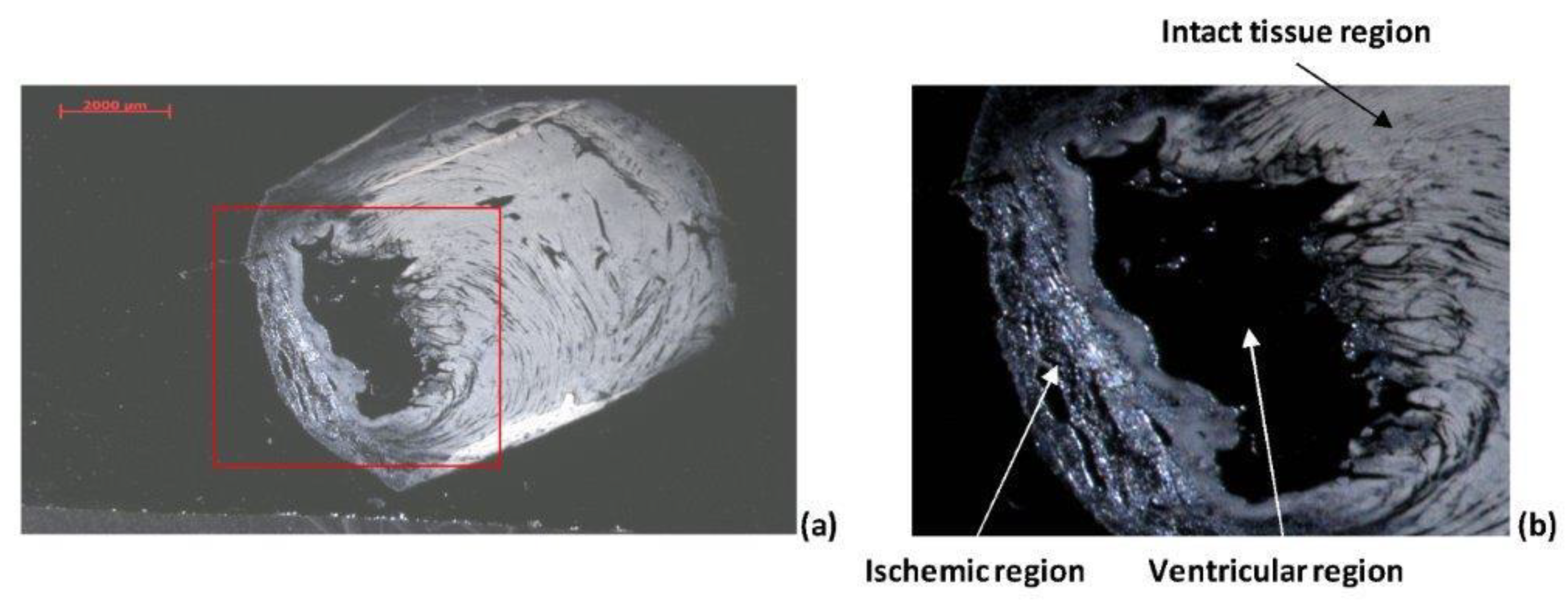
2.2.1. Optical Analysis
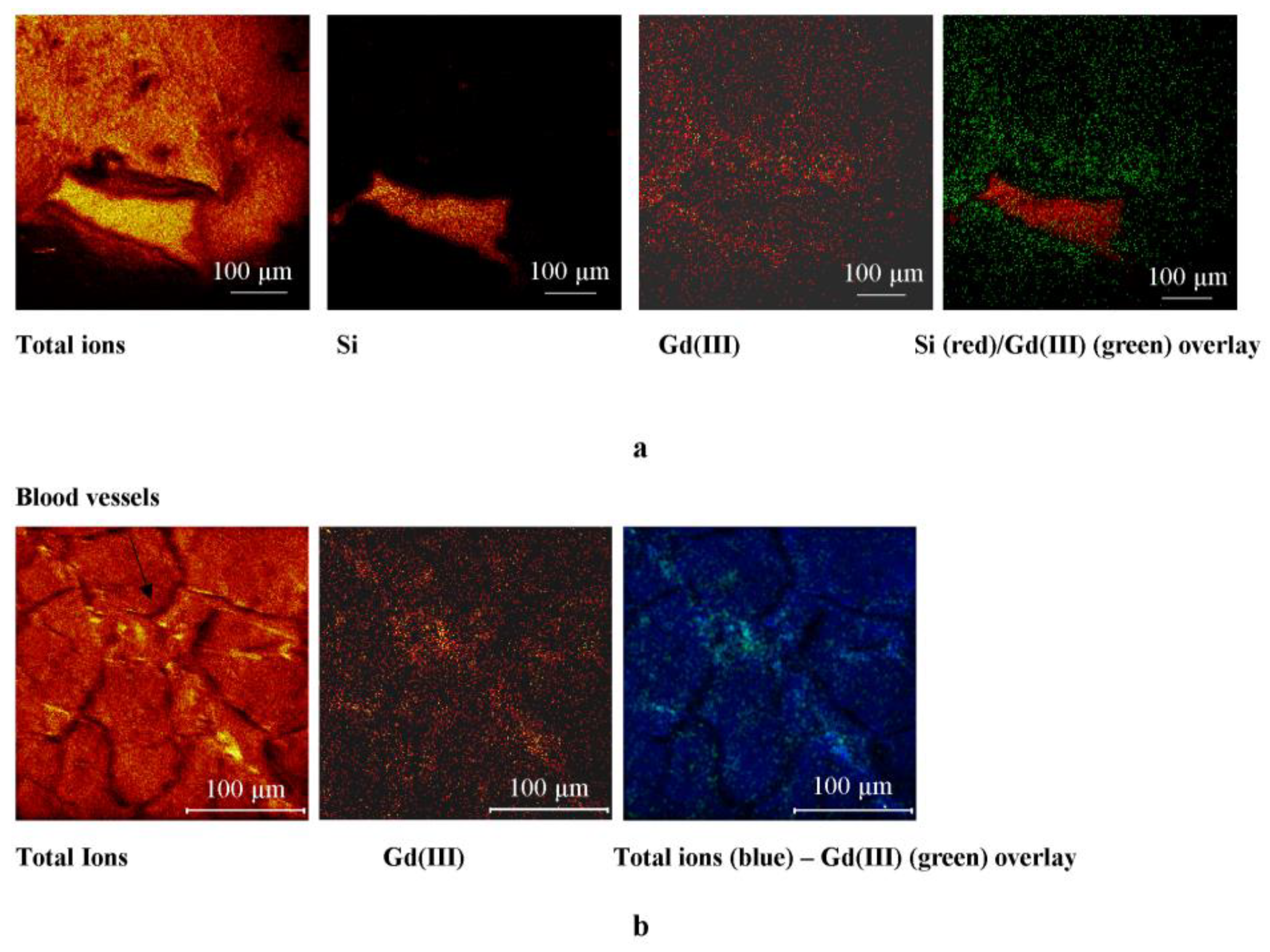
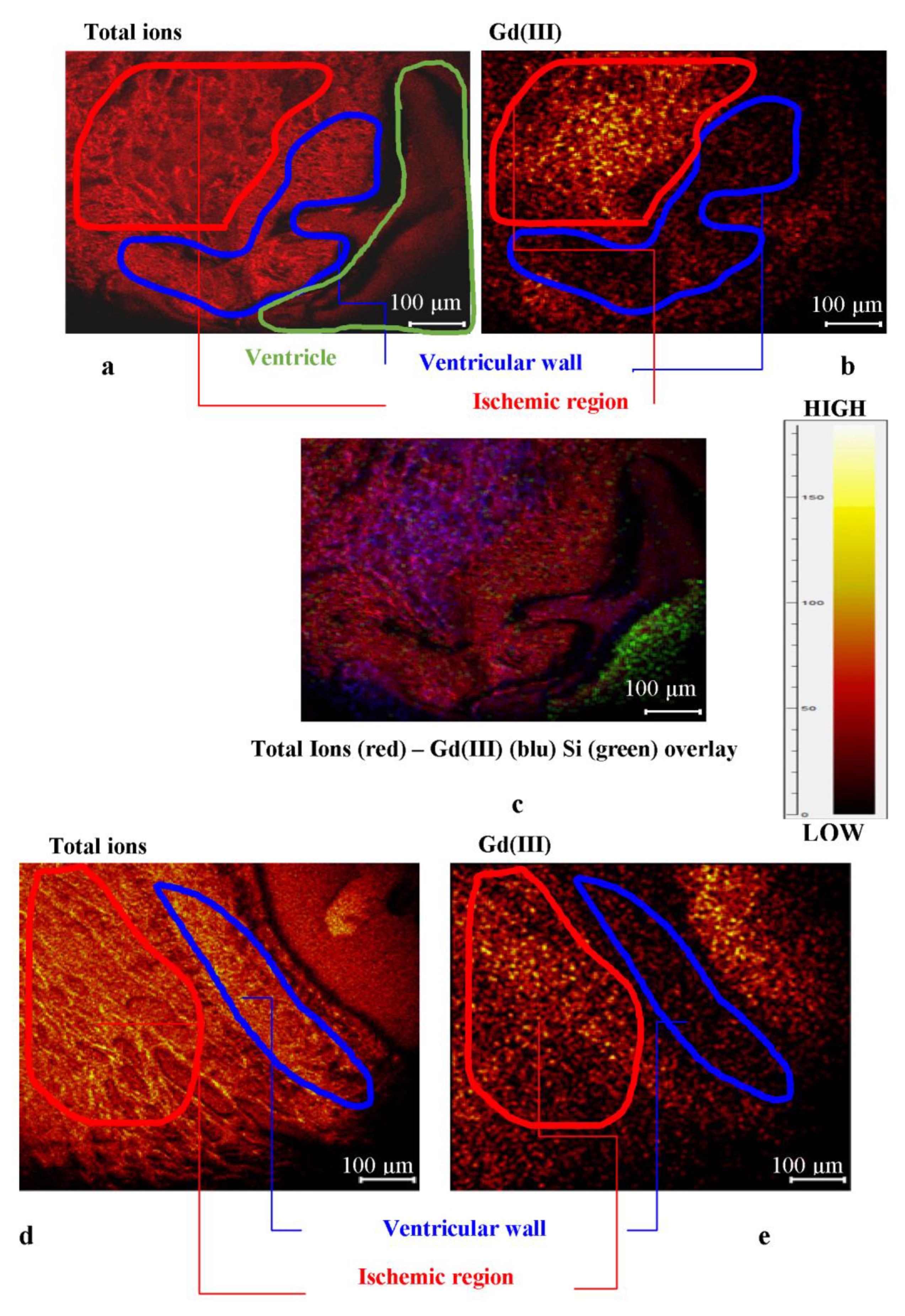
2.2.2. ToF-SIMS Analysis of Heart Tissue Sections: Chemical Maps of Gd(III) Distribution
2.2.3. Physiological Heart Tissue Sections
2.2.4. Ischemic Heart Tissue Sections
3. Materials and Methods
3.1. Animal Models and Isolated Heart Preparation
3.2. Sample Preparation
3.3. Optical Microscopy Measurements
3.4. ToF-SIMS Measurements
3.5. Relaxation Rate Measurements
3.6. Determination of Gadolinium Content in Homogenized ex vivo Langendorff Isolated Rat Hearts
3.7. Statistical Data Treatment
4. Conclusions
Author Contributions
Funding
Ethical issues
Conflicts of Interest
References
- Vickerman, J.C.; Briggs, D. ToF-SIMS: Surface Analysis by mass Spectrometry, 2nd ed.; IM Publications and Surface Spectra Limited: Chichester, UK, 2003. [Google Scholar]
- Malm, J.; Giannaras, D.; Riehle, M.O.; Gadegaard, N.; Sjövall, P. Fixation and drying protocols for the preparation of cell samples for time-of-flight secondary ion mass spectrometry analysis. Anal. Chem. 2009, 81, 7197–7205. [Google Scholar] [CrossRef]
- Kailas, L.; Audinot, J.-N.; Migeon, H.-N.; Bertrand, P. ToF-SIMS molecular characterization and nano-SIMS imaging of submicron domain formation at the surface of PS/PMMA blend and copolymer thin films. Appl. Surf. Sci. 2004, 231, 289–295. [Google Scholar] [CrossRef]
- Piras, F.M.; Di Mundo, R.; Fracassi, F.; Magnani, A. Silicon nitride and oxynitride films deposited from organosilicon plasmas: ToF-SIMS characterization with multivariate analysis. Surf. Coat. Technol. 2008, 202, 1606–1614. [Google Scholar] [CrossRef]
- Leone, G.; Consumi, M.; Tognazzi, A.; Magnani, A. Realisation and chemical characterisation of a model system for saccharide-based biosensor. Thin Solid Films 2010, 519, 462–470. [Google Scholar] [CrossRef]
- Leone, G.; Consumi, M.; Lamponi, S.; Magnani, A. Combination of Static Time of Flight Secondary Ion Mass Spectrometry and InfraRed Reflection-Adsorption Spectroscopy for the characterisation of a four steps built-up carbohydrate array. Appl. Surf. Sci. 2012, 258, 6302–6315. [Google Scholar] [CrossRef]
