Long Noncoding RNA and Protein Interactions: From Experimental Results to Computational Models Based on Network Methods
Abstract
1. Introduction
2. A Brief Introduction to the Relevant Databases Used for Analyzing LncRNA–Protein Interactions
3. A Brief Introduction of Experimental Approaches and Computational Approaches Based on Machine Learning to Study LncRNA–Protein Interactions
3.1. LncRNA–Protein Interactions: From Experimental Approaches to Computational Models Based on High-Throughput Experiments
3.2. LncRNA–Protein Interactions: From Experimental Results to Computational Models Based on Machine Learning
4. Computational Models for LncRNA–Protein Interaction Prediction Based on Biological Networks
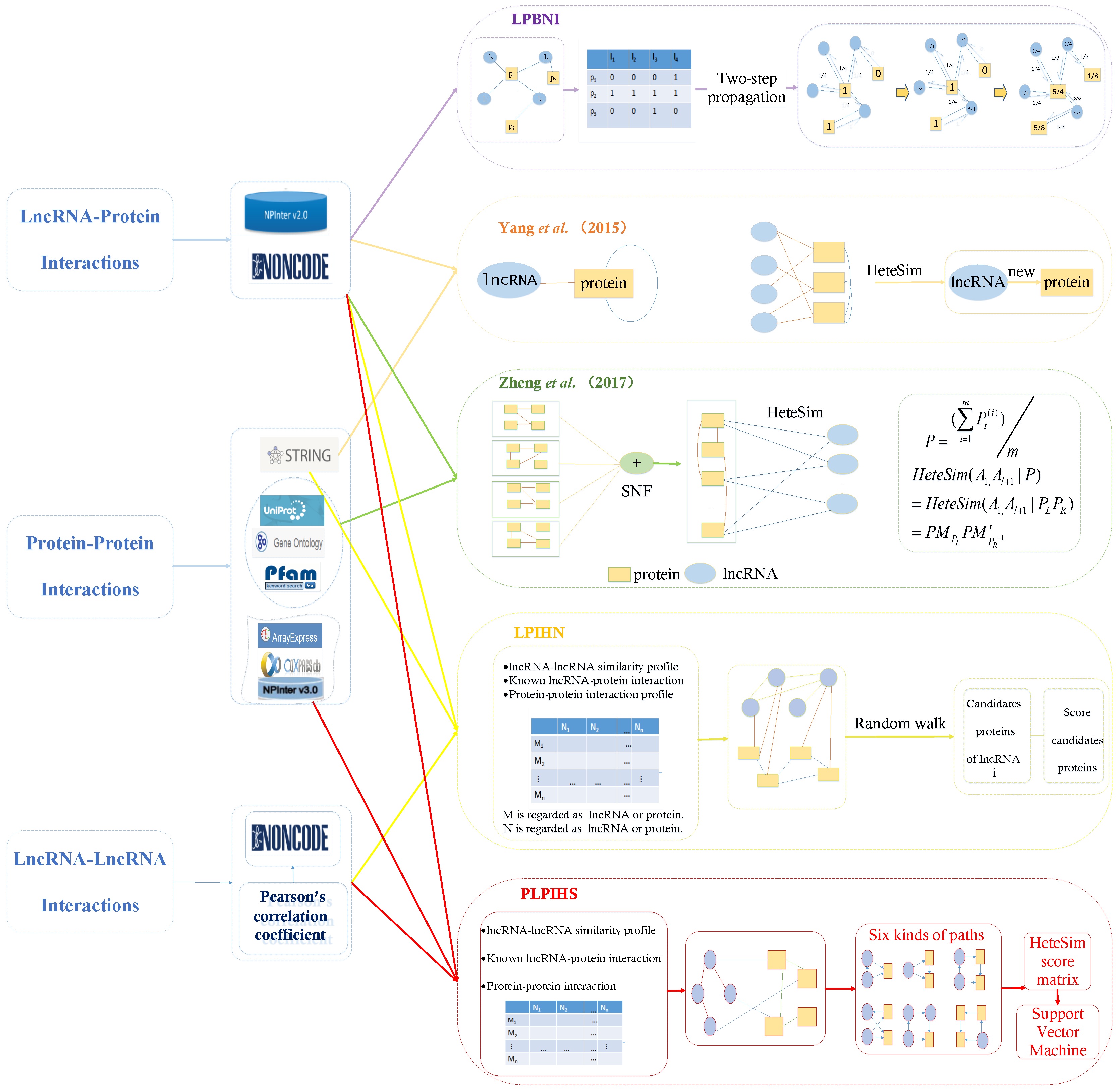
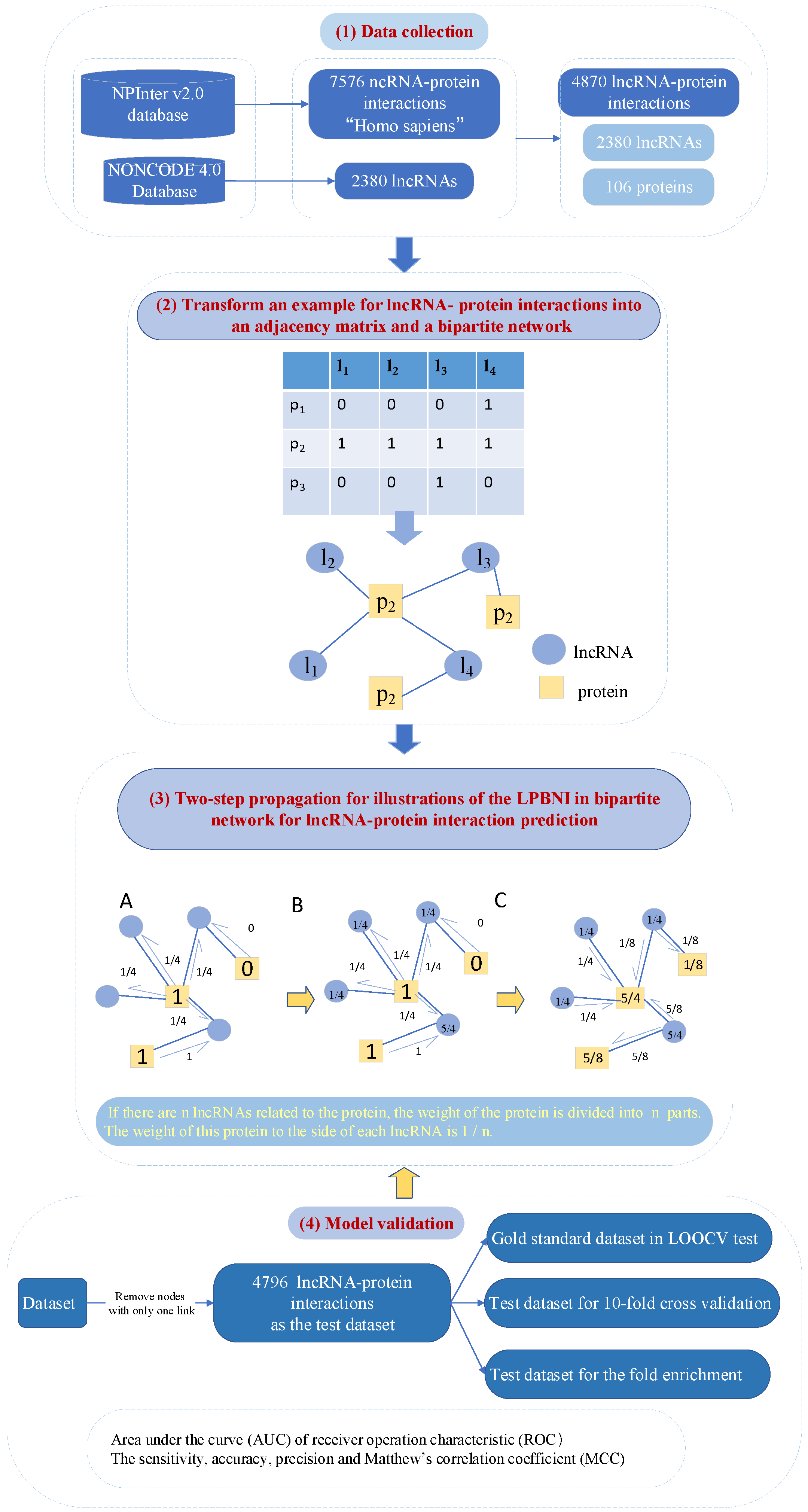
4.1. LPBNI: A Bipartite Network-Based Method for the Prediction of LncRNA–Protein Interactions
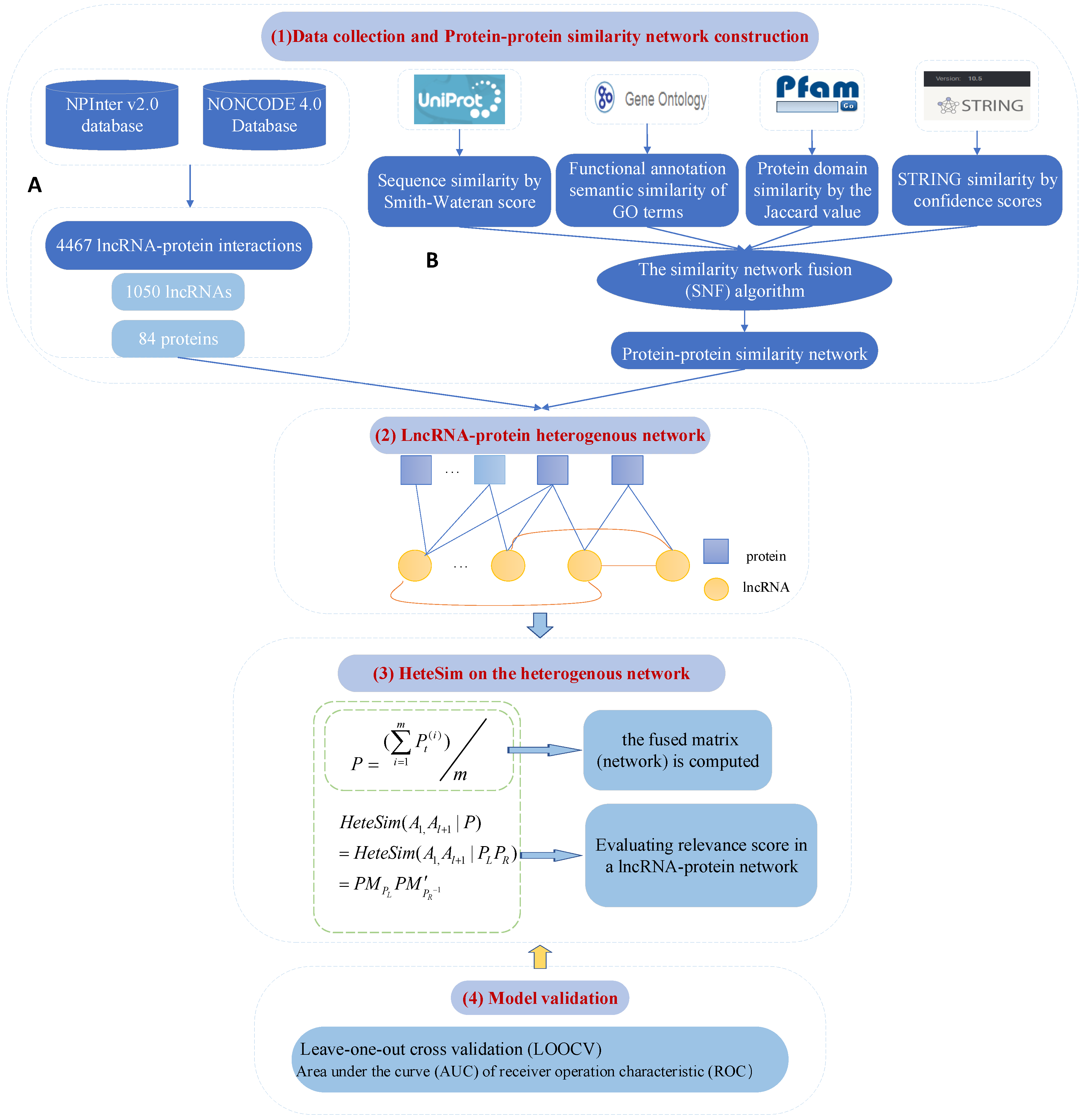
4.2. Fusing Multiple Protein–Protein Similarity Networks to Effectively Predict LncRNA–Protein Interactions
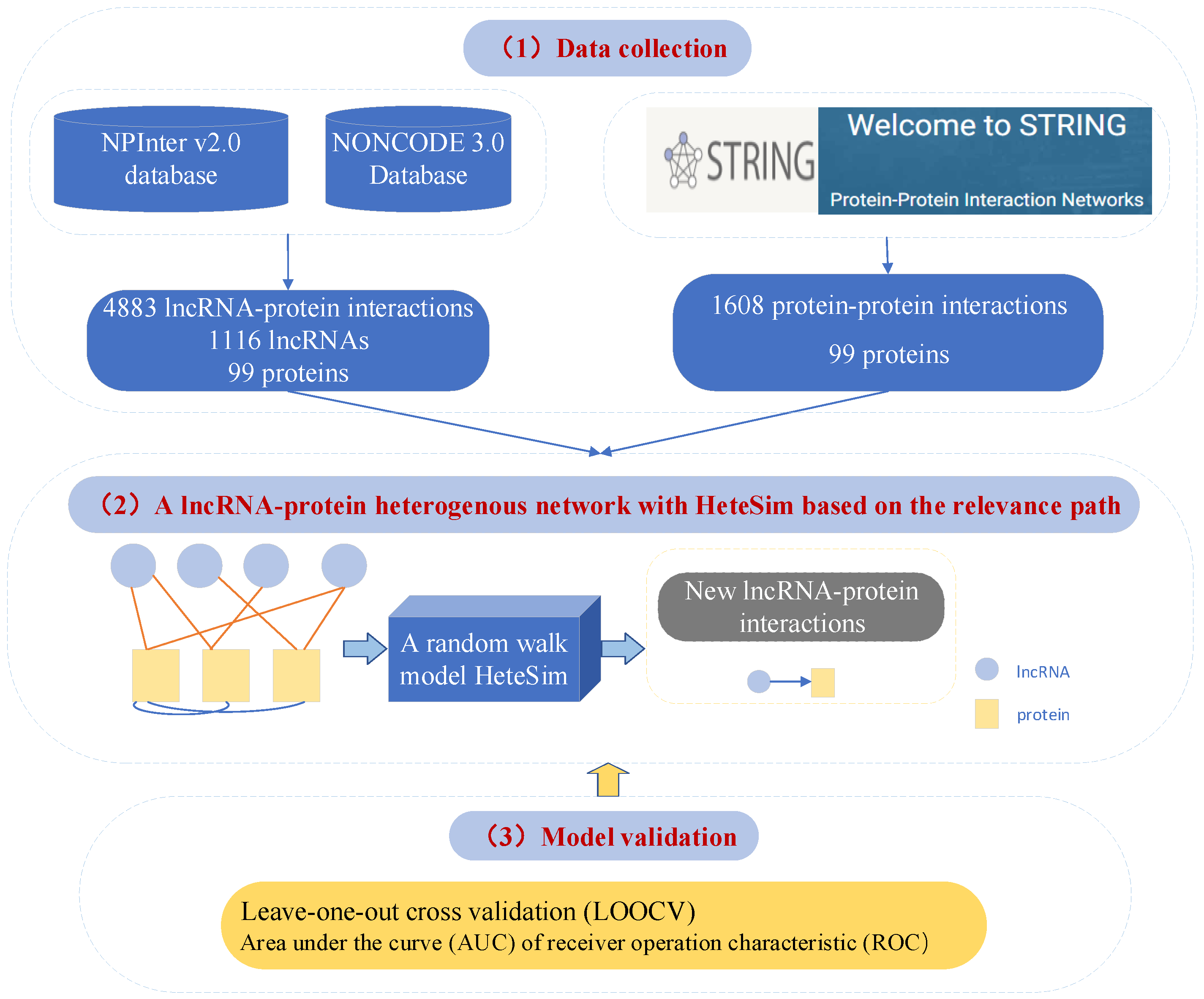
4.3. Prediction of Interactions between lncRNA and Protein by Using Relevance Search in a Heterogeneous LncRNA–Protein Network
4.4. LPIHN: LncRNA–Protein Interaction Prediction Based on Heterogeneous Network Models
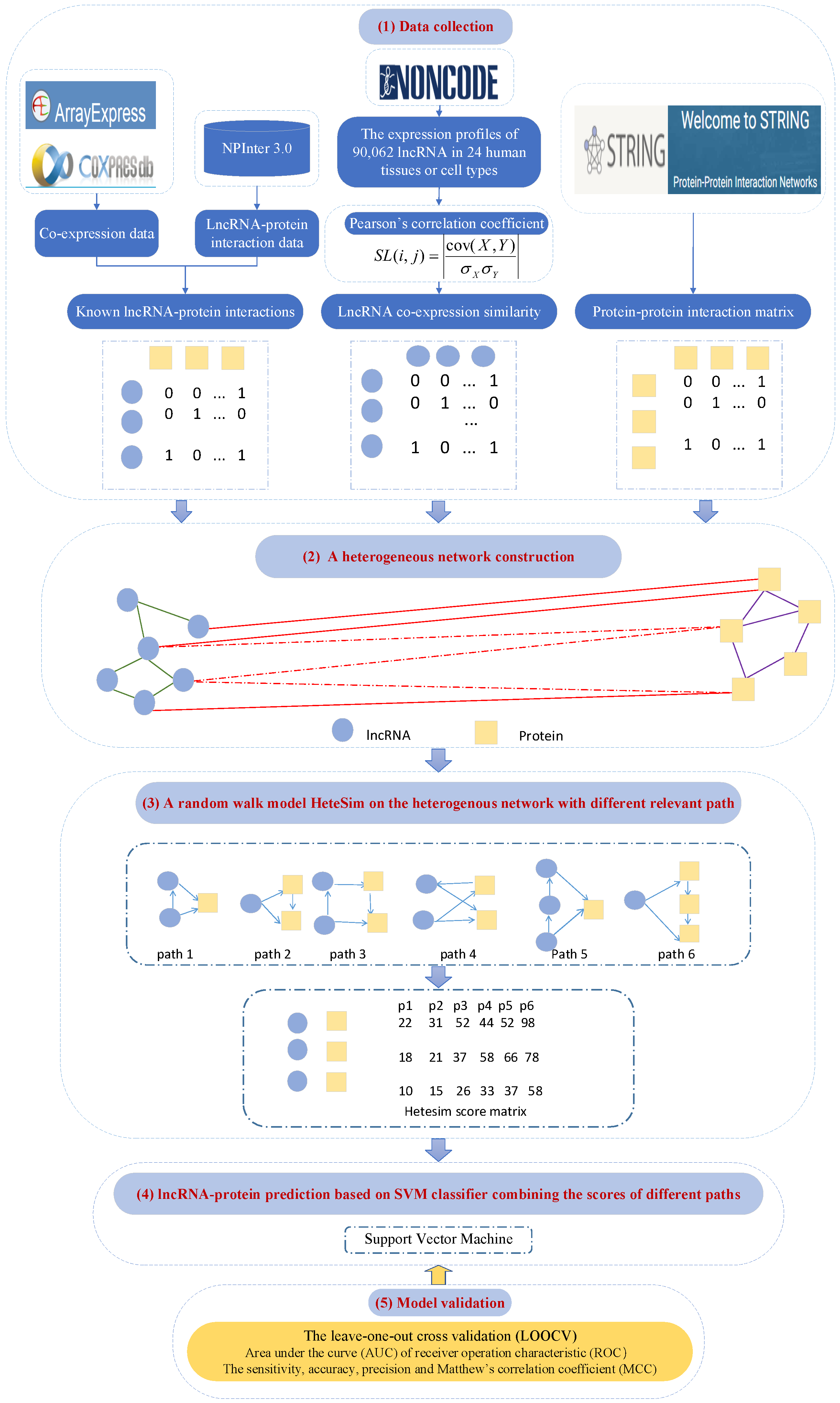
4.5. PLPIHS: Prediction of LncRNA–Protein Interactions Using HeteSim Scores Based on Heterogeneous Networks
5. Results of Comparisons of the Network-Based Models for Predicting LncRNA–Protein Interactions
6. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Nakaya, H.I.; Amaral, P.P.; Louro, R.; Lopes, A.; Fachel, A.A.; Moreira, Y.B.; El-Jundi, T.A.; da Silva, A.M.; Reis, E.M.; Verjovski-Almeida, S. Genome mapping and expression analyses of human intronic noncoding RNAs reveal tissue-specific patterns and enrichment in genes related to regulation of transcription. Genome Biol. 2007, 8, R43. [Google Scholar] [CrossRef] [PubMed]
- Guttman, M.; Amit, I.; Garber, M.; French, C.; Lin, M.F.; Feldser, D.; Huarte, M.; Zuk, O.; Carey, B.W.; Cassady, J.P.; et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 2009, 458, 223–227. [Google Scholar] [CrossRef] [PubMed]
- Guttman, M.; Garber, M.; Levin, J.Z.; Donaghey, J.; Robinson, J.; Adiconis, X.; Fan, L.; Koziol, M.J.; Gnirke, A.; Nusbaum, C.; et al. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat. Biotechnol. 2010, 28, 503–510. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.C.; Chang, H.Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell 2011, 43, 904–914. [Google Scholar] [CrossRef] [PubMed]
- Lu, Q.; Ren, S.; Lu, M.; Zhang, Y.; Zhu, D.; Zhang, X.; Li, T. Computational prediction of associations between long non-coding RNAs and proteins. BMC Genom. 2013, 14. [Google Scholar] [CrossRef]
- Zhao, T.; Xu, J.; Liu, L.; Bai, J.; Xu, C.; Xiao, Y.; Li, X.; Zhang, L. Identification of cancer-related lncRNAs through integrating genome, regulome and transcriptome features. Mol. BioSyst. 2015, 11, 126–136. [Google Scholar] [CrossRef]
- Wilusz, J.E.; Sunwoo, H.; Spector, D.L. Long noncoding RNAs: Functional surprises from the RNA world. Genes Dev. 2009, 23, 1494–1504. [Google Scholar] [CrossRef] [PubMed]
- Managadze, D.; Rogozin, I.B.; Chernikova, D.; Shabalina, S.A.; Koonin, E.V. Negative correlation between expression level and evolutionary rate of long intergenic noncoding RNAs. Genome Biol. Evol. 2011, 3, 1390–1404. [Google Scholar] [CrossRef]
- International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921. [Google Scholar] [CrossRef]
- Qureshi, I.A.; Mattick, J.S.; Mehler, M.F. Long non-coding RNAs in nervous system function and disease. Brain Res. 2010, 1338, 20–35. [Google Scholar] [CrossRef]
- Liao, Q.; Liu, C.; Yuan, X.; Kang, S.; Miao, R.; Xiao, H.; Zhao, G.; Luo, H.; Bu, D.; Zhao, H.; et al. Large-scale prediction of long non-coding RNA functions in a coding-non-coding gene co-expression network. Nucleic Acids Res. 2011, 39, 3864–3878. [Google Scholar] [CrossRef] [PubMed]
- Moran, V.A.; Perera, R.J.; Khalil, A.M. Emerging functional and mechanistic paradigms of mammalian long non-coding RNAs. Nucleic Acids Res. 2012, 40, 6391–6400. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.J.; Fu, H.J.; Wu, Y.G.; Zheng, X.F. Function of lncRNAs and approaches to lncRNA-protein interactions. Sci. China Life Sci. 2013, 56, 876–885. [Google Scholar] [CrossRef] [PubMed]