- Leone, G.; Bidini, A.; Lamponi, S.; Magnani, A. States of water, surface and rheological characterisation of a new biohydrogel as articular cartilage substitute. Polym. Adv. Technol. 2013, 24, 824–833. [Google Scholar] [CrossRef]
- Passarelli, M.K.; Ewing, A.G.; Winograd, N. Single-Cell Lipidomics: Characterizing and Imaging Lipids on the Surface of Individual Aplysia californica Neurons with Cluster Secondary Ion Mass Spectrometry. Anal. Chem. 2013, 85, 2231–2238. [Google Scholar] [CrossRef]
- Volandri, G.; Menichetti, L.; Matteucci, M.; Kusmic, C.; Consumi, M.; Magnani, A.; L’Abbate, A.; Landini, L.; Positano, V. An image formation model for Secondary Ion Mass Spectrometry imaging of biological tissue samples. Appl. Surf. Sci. 2010, 257, 1267–1275. [Google Scholar] [CrossRef]
- Piras, F.; Dettori, M.F.; Magnani, A. ToF-SIMS PCA analysis of Myrtus communis L. Appl. Surf. Sci. 2009, 255, 7805–7811. [Google Scholar] [CrossRef]
- Nakano, S.; Yokoyama, Y.; Aoyagi, S.; Himi, N.; Fletcher, J.S.; Lockyer, N.P.; Henderson, A.; Vickerman, J.C. Evaluation of biomolecular distributions in rat brain tissues by means of ToF-SIMS using a continuous beam of Ar clusters. Biointerphases 2016, 11, 02A307. [Google Scholar] [CrossRef]
- Barreto, G.; Soininen, A.; Sillat, T.; Konttinen, Y.T.; Kaivosoja, E. Sample processing, protocol, and statistical analysis of the time-of-flight secondary ion mass spectrometry (ToF-SIMS) of protein, cell, and tissue samples. Methods Mol. Biol. 2014, 1142, 177–188. [Google Scholar]
- Römpp, A.; Both, J.P.; Brunelle, A.; Heeren, R.M.; Laprévote, O.; Prideaux, B.; Seyer, A.; Spengler, B.; Stoeckli, M.; Smith, D.F. Mass spectrometry imaging of biological tissue: An approach for multicenter studies. Anal. Bioanal. Chem. 2015, 407, 2329–2335. [Google Scholar] [CrossRef]
- Touboul, D.; Roy, S.; Germain, D.P.; Chaminade, P.; Brunelle, A.; Laprévote, O. MALDI-TOF and cluster TOF-SIMS imaging in Fabry disease biomarkers. Int. J. Mass Spectrom. 2007, 260, 158–165. [Google Scholar] [CrossRef]
- Khatib-Shahidi, S.; Andersson, M.; Herman, J.L.; Gillespie, T.A.; Caprioli, R.M. Direct Molecular Analysis of Whole-Body Animal Tissue Sections by Imaging MALDI Mass Spectrometry. Anal. Chem. 2006, 78, 6448–6456. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Lu, X.; Kulp, K.S.; Knize, M.G.; Berman, E.S.F.; Nelson, E.J.; Felton, J.S.; Wu, K.J.J. Imaging and differentiation of mouse embryo tissues by ToF-SIMS. Int. J. Mass Spectrom. 2007, 260, 137–145. [Google Scholar] [CrossRef]
- Bich, C.; Touboul, D.; Brunelle, A. Biomedical studies by TOF-SIMS imaging. Biointerphases 2015, 10, 018901. [Google Scholar] [CrossRef]
- Abraham, J.L.; Chandra, S.; Thakra, C.; Abraham, J.M. SIMS imaging of gadolinium isotopes in tissue from Nephrogenic Systemic Fibrosis patients: Release of free Gd from magnetic resonance imaging (MRI) contrast agents. Appl. Surf. Sci. 2008, 255, 1181–1184. [Google Scholar] [CrossRef]
- Kimmich, R.; Anoardo, E. Field-cycling NMR relaxometry. Prog. Nucl. Magn. Reson. Spectrosc. 2004, 44, 257–320. [Google Scholar] [CrossRef]
- Schühle, D.T.; Schatz, J.; Laurent, S.; Vander Elst, L.; Muller, R.N.; Stuart, M.C.; Peters, J.A. Calix[4]arenes as molecular platforms for magnetic resonance imaging (MRI) contrast agents. Chemistry 2009, 15, 3290–3296. [Google Scholar] [CrossRef] [PubMed]
- Urbanczyk-Pearson, L.M.; Femia, F.J.; Smith, J.; Parigi, G.; Luchinat, C.; Meade, T.J. Mechanistic investigation of β-galactosidase-activated MR contrast agents. Inorg. Chem. 2008, 47, 56–68. [Google Scholar] [CrossRef] [PubMed]
- Caravan, P.; Parigi, G.; Chasse, J.M.; Cloutier, N.; Ellison, J.J.; Lauffer, R.B.; Luchinat, C.; McDermid, S.A.; Spiller, M.; McMurry, T.J. Albumin binding, relaxivity and water exchange kinetics of the diastereoisomers of MS-325 a gadolinium(III) based magnetic resonance angiography contrast agent. Inorg. Chem. 2007, 46, 6632–6639. [Google Scholar] [CrossRef]
- Tamasi, G.; Bonechi, C.; Rossi, C.; Cini, R.; Magnani, A. Simulating the active sites of copper-trafficking proteins. Density functional structural and spectroscopy studies on copper(I) complexes with thiols, carboxylato, amide and phenol ligands. J. Coord. Chem. 2016, 69, 404–424. [Google Scholar] [CrossRef]
- Diakova, G.; Goddard, Y.A.; Korb, J.P.; Bryant, R.G. Changes in protein structure and dynamics as a function of hydration from 1H second moments. J. Magn. Reson. 2007, 189, 166–172. [Google Scholar] [CrossRef] [PubMed]