- Mercer, T.R.; Dinger, M.E.; Mattick, J.S. Insights into functions. Nat. Rev. Genet. 2009, 10, 155–159. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wu, Z.; Fu, X.; Han, W. LncRNAs: Insights into their function and mechanics in underlying disorders. Mutat. Res./Rev. Mutat. Res. 2014, 762, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Wang, Z.; Wang, D.; Qiu, C.; Liu, M.; Chen, X.; Zhang, Q.; Yan, G.; Cui, Q. LncRNADisease: A database for long-non-coding RNA-associated diseases. Nucleic Acids Res. 2013, 41, 983–986. [Google Scholar] [CrossRef] [PubMed]
- Ning, S.; Zhang, J.; Wang, P.; Zhi, H.; Wang, J.; Liu, Y.; Gao, Y.; Guo, M.; Yue, M.; Wang, L.; Li, X. Lnc2Cancer: A manually curated database of experimentally supported lncRNAs associated with various human cancers. Nucleic Acids Res. 2016, 44, D980–D985. [Google Scholar] [CrossRef]
- Ray, D.; Kazan, H.; Chan, E.T.; Castillo, L.P.; Chaudhry, S.; Talukder, S.; Blencowe, B.J.; Morris, Q.; Hughes, T.R. Rapid and systematic analysis of the RNA recognition specificities of RNA-binding proteins. Nat. Biotechnol. 2009, 27, 667–670. [Google Scholar] [CrossRef]
- Keene, J.D.; Komisarow, J.M.; Friedersdorf, M.B. RIP-Chip: The isolation and identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes from cell extracts. Nat. Protoc. 2006, 1, 302–307. [Google Scholar] [CrossRef] [PubMed]
- Licatalosi, D.D.; Mele, A.; Fak, J.J.; Ule, J.; Kayikci, M.; Chi, S.W.; Clark, T.A.; Schweitzer, A.C.; Blume, J.E.; Wang, X.; et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature 2008, 456, 464–469. [Google Scholar] [CrossRef]
- Hafner, M.; Landthaler, M.; Burger, L.; Khorshid, M.; Hausser, J.; Berninger, P.; Rothballer, A.; Ascano, M.; Jungkamp, A.C.; Munschauer, M.; et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 2010, 141, 129–141. [Google Scholar] [CrossRef]
- Pietrosanto, M.; Mattei, E.; Helmer-citterich, M. A novel method for the identification of conserved structural patterns in RNA: From small scale to high-throughput applications. Nucleic Acids Res. 2016, 44, 8600–8609. [Google Scholar] [CrossRef]
- Polishchuk, M.; Paz, I.; Kohen, R.; Mesika, R.; Yakhini, Z. A combined sequence and structure based method for discovering enriched motifs in RNA from in vivo binding data. Methods 2017, 118–119, 73–81. [Google Scholar] [CrossRef]
- Muppirala, U.K.; Honavar, V.G.; Dobbs, D. Predicting RNA-protein interactions using only sequence information. BMC Bioinform. 2011, 12, 489. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, X.; Liu, Z.P.; Huang, Q.; Wang, Y.; Xu, D.; Zhang, X.S.; Chen, R.; Chen, L. De novo prediction of RNA–protein interactions from sequence information. Mol. BioSyst. 2013, 9, 133–142. [Google Scholar] [CrossRef]
- Bellucci, M.; Agostini, F.; Masin, M.; Tartaglia, G.G. Predicting protein associations with long noncoding RNAs. Nat. Methods 2011, 8, 444–445. [Google Scholar] [CrossRef]
- Suresh, V.; Liu, L.; Adjeroh, D.; Zhou, X. RPI-Pred: Predicting ncRNA-protein interaction using sequence and structural information. Nucleic Acids Res. 2015, 43, 1370–1379. [Google Scholar] [CrossRef]
- Akbaripour-Elahabad, M.; Zahiri, J.; Rafeh, R.; Eslami, M.; Azari, M. rpiCOOL: A tool for in silico RNA-protein interaction detection using random forest. J. Theor. Biol. 2016, 402, 1–8. [Google Scholar] [CrossRef]
- Pan, X.; Fan, Y.X.; Yan, J.; Shen, H.B. IPMiner: Hidden ncRNA-protein interaction sequential pattern mining with stacked autoencoder for accurate computational prediction. BMC Genom. 2016, 17, 1–14. [Google Scholar] [CrossRef]
- Yang, C.; Yang, L.; Zhou, M.; Xie, H.; Zhang, C.; Wang, M.D.; Zhu, H. LncADeep: An ab initio lncRNA identification and functional annotation tool based on deep learning. Bioinformatics 2018, 34, 3825–3834. [Google Scholar] [CrossRef]
- Ge, M.; Li, A.; Wang, M. A Bipartite Network-based Method for Prediction of Long Non-coding RNA-protein Interactions. Genom. Proteom. Bioinform. 2016, 14, 62–71. [Google Scholar] [CrossRef]
- Zheng, X.; Wang, Y.; Tian, K.; Zhou, J.; Guan, J.; Luo, L.; Zhou, S. Fusing multiple protein-protein similarity networks to effectively predict lncRNA-protein interactions. BMC Bioinform. 2017, 18, 420. [Google Scholar] [CrossRef]
- Yang, J.; Li, A.; Ge, M.; Wang, M. Prediction of interactions between lncRNA and protein by using relevance search in a heterogeneous lncRNA-protein network. In Proceedings of the 2015 34th Chinese Control Conference (CCC), Hangzhou, China, 28–30 July 2015; pp. 8540–8544. [Google Scholar] [CrossRef]
- Li, A.; Ge, M.; Zhang, Y.; Peng, C.; Wang, M. Predicting long noncoding RNA and protein interactions using heterogeneous network model. BioMed Res. Int. 2015, 2015, 1–11. [Google Scholar] [CrossRef]
- Xiao, Y.; Zhang, J.; Deng, L. Prediction of lncRNA-protein interactions using HeteSim scores based on heterogeneous networks. Sci. Rep. 2017, 7, 1–12. [Google Scholar] [CrossRef]
- Emmert-Streib, F.; Tripathi, S.; Simoes, R.d.M.; Hawwa, A.F.; Dehmer, M. The human disease network. Syst. Biomed. 2013, 1, 20–28. [Google Scholar] [CrossRef]
- Bauer, S.; Horn, D.; Robinson, P.N. Walking the interactome for prioritization of candidate disease genes. AJHG 2008, 82, 949–958. [Google Scholar] [CrossRef]
- Barabási, A.L.; Gulbahce, N.; Loscalzo, J. Network medicine: A network-based approach to human disease. Nat. Rev. Genet. 2011, 12, 56–68. [Google Scholar] [CrossRef]
- Chen, X.; Yan, C.C.; Zhang, X.; You, Z.H. Long non-coding RNAs and complex diseases: From experimental results to computational models. Brief. Bioinform. 2016, 18, bbw060. [Google Scholar] [CrossRef]
- Chen, X.; Yan, G.Y. Novel human lncRNA-disease association inference based on lncRNA expression profiles. Bioinformatics 2013, 29, 2617–2624. [Google Scholar] [CrossRef]
- Chen, X.; Sun, Y.z.; Guan, N.n.; Qu, J.; Huang, Z.a.; Zhu, Z.x.; Li, J.q. Computational models for lncRNA function prediction and functional similarity calculation. Brief. Functional Genom. 2019, 18, 58–82. [Google Scholar] [CrossRef]
- Liu, Y.; Zeng, X.; He, Z.; Zou, Q. Inferring microRNA-disease associations by random walk on a heterogeneous network with multiple data sources. IEEE/ACM Trans. Comput. Biol. Bioinf. 2017, 14, 905–915. [Google Scholar] [CrossRef]
- Zeng, X.; Liu, L.; Lu, L. Prediction of potential disease-associated microRNAs using structural perturbation method. Bioinformatics 2018, 34, 2425–2432. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.; Zhang, X.; Zou, Q. Integrative approaches for predicting microRNA function and prioritizing disease-related microRNA using biological interaction networks. Brief. Bioinform. 2016, 17, 193–203. [Google Scholar] [CrossRef]
- Chen, X.; Wang, L.; Qu, J.; Guan, N.n.; Li, J.-Q. Predicting miRNA-disease association based on inductive matrix completion. Bioinformatics 2018, 34, 4256–4265. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Xie, D.; Wang, L.; Zhao, Q.; You, Z.h.; Liu, H. Systems biology BNPMDA: Bipartite network projection for MiRNA-disease association prediction. Bioinformatics 2018, 34, 3178–3186. [Google Scholar] [CrossRef]
- Chen, X.; Yin, J.; Qu, J.; Huang, L. MDHGI: Matrix Decomposition and Heterogeneous Graph Inference for miRNA-disease association prediction. PLoS Comput. Biol. 2018, 1–25. [Google Scholar] [CrossRef]
- Chen, X.; Huang, L. LRSSLMDA: Laplacian regularized sparse subspace learning for MiRNA-disease association prediction. PLoS Comput. Biol. 2017, 1–29. [Google Scholar] [CrossRef]
- Yao, Q.; Xu, Y.; Yang, H.; Shang, D.; Zhang, C.; Zhang, Y.; Sun, Z.; Shi, X.; Feng, L.; Han, J.; Su, F.; Li, C.; Li, X. Global prioritization of disease candidate metabolites based on a multi-omics composite network. Sci. Rep. 2015, 5, 1–14. [Google Scholar] [CrossRef]
- Chen, X.; Yan, C.C.; Zhang, X.; Zhang, X.; Dai, F. Drug-target interaction prediction: Databases, web servers and computational models. Brief. Bioinform. 2016, 17, 696–712. [Google Scholar] [CrossRef]
- Chen, X.; Guan, N.n.; Sun, Y.z.; Li, J.Q.; Qu, J. MicroRNA-small molecule association identification: From experimental results to computational models. Brief. Bioinform. 2018, 16, 1–15. [Google Scholar] [CrossRef]
- Qu, J.; Chen, X.; Sun, Y.Z.; Li, J.Q.; Ming, Z. Inferring potential small molecule-miRNA association based on triple layer heterogeneous network. J. Cheminform. 2018, 10, 30. [Google Scholar] [CrossRef] [PubMed]
- Consortium, T.R. RNAcentral: An international database of ncRNA sequences. Nucleic Acids Res. 2015, 43, D123–D129. [Google Scholar] [CrossRef]
- Zhao, Y.; Li, H.; Fang, S.; Kang, Y.; Wu, W.; Hao, Y.; Li, Z.; Bu, D.; Sun, N.; Zhang, M.Q.; Chen, R. NONCODE 2016: An informative and valuable data source of long non-coding RNAs. Nucleic Acids Res. 2016, 44, D203–D208. [Google Scholar] [CrossRef]
- Cui, T.; Zhang, L.; Huang, Y.; Yi, Y.; Tan, P.; Zhao, Y.; Hu, Y.; Xu, L.; Li, E.; Wang, D. MNDR v2.0: An updated resource of ncRNA-disease associations in mammals. Nucleic Acids Res. 2018, 46, 371–374. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.l.; Li, J.h.; Wu, J.; Sun, W.j.; Liu, S.; Wang, Z.l.; Zhou, H. deepBase v2.0: Identification, expression, evolution and function of small RNAs, LncRNAs and circular RNAs from deep-sequencing data. Nucleic Acids Res. 2016, 44, 196–202. [Google Scholar] [CrossRef]