- Tamasi, G.; Corsini, M.; Cini, R. Synthesis, and electrochemical and density functional studies of new copper(II)- and manganese(II) oxicam drugs. Redox potentials and MOs compatible with SOD-like activity and unusual six-membered rings of water molecules bridging complex units. Z. Anorg. Allg. Chem. 2014, 640, 952–961. [Google Scholar] [CrossRef]
- Tamasi, G.; Cini, R. Ruthenium complexes as nitric oxide donors and scavengers. Synthesis and crystal and molecular structure for mer, trans-[RuIICl3(NO+) (N-4-ethylisonicotinate)2], and mer, trans-[RuIIICl3(N-CH3CN) (N-4-ethylisonicotinate)2] as obtained via UV-photochemical activation of RuII(NO+)3+-core parent complex in acetonitrile solution. J. Mol. Struct. 2013, 1048, 27–32. [Google Scholar]
- Tamasi, G.; Mangani, A.; Cini, R. Copper(I)-alkyl sulfide and-cysteine tri-nuclear clusters as models for metallo proteins: A structural density functional analysis. J. Biomol. Struct. Dyn. 2012, 30, 728–751. [Google Scholar] [CrossRef] [PubMed]
- Intini, F.P.; Cini, R.; Tamasi, G.; Hursthouse, M.B.; Marzilli, L.G.; Natile, G. X-ray structural characterization of the bis-guanine derivative of a cisplatin analogue having just one proton on each coordinated nitrogen and a head-to-head conformation: [Pt(±)-N, N′-dimethyl-2,3-diaminobutane(9-ethyl-guanine)2]dinitrate. Inorg. Chem. 2010, 49, 7853–7860. [Google Scholar] [CrossRef]
- Tamasi, G.; Defazio, S.; Chiasserini, L.; Sega, A.; Cini, R. Ruthenium-Thiobase complexes. Synthesis, spectroscopy, density functional studies for trans,cis,cis-[RuII(AsPh3)2(N,S-2-thiopyrimidinato)2] and structural analysis of selected weak C-H…N and C-H…S interactions. Inorg. Chim. Acta 2009, 362, 1011–1021. [Google Scholar] [CrossRef]
- Bonechi, C.; Collodel, G.; Donati, A.; Martini, S.; Moretti, E.; Rossi, C. Discrimination of human semen specimens by NMR data, sperm parameters and PCA. Syst. Biol. Reprod. Med. 2015, 61, 1–7. [Google Scholar] [CrossRef]
- Bonechi, C.; Donati, A.; Tamasi, G.; Leone, G.; Consumi, M.; Rossi, C.; Lamponi, S.; Magnani, A. Protective effect of quercetin and rutin encapsulated liposomes on induced oxidative stress. Biophys. Chem. 2018, 233, 55–63. [Google Scholar] [CrossRef]