- Dinger, M.E.; Pang, K.C.; Mercer, T.R.; Crowe, M.L.; Grimmond, S.M.; Mattick, J.S. NRED: A database of long noncoding RNA expression. Nucleic Acids Res. 2009, 37, 122–126. [Google Scholar] [CrossRef]
- Zhou, K.R.; Liu, S.; Sun, W.j.; Zheng, L.l.; Zhou, H.; Yang, J.h.; Qu, L.h. ChIPBase v2.0: Decoding transcriptional regulatory networks of non-coding RNAs and protein-coding genes from ChIP-seq data. Nucleic Acids Res. 2017, 45, 43–50. [Google Scholar] [CrossRef]
- Bhattacharya, A.; Ziebarth, J.D.; Cui, Y. SomamiR: A database for somatic mutations impacting microRNA function in cancer. Nucleic Acids Res. 2013, 41, 977–982. [Google Scholar] [CrossRef]
- Jiang, Q.; Ma, R.; Wang, J.; Wu, X.; Jin, S.; Peng, J.; Tan, R.; Zhang, T.; Li, Y.; Wang, Y. LncRNA2Function: A comprehensive resource for functional investigation of human lncRNAs based on RNA-seq data. BMC Genom. 2015, 16, 1–11. [Google Scholar] [CrossRef]
- Ning, S.; Yue, M.; Wang, P.; Liu, Y.; Zhi, H.; Zhang, Y.; Zhang, J.; Gao, Y.; Guo, M.; Zhou, D.; et al. LincSNP 2.0: An updated database for linking disease-associated SNPs to human long non-coding RNAs and their TFBSs. Nucleic Acids Res. 2017, 45, 74–78. [Google Scholar] [CrossRef]
- Gong, J.; Liu, W.; Zhang, J.; Miao, X.; Guo, A.Y. lncRNASNP: A database of SNPs in lncRNAs and their potential functions in human and mouse. Nucleic Acids Res. 2015, 43, 181–186. [Google Scholar] [CrossRef] [PubMed]
- Volders, P.J.; Helsens, K.; Wang, X.; Menten, B.; Martens, L.; Gevaert, K.; Vandesompele, J.; Mestdagh, P. LNCipedia: A database for annotated human IncRNA transcript sequences and structures. Nucleic Acids Res. 2013, 41, 246–251. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Zhang, J.; Zhou, Z.; Wang, L.; Liu, Y.; Liu, Y. ALDB: A domestic-animal long noncoding RNA database. PLoS ONE 2015, 10, e0124003. [Google Scholar] [CrossRef]
- Park, C.; Yu, N.; Choi, I.; Kim, W.; Lee, S. Databases and ontologies lncRNAtor: A comprehensive resource for functional investigation of long non-coding RNAs. Bioinformatics 2014, 30, 2480–2485. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Bai, J.; Wu, A.; Wang, Y.; Zhang, J.; Wang, Z.; Li, Y.; Xu, J.; Li, X. Co-LncRNA: Investigating the lncRNA combinatorial effects in GO annotations and KEGG pathways based on human RNA-Seq data. Database 2015, 2015, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Chan, W.L.; Huang, H.D.; Chang, J.G. lncRNAMap: A map of putative regulatory functions in the long non-coding transcriptome. Comput. Biol. Chem. 2014, 50, 41–49. [Google Scholar] [CrossRef]
- Gong, J.; Liu, C.; Liu, W.; Xiang, Y.; Diao, L.; Guo, A.y.; Han, L. LNCediting: A database for functional effects of RNA editing in lncRNAs. Nucleic Acids Res. 2017, 45, 79–84. [Google Scholar] [CrossRef]
- Paraskevopoulou, M.D.; Georgakilas, G.; Kostoulas, N.; Reczko, M.; Maragkakis, M.; Dalamagas, T.M.; Hatzigeorgiou, A.G. DIANA-LncBase: Experimentally verified and computationally predicted microRNA targets on long non-coding RNAs. Nucleic Acids Res. 2013, 41, 239–245. [Google Scholar] [CrossRef]
- Jiang, Q.; Wang, J.; Wang, Y.; Ma, R.; Wu, X.; Li, Y. TF2LncRNA: Identifying common transcription factors for a list of lncRNA genes from ChIP-Seq data. BioMed Res. Int. 2014, 2014. [Google Scholar] [CrossRef]
- Xu, Y.; Li, F.; Wu, T.; Xu, Y.; Yang, H.; Dong, Q. LncSubpathway: A novel approach for identifying dysfunctional subpathways associated with risk lncRNAs by integrating lncRNA and mRNA expression profiles and pathway topologies. Oncotarget 2017, 8, 15453–15469. [Google Scholar] [CrossRef]
- Jiang, Q.; Wang, J.; Wu, X.; Ma, R.; Zhang, T.; Jin, S.; Han, Z.; Tan, R.; Peng, J.; Liu, G.; Li, Y.; Wang, Y. LncRNA2Target: A database for differentially expressed genes after IncRNA knockdown or overexpression. Nucleic Acids Res. 2015, 43, D193–D196. [Google Scholar] [CrossRef]
- Zhou, Z.; Shen, Y.; Khan, M.R.; Li, A. Original article LncReg: A reference resource for lncRNA-associated regulatory networks. Database 2015, 2015, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Quek, X.C.; Thomson, D.W.; Maag, J.L.V.; Bartonicek, N.; Signal, B.; Clark, M.B.; Gloss, B.S.; Dinger, M.E. lncRNAdb v2.0: Expanding the reference database for functional long noncoding RNAs. Nucleic Acids Res. 2015, 43, D168–D173. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.; Wu, W.; Xie, C.; Zhao, G.; Zhao, Y.; Chen, R. NPInter v2.0: An updated database of ncRNA interactions. Nucleic Acids Res. 2014, 42, 104–108. [Google Scholar] [CrossRef] [PubMed]
- Lewis, B.A.; Walia, R.R.; Terribilini, M.; Ferguson, J.; Zheng, C.; Honavar, V.; Dobbs, D. PRIDB: A protein- RNA interface database. Nucleic Acids Res. 2011, 39, 277–282. [Google Scholar] [CrossRef] [PubMed]
- Sussman, J.L.; Lin, D.; Jiang, J.; Manning, N.O.; Prilusky, J.; Ritter, O.; Abola, E.E. Protein Data Bank (PDB): Database of three-dimensional structural information of biological macromolecules. Acta Crystallogr. Sect. D Biol. Crystallogr. 1998, 54, 1078–1084. [Google Scholar] [CrossRef]
- Li, J.H.; Liu, S.; Zhou, H.; Qu, L.H.; Yang, J.H. starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014, 42, D92–D97. [Google Scholar] [CrossRef]
- Narayanan, B.C.; Westbrook, J.; Ghosh, S.; Petrov, A.I.; Sweeney, B.; Zirbel, C.L.; Leontis, N.B.; Berman, H.M. The nucleic acid database: New features and capabilities. Nucleic Acids Res. 2014, 42, 114–122. [Google Scholar] [CrossRef]
- Chen, X. Predicting lncRNA-disease associations and constructing lncRNA functional similarity network based on the information of miRNA. Sci. Rep. 2015, 5, 1–12. [Google Scholar] [CrossRef]
- Jimeno-Yepes, A.J.; Sticco, J.C.; Mork, J.G.; Aronson, A.R. GeneRIF indexing: Sentence selection based on machine learning. BMC Bioinform. 2013, 14, 1–11. [Google Scholar] [CrossRef]
- Zhong, Y.; Xuan, P.; Wang, X.; Zhang, T.; Li, J.; Liu, Y.; Zhang, W. A non-negative matrix factorization based method for predicting disease-associated miRNAs in miRNA-disease bilayer network. Bioinformatics 2018, 34, 267–277. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Qiu, C.; Tu, J.; Geng, B.; Yang, J.; Jiang, T.; Cui, Q. HMDD v2.0: A database for experimentally supported human microRNA and disease associations. Nucleic Acids Res. 2014, 42, 1070–1074. [Google Scholar] [CrossRef]
- Jiang, Q.; Wang, Y.; Hao, Y.; Juan, L.; Teng, M.; Zhang, X.; Li, M.; Wang, G.; Liu, Y. miR2Disease: A manually curated database for microRNA deregulation in human disease. Nucleic Acids Res. 2009, 37, 98–104. [Google Scholar] [CrossRef]
- Xie, B.; Ding, Q.; Han, H.; Wu, D. MiRCancer: A microRNA-cancer association database constructed by text mining on literature. Bioinformatics 2013, 29, 638–644. [Google Scholar] [CrossRef]
- Chou, C.H.; Chang, N.W.; Shrestha, S.; Hsu, S.D.; Lin, Y.L.; Lee, W.H.; Yang, C.D.; Hong, H.C.; Wei, T.Y.; Tu, S.J.; et al. miRTarBase 2016: Updates to the experimentally validated miRNA-target interactions database. Nucleic Acids Res. 2016, 44, D239–D247. [Google Scholar] [CrossRef] [PubMed]
- Dimmer, E.C.; Huntley, R.P.; Alam-Faruque, Y.; Sawford, T.; O’Donovan, C.; Martin, M.J.; Bely, B.; Browne, P.; Chan, W.M.; Eberhardt, R.; et al. The UniProt-GO annotation database in 2011. Nucleic Acids Res. 2012, 40, 565–570. [Google Scholar] [CrossRef] [PubMed]
- Piñero, J.; Bravo, Á.; Queralt-Rosinach, N.; Gutiérrez-Sacristán, A.; Deu-Pons, J.; Centeno, E.; García-García, J.; Sanz, F.; Furlong, L.I. DisGeNET: A comprehensive platform integrating information on human disease- associated genes and variants. Nucleic Acids Res. 2017, 45, D833–D839. [Google Scholar] [CrossRef]
- Law, V.; Knox, C.; Djoumbou, Y.; Jewison, T.; Guo, A.C.; Liu, Y.; MacIejewski, A.; Arndt, D.; Wilson, M.; Neveu, V.; et al. DrugBank 4.0: Shedding new light on drug metabolism. Nucleic Acids Res. 2014, 42, 1091–1097. [Google Scholar] [CrossRef]
- Wishart, D.S.; Jewison, T.; Guo, A.C.; Wilson, M.; Knox, C.; Liu, Y.; Djoumbou, Y.; Mandal, R.; Aziat, F.; Dong, E.; et al. HMDB 3.0—The human metabolome database in 2013. Nucleic Acids Res. 2013, 41, 801–807. [Google Scholar] [CrossRef] [PubMed]
- Mattingly, C.J.; Colby, G.T.; Forrest, J.N.; Boyer, J.L. The Comparative Toxicogenomics Database (CTD). Environ. Health Perspect. 2003. [Google Scholar] [CrossRef]
- Kuhn, M.; Letunic, I.; Jensen, L.J.; Bork, P. The SIDER database of drugs and side effects. Nucleic Acids Res. 2016, 44, D1075–D1079. [Google Scholar] [CrossRef] [PubMed]
- Gruber, A.R.; Lorenz, R.; Bernhart, S.H.; Neuböck, R.; Hofacker, I.L. The Vienna RNA websuite. Nucleic Acids Res. 2008, 36, 70–74. [Google Scholar] [CrossRef]
- Shi, C.; Kong, X.; Huang, Y.; Yu, P.S.; Wu, B. HeteSim: A general framework for relevance measure in heterogeneous networks. IEEE Trans. Knowl. Data Eng. 2014, 26, 2479–2492. [Google Scholar] [CrossRef]
- Wang, B.; Mezlini, A.M.; Demir, F.; Fiume, M.; Tu, Z.; Brudno, M.; Haibe-Kains, B.; Goldenberg, A. Similarity network fusion for aggregating data types on a genomic scale. Nat. Methods 2014, 11, 333–337. [Google Scholar] [CrossRef]