- Leone, G.; Consumi, M.; Pepi, S.; Lamponi, S.; Bonechi, C.; Tamasi, G.; Donati, A.; Rossi, C.; Magnani, A. Alginate-Gelatin formulation to modify lovastatin release profile from red yeast rice for hypercholesterolemia treatment. Ther. Deliv. 2017, 8, 843–854. [Google Scholar] [CrossRef]
- Leone, G.; Consumi, M.; Pepi, S.; Lamponi, S.; Bonechi, C.; Tamasi, G.; Donati, A.; Rossi, C.; Magnani, A. New formulations to enhance lovastatin release from red yeast rice (RYR). J. Drug Deliv. Sci. Technol. 2016, 36, 110–119. [Google Scholar] [CrossRef]
- Pontillo, N.; Ferraro, G.; Messori, L.; Tamasi, G.; Merlino, A. Ru-Based CO releasing molecules with azole ligands: Interaction with proteins and the CO release mechanism disclosed by X-ray crystallography. Dalton Trans. 2017, 46, 9621–9629. [Google Scholar] [CrossRef] [PubMed]
- Barone, C.R.; Cini, R.; Pinto, S.; Di Masi, N.G.; Maresca, L.; Natile, G.; Tamasi, G. Coupling of cationic olefin complexes of platinum(II) with potential ambident nucleophiles. Inorg. Chim. Acta. 2010, 363, 205–212. [Google Scholar] [CrossRef]
- Tamasi, G.; Carpini, A.; Valensin, D.; Messori, L.; Pratesi, A.; Scaletti, F.; Jakupec, M.; Keppler, B. Ru(CO)x-core complexes with selected azoles: Synthesis, X-ray structure, spectroscopy, DFT analysis and evaluation of cytotoxic activity against human cancer cells. Polyhedron 2014, 81, 227–237. [Google Scholar] [CrossRef]
- Tamasi, G.; Merlino, A.; Scaletti, F.; Heffeter, P.; Legin, A.A.; Jakupec, M.A.; Berger, W.; Messori, L.; Keppler, B.K.; Cini, R. {Ru(CO)x}-Core complexes with benzimidazole ligands: Synthesis, X-ray structure and evaluation of anticancer activity in vivo. Dalton Trans. 2017, 46, 3025–3040. [Google Scholar] [CrossRef]
- Tamasi, G.; Bonechi, C.; Donati, A.; Cini, R.; Magnani, A. Analytical and structural investigation via infrared spectroscopy and density functional methods of cuprous complexes of the antioxidant tripeptide glutathione (GSH). Synthesis and characterization of a novel CuI-GSH compound. Inorg. Chim. Acta 2018, 470, 158–171. [Google Scholar] [CrossRef]