- Zhou, T.; Ren, J.; Medo, M.; Zhang, Y.C. Bipartite network projection and personal recommendation. Phys. Rev. E 2007, 76, 1–7. [Google Scholar] [CrossRef]
- Chen, X.; Ba, Y.; Ma, L.; Cai, X.; Yin, Y.; Wang, K.; Guo, J.; Zhang, Y.; Chen, J.; Guo, X.; et al. Characterization of microRNAs in serum: A novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008, 18, 997–1006. [Google Scholar] [CrossRef]
- Wang, F.; Zheng, Z.; Guo, J.; Ding, X. Correlation and quantitation of microRNA aberrant expression in tissues and sera from patients with breast tumor. Gynecol. Oncol. 2010, 119, 586–593. [Google Scholar] [CrossRef]
- Ganegoda, G.U.; Wang, J.X.; Wu, F.X.; Li, M. Prioritization of candidate genes based on disease similarity and protein’s proximity in PPI networks. In Proceedings of the 2013 IEEE International Conference on Bioinformatics and Biomedicine, IEEE BIBM 2013, Shanghai, China, 18–21 December 2013; pp. 103–108. [Google Scholar] [CrossRef]
- Tang, X.; Wang, J.; Zhong, J.; Pan, Y. Predicting essential proteins basedon weighted degree centrality. IEEE/ACM Trans. Comput. Biol. Bioinform. 2014, 11, 407–418. [Google Scholar] [CrossRef]
- Li, M.; Zheng, R.; Zhang, H.; Wang, J.; Pan, Y. Effective identification of essential proteins based on priori knowledge, network topology and gene expressions. Methods 2014, 67, 325–333. [Google Scholar] [CrossRef]
- Li, M.; Zhang, H.; Wang, J.x.; Pan, Y. A new essential protein discovery method based on the integration of protein-protein interaction and gene expression data. BMC Syst. Biol. 2012, 6, 15. [Google Scholar] [CrossRef]
- Shang, D.; Yang, H.; Xu, Y.; Yao, Q.; Zhou, W.; Shi, X.; Han, J.; Su, F.; Su, B.; Zhang, C.; et al. A global view of network of lncRNAs and their binding proteins. Mol. BioSyst. 2015, 11, 656–663. [Google Scholar] [CrossRef] [PubMed]
- Franceschini, A.; Szklarczyk, D.; Frankild, S.; Kuhn, M.; Simonovic, M.; Roth, A.; Lin, J.; Minguez, P.; Bork, P.; von Mering, C.; et al. STRING v9.1: Protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2012, 41, D808–D815. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.; Liao, Y.; Liu, Y.; Zou, Q. Prediction and validation of disease genes using HeteSim scores. IEEE/ACM Trans. Comput. Biol. Bioinform. 2017, 14, 687–695. [Google Scholar] [CrossRef] [PubMed]
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martin, D.; Merkel, A.; Knowles, D.G.; et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012, 1775–1789. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef]
- Kohavi, R. A study of cross-validation and bootstrap for accuracy estimation and model selection a study of cross-validation and bootstrap for accuracy estimation and model selection. In Proceedings of the 14th International Joint Conference on Artificial Intelligence, Montreal, QC, Canada, 20–25 August 1995. [Google Scholar]

| Database | Description | Availability |
|---|---|---|
| ncRNA database (Especially lncRNAs): | ||
| NONCODE [54] | Comprehensive knowledge database of non-coding RNAs, including lncRNAs from 17 species, and predicted/validated lncRNA–disease relationships. | http://www.noncode.org |
| MNDR [55] | Database of ncRNA–disease associations in mammals. | http://www.rna-society.org/mndr |
| deepBase [56] | Database for identification, expression, evolution and function of small RNAs, lncRNAs and circular RNAs from deep-sequencing data. | http://rna.sysu.edu.cn/deepBase |
| NRED [57] | Database integrating annotated human and mouse ncRNA expression data from various resources. | http://nred.matticklab.com |
| ChIPBase [58] | Database on the transcriptional regulation of ncRNAs based on ChIP-sequencing data. | http://rna.sysu.edu.cn/chipbase |
| SomamiR [59] | Cancer somatic mutations with altering microRNA–ceRNA interactions. | http://compbio.uthsc.edu/SomamiR |
| LncRNA2Function [60] | Functional annotations and expression profiles (RNAseq) of human lncRNAs. | http://mlg.hit.edu.cn/lncrna2function |
| LincSNP [61] | A database containing human lncRNAs information about linking disease related SNPs. | http://bioinfo.hrbmu.edu.cn/LincSNP |
| LncRNA-SNP [62] | A database of SNPs in lncRNAs and their predicted effects in human and mouse. | http://bioinfo.life.hust.edu.cn/lncRNASNP |
| LNCipedia [63] | A database for annotated human lncRNA transcript sequences and structures. | http://www.lncipedia.org |
| ALDB [64] | A farm livestock lncRNA database. | http://res.xaut.edu.cn/aldb/index.jsp |
| lncRNAtor [65] | A database for functional investigation of lncRNAs that encompasses annotation, sequence analysis, gene expression, protein binding and phylogenetic conservation. | http://lncrnator.ewha.ac.kr |
| Co-LncRNA [66] | A web-sever containing effects of lncRNAs in GO functions and KEGG pathways based on co-expressed genes. | http://www.bio-bigdata.com/Co-LncRNA |
| Lnc2Cancer [17] | A database for experimentally validated associations between lncRNAs and cancers. | http://www.bio-bigdata.net/lnc2cancer |
| LncRNADisease [16] | A database for experimentally validated lncRNA-associated diseases. | http://www.cuilab.cn/lncrnadisease |
| lncRNAMap [67] | A map of putative regulatory functions in the long non-coding transcriptome. | http://lncRNAMap.mbc.nctu.edu.tw/ |
| TANRIC [34] | A web-resource for interactive exploration of lncRNAs in cancer. | http://ibl.mdanderson.org/tanric/_design/basic/index.html |
| LncRNA ontology [64] | A web-resource for inferring lncRNA functions based on chroma-tin states and expression patterns. | http://www.bio-bigdata.com/lncrnaontology/ |
| LNCediting [68] | A database for functional effects of RNA editing in lncRNAs. | http://bioinfo.life.hust.edu.cn/LNCediting/ |
| LncBase [69] | A database of interactions between miRNAs and lncRNAs. | http://www.microrna.gr/LncBase |
| TF2LncRNA [70] | A Web-resource for the identification of common transcription factors for a list of lncRNA genes. | http://mlg.hit.edu.cn/tf2lncrna |
| LncSubpathway [71] | A web server for the identification of dysfunctional subpathways associated with risk lncRNAs. | http://www.bio-bigdata.com/lncSubpathway/ |
| LncRNA2Target [72] | A database of differentially expressed genes after lncRNA knock-down or overexpression. | http://lncrna2target.org |
| LncReg [73] | A reference resource for lncRNA-associated regulatory networks. | http://bioinformatics.ustc.edu.cn/lncreg/ |
| lncRNAdb [74] | An annotation database of eukaryotic lncRNAs. | http://www.lncrnadb.org/ |
| Database information on proteins or microRNAs that may be associated with lncRNAs: | ||
| NPInter [75] | Database of noncoding RNA-associated interactions. | http://www.bioinfo.org/NPInter |
| PRIDB [76] | Comprehensive database of RNA–protein interfaces extracted from complexes in the PDB. | http://bindr.gdcb.iastate.edu/PRIDB |
| PDB [77] | A database of experimentally determined three-dimensional structures of proteins, nucleic acids and other biomolecules. | http://www.rcsb.org/ |
| StarBase v 2.0 [78] | A database of experimentally supported interactions from RBPs, mRNAs, miRNAs, RNAs, proteins and so on. | http://starbase.sysu.edu.cn/ |
| Nucleic acid database (NDB) [79] | A database about three-dimensional nucleic acid structures and their complexes, geometric data, structure information. | http://ndbserver.rutgers.edu/ |
| Name | Samples | Interactions | Source |
|---|---|---|---|
| LncRNA–Disease | 804 × 288 | 1454 | LncRNADisease [16], Lnc2Cancer [17] |
| LncRNA–LncRNA | 1114 × 1114 | 1,179,256 | LFSCM [80] |
| LncRNA–microRNA | 1127 × 277 | 10,198 | StarBase v2.0 [78] |
| LncRNA–Gene | 240 × 15,527 | 6186 | LncRNA2Target [72] |
| LncRNA–GO | 240 × 6428 | 3094 | GeneRIF [81] |
| MicroRNA–MicroRNA | 271 × 271 | 24,062 | Zhong et al. [82] |
| MicroRNA–Disease | 1080 × 592 | 11,835 | HMDD [83], miR2Disease [84], miRCancer [85] |
| MicroRNA–Gene | 495 × 15,527 | 135,852 | miRTarBase [86] |
| MicroRNA–Target | 495 × 15,527 | 135,852 | miRTarBase [86] |
| Gene–Gene | 16,785 × 16,785 | 1,515,370 | Yao et al. [49] |
| Gene–Metabolite | 12,342 × 3278 | 192,763 | Yao et al. [49] |
| Metabolite–Metabolite | 3764 × 3764 | 74,667 | Yao et al. [49] |
| Gene–GO | 15,527 × 6428 | 1,191,503 | GO Annotation [87] |
| Gene–Disease | 1715 × 1886 | 2603 | DisGeNET [88] |
| Gene–Drug | 155,275 × 8283 | 3760 | DrugBank [89] |
| Metabolite–Disease | 388 × 149 | 664 | HMDB [90] |
| Drug–Disease | 15,527 × 412 | 115,317 | CTD [91] |
| Drug–Drug | 8283 × 8283 | 453,436 | DrugBank [89] |
| Drug–Side-effects | 1430 × 5880 | 140,064 | SIDER [92] |
| Disease–Disease | 5080 × 5080 | 20,280,092 | Yao et al. [49] |
| CatRAPID [26] | RPISeq [24] | De novo [25] | LncPro [5] | RPI-Pred [27] | rpiCOOL [28] | IPMiner [29] | lncADeep [30] | ||
|---|---|---|---|---|---|---|---|---|---|
| Feature | RNA Sequence | √ | √ | √ | √ | √ | √ | ||
| Protein Sequence | √ | √ | √ | √ | √ | √ | |||
| 3D Structure(RNA) | √ | ||||||||
| 3D Structure (protein) | √ | ||||||||
| The secondary structure (RNA) | √ | √ | |||||||
| The secondary structure(protein) | √ | ||||||||
| Hydrogen-Bonding Propensities | √ | √ | |||||||