- Bonechi, C.; Martini, S.; Rossi, C. Interaction Study of Bioactive Molecules with Fibrinogen and Human Platelets Determined by 1H-NMR Relaxation Experiments. Bioorg. Med. Chem. 2009, 17, 1630–1635. [Google Scholar] [CrossRef] [PubMed]
- Martini, S.; Bonechi, C.; Rossi, C. Interaction between vine pesticides and serum albumin studied by nuclear spin relaxation data. J. Agric. Food Chem. 2010, 58, 10705–10709. [Google Scholar] [CrossRef]
- Bonechi, C.; Martini, S.; Rossi, C. Interaction study of indigo carmine with albumin and dextran by NMR Relaxation. J. Mater. Sci. 2011, 46, 2541–2547. [Google Scholar] [CrossRef]
- Bonechi, C.; Lamponi, S.; Donati, A.; Tamasi, G.; Consumi, M.; Leone, G.; Rossi, C.; Magnani, A. Effect of Resveratrol on Platelet Aggregation by Fibrinogen Protection. Biophys. Chem. 2017, 222, 41–48. [Google Scholar] [CrossRef] [PubMed]
- Martini, S.; Bonechi, C.; Foletti, A.; Rossi, C. Water-Protein Interactions the Secret of Protein Flexibility. Sci. World J. 2013. [Google Scholar] [CrossRef] [PubMed]
- Narmani, A.; Farhood, B.; Haghi-aminjan, H.; Mortezazadeh, T.; Aliasgharzadeh, A.; Mohseni, M.; Najafi, M. Gadolinium nanoparticles as diagnostic and therapeutic agents: Their delivery systems in magnetic resonance imaging and neutron capture therapy. J. Drug Deliv. Sci. Technol. 2018. [Google Scholar] [CrossRef]
- Podgórna, K.; Szczepanowicz, K.; Piotrowski, M.; Gajdošová, M.; Štěpánek, F.; Warszyński, P. Gadolinium alginate nanogels for theranostic applications. Colloids Surf. B 2017, 153, 183–189. [Google Scholar] [CrossRef]
- Fretellier, N.; Salhi, M.; Schroeder, J.; Siegmund, H.; Chevalier, T.; Bruneval, P.; Jestin-Mayer, G.; Delaloge, F.; Factor, C.; Mayer, J.-F.; et al. Distribution profile of gadolinium in gadolinium chelate-treated renally-impaired rats: Role of pharmaceutical formulation. Eur. J. Pharm. Sci. 2015, 72, 46–56. [Google Scholar] [CrossRef] [PubMed]
- Bort, G.; Catoen, S.; Borderies, H.; Kebsi, A.; Ballet, S.; Louin, G.; Port, M.; Ferroud, C. Gadolinium-based contrast agents targeted to amyloid aggregates for the early diagnosis of Alzheimer’s disease by MRI. Eur. J. Med. Chem. 2014, 87, 843–861. [Google Scholar] [CrossRef] [PubMed]