| van der Waals’ Propensities | √ | √ | |||||||
| Classifier | Random Forest | √ | √ | √ | |||||
| Naive Bayesian | √ | ||||||||
| Extended NB | √ | ||||||||
| SVM | √ | √ | |||||||
| Fisher’s linear | √ | ||||||||
| automatic encoder | √ | ||||||||
| deep neural network | √ | ||||||||
| p-values | √ | √ | |||||||
| Web server or offline package | √ | √ | √ | √ | √ | √ | √ | ||
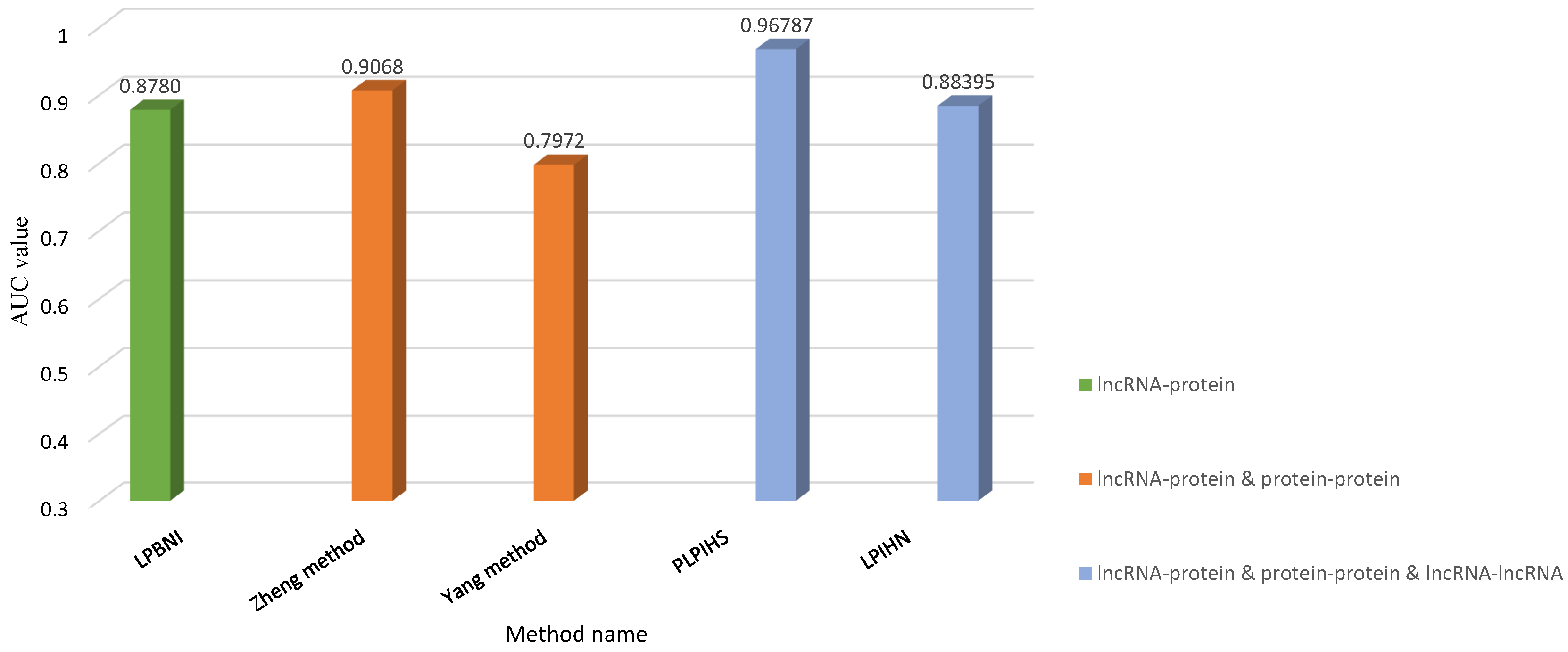
| Method | Dataset | Algorithm | AUC | |
|---|---|---|---|---|
| LPBNI [31] | LPI | 4870 lncRNA–protein interactions from NPInter database (2380 lncRNAs and 106 proteins) | Bipartite Network | 0.8780 |
| PPI | × | |||
| LLI | × | |||
| Yang et al. [33] | LPI | 4883 lncRNA–protein interactions from NPInter database (1116 lncRNAs and 99 proteins) | A random walk model HeteSim | 0.7972 |
| PPI | 1608 protein–protein interactions from STRING database | |||
| LLI | × | |||
| LPIHN [34] | LPI | 10232 lncRNA–protein interactions from NPInter database (1113 lncRNAs and 99 proteins) | Random Walk with Restart | 0.8839 |
| PPI | 804 protein–protein interactions from STRING database | |||
| LLI | lncRNA expression similarity from NONCODE 4.0 database (1113 lncRNA expression profiles) | |||
| Zheng et al. [32] | LPI | 4467 lncRNA–protein interactions from NPInter database (1050 lncRNAs and 84 proteins) | SNF; A random walk model HeteSim | 0.9068 |
| PPI | Sequence similarity from UniProt database; Functional annotation similarity from GO database; Protein domain similarity from Pfam database; STRING similarity from STRING database; | |||
| LLI | × | |||
| PLPIHS [35] | LPI | lncRNA–protein interactions from GENCODE Release 24 (15941 lncRNAs and 20284 proteins) Co-expression data from COXPRESdb; Co-expression data from ArrayExpress and GEO; lncRNA–protein interactions from NPInter database; | SVM; A random walk model HeteSim | 0.9678 |
| PPI | Protein–protein interactions from STRING database | |||
| LLI | lncRNA co-expression similarity from NONCODE database (lncRNA expression profiles) | |||
| Method | Measure for the Evaluation | Test Dataset | Measurement or Illustration | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LOOCV | Precision Versus | Fold Enrichment | AUC | SPE | ACC | PRE | MCC | REC | F1-Score | SEN | ||
| LPBNI [31] | √ | √ | 4870 lncRNA–protein interactions from NPInter v2.0 | 0.878 | 0.99 | 0.873 | 0.852 | 0.449 | − | − | 0.288 | |
| 0.95 | 0.880 | 0.681 | 0.534 | 0.532 | ||||||||
| Zheng et al. [32] | √ | 4467 lncRPIs, including 1050 lncRNAs and 84 proteins | AUC values of 15 settings: Seqs-0.8565, Pfam-0.8459, GO-0.8584, STRING-0.7972; Seqs+Pfam-0.8689, Seqs+GO-0.8626, Seqs+STRING-0.8762, Pfam+GO-0.8677, Pfam+STRING-0.8977, and GO+STRING-0.8814; Seqs+Pfam+GO-0.8704, Seqs+Pfam+STRING-0.9023, Seqs+GO+STRING-0.8904, Pfam+GO+STRING-0.9066; Seqs+Pfam+GO+STRING-0.9068. | |||||||||
| Yang et al. [33] | √ | MALAT1 with all 99 proteins | 0.955 | − | − | − | − | − | − | − | ||
| AK0951949 with all 99 proteins | 0.973 | |||||||||||
| LPIHN [34] | √ | √ | √ | The test dataset is the interaction of each known lncRNA–protein, and the rest is used as training dataset. | 0.96 | √ | √ | √ | √ | √ | √ | |
| PLPIHS [35] | √ | The remaining positive samples found in the 0.9 network had 2000 lncRNA–protein interactions and the same number of negative interactions in the 0.3 network | 0.879 | − | √ | √ | √ | √ | √ | |||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, H.; Liang, Y.; Han, S.; Peng, C.; Li, Y. Long Noncoding RNA and Protein Interactions: From Experimental Results to Computational Models Based on Network Methods. Int. J. Mol. Sci. 2019, 20, 1284. https://doi.org/10.3390/ijms20061284
Zhang H, Liang Y, Han S, Peng C, Li Y. Long Noncoding RNA and Protein Interactions: From Experimental Results to Computational Models Based on Network Methods. International Journal of Molecular Sciences. 2019; 20(6):1284. https://doi.org/10.3390/ijms20061284
Chicago/Turabian StyleZhang, Hui, Yanchun Liang, Siyu Han, Cheng Peng, and Ying Li. 2019. "Long Noncoding RNA and Protein Interactions: From Experimental Results to Computational Models Based on Network Methods" International Journal of Molecular Sciences 20, no. 6: 1284. https://doi.org/10.3390/ijms20061284
APA StyleZhang, H., Liang, Y., Han, S., Peng, C., & Li, Y. (2019). Long Noncoding RNA and Protein Interactions: From Experimental Results to Computational Models Based on Network Methods. International Journal of Molecular Sciences, 20(6), 1284. https://doi.org/10.3390/ijms20061284