- Fingerhut, S.; Niehoff, A.C.; Sperling, M.; Jeibmann, A.; Paulus, W.; Niederstadt, T.; Allkemper, T.; Heindel, W.; Holling, M.; Karst, U. Spatially resolved quantification of gadolinium deposited in the brain of a patient treated with gadolinium-based contrast agents. J. Trace Elem. Med. Biol. 2018, 45, 125–130. [Google Scholar] [CrossRef]
- Ramalho, J.; Ramalho, M.; Jay, M.; Burke, L.M.; Semelka, R.C. Gadolinium toxicity and treatment. Magn. Reson. Imaging 2016, 34, 1394–1398. [Google Scholar] [CrossRef]
- Young, J.R.; Orosz, I.; Franke, M.A.; Kim, H.J.; Woodworth, D.; Ellingson, B.M.; Salamon, N.; Pope, W.B. Gadolinium deposition in the paediatric brain:T1-weighted hyperintensity within the dentate nucleus following repeated gadolinium-based contrast agent administration. Clin. Radiol. 2018, 73, 290–295. [Google Scholar] [CrossRef]
- Korb, J.-P.; Diakova, G.; Goddard, Y.; Bryant, R.G. Relaxation of protons by radicals in rotationally immobilized proteins. J. Magn. Reson. 2007, 186, 176–181. [Google Scholar] [CrossRef] [PubMed]
- Goddard, Y.A.; Korb, J.-P.; Bryant, R.G. Water Molecule Contributions to Proton Spin-Lattice Relaxation in Rotationally Immobilized Protein. J. Magn. Reson. 2009, 199, 68–74. [Google Scholar] [CrossRef] [PubMed]
- Tweedle, M.F. Gadolinium deposition: I sit chelated or dissociated gadolinium? How can we tell? Magn. Reson. Imaging 2016, 34, 1377–1382. [Google Scholar] [CrossRef] [PubMed]
- Helm, L. Relaxivity in paramagnetic systems: Theory and mechanisms. Prog. Nucl. Magn. Reson. Spectrosc. 2006, 49, 45–64. [Google Scholar] [CrossRef]
- Toth, E.; Helm, L.; Merbach, A.E. Relaxivity of MRI Contrast Agents. Top. Curr. Chem. 2002, 221, 61–101. [Google Scholar]
- Taupitz, M.; Stolzenburg, N.; Ebert, M.; Scnorr, J.; Hauptmann, R.; Kratz, H.; Hamm, B.; Wagner, S. Gadolinium-containing magnetic resonance contrast media: investigation on the possible transchelation of Gd3+ to the glycosaminoglycan heparin. Contrast Media Mol. Imaging 2013, 8, 108–116. [Google Scholar] [CrossRef] [PubMed]
- Steliopoulos, P. Extension of the standard addition method by blank addition. MethodsX 2015, 3, 353–359. [Google Scholar] [CrossRef]
- Pedersen, M.; Shi, Y.; Anderson, P.; Stødkilde-Jørgensen, H.; Djurhuus, J.C.; Gordon, I.; Frøkiaer, J. Quantitation of differential renal blood flow and renal function using dynamic contrast-enhanced MRI in rats. Magn Reson Med. 2004, 51, 510–517. [Google Scholar] [CrossRef] [PubMed]
- Moriyama, Y.; Saito, S.; Kobayashi, S.; Ogihara, R.; Koto, D.; Kitamura, A.; Matsushita, T.; Nishiura, M.; Murase, K. Evaluation of concanavalin A-induced acute liver injury in rats using an empirical mathematical model and dynamic contrast-enhanced MR imaging with Gd-EOB-DTPA. Magn. Reson. Med. Sci. 2012, 11, 53–60. [Google Scholar] [CrossRef]
- Foster, R.J.; Damion, R.A.; Baboolal, T.G.; Smye, S.W.; Ries, M.E. A nuclear magnetic resonance study of water in aggrecan solutions. R. Soc. Open Sci. 2016, 3, 150705. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bonechi, C.; Consumi, M.; Matteucci, M.; Tamasi, G.; Donati, A.; Leone, G.; Menichetti, L.; Kusmic, C.; Rossi, C.; Magnani, A. Distribution of Gadolinium in Rat Heart Studied by Fast Field Cycling Relaxometry and Imaging SIMS. Int. J. Mol. Sci. 2019, 20, 1339. https://doi.org/10.3390/ijms20061339
Bonechi C, Consumi M, Matteucci M, Tamasi G, Donati A, Leone G, Menichetti L, Kusmic C, Rossi C, Magnani A. Distribution of Gadolinium in Rat Heart Studied by Fast Field Cycling Relaxometry and Imaging SIMS. International Journal of Molecular Sciences. 2019; 20(6):1339. https://doi.org/10.3390/ijms20061339
Chicago/Turabian StyleBonechi, Claudia, Marco Consumi, Marco Matteucci, Gabriella Tamasi, Alessandro Donati, Gemma Leone, Luca Menichetti, Claudia Kusmic, Claudio Rossi, and Agnese Magnani. 2019. "Distribution of Gadolinium in Rat Heart Studied by Fast Field Cycling Relaxometry and Imaging SIMS" International Journal of Molecular Sciences 20, no. 6: 1339. https://doi.org/10.3390/ijms20061339
APA StyleBonechi, C., Consumi, M., Matteucci, M., Tamasi, G., Donati, A., Leone, G., Menichetti, L., Kusmic, C., Rossi, C., & Magnani, A. (2019). Distribution of Gadolinium in Rat Heart Studied by Fast Field Cycling Relaxometry and Imaging SIMS. International Journal of Molecular Sciences, 20(6), 1339. https://doi.org/10.3390/ijms20061339









