Efficient Genome Editing Using CRISPR/Cas9 Technology in Chicory
Abstract
1. Introduction
2. Results
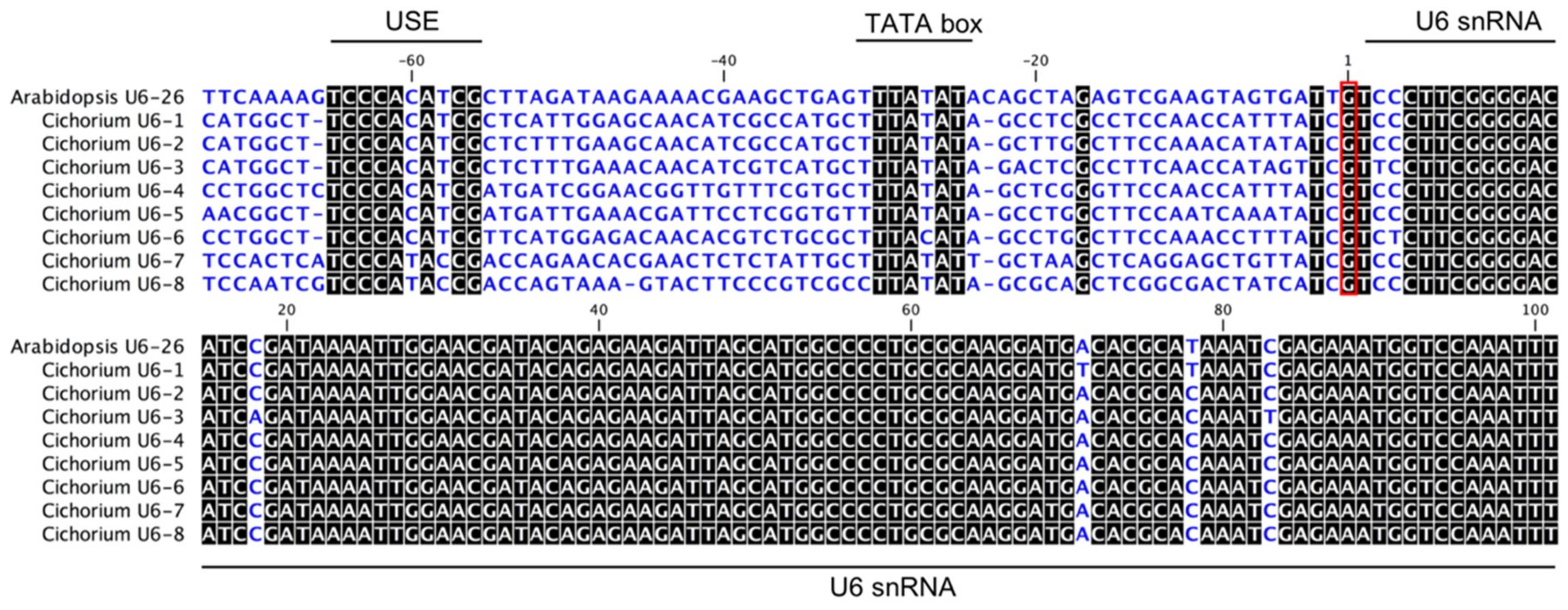
2.1. Prediction of U6 Promoter Sequences in Chicory
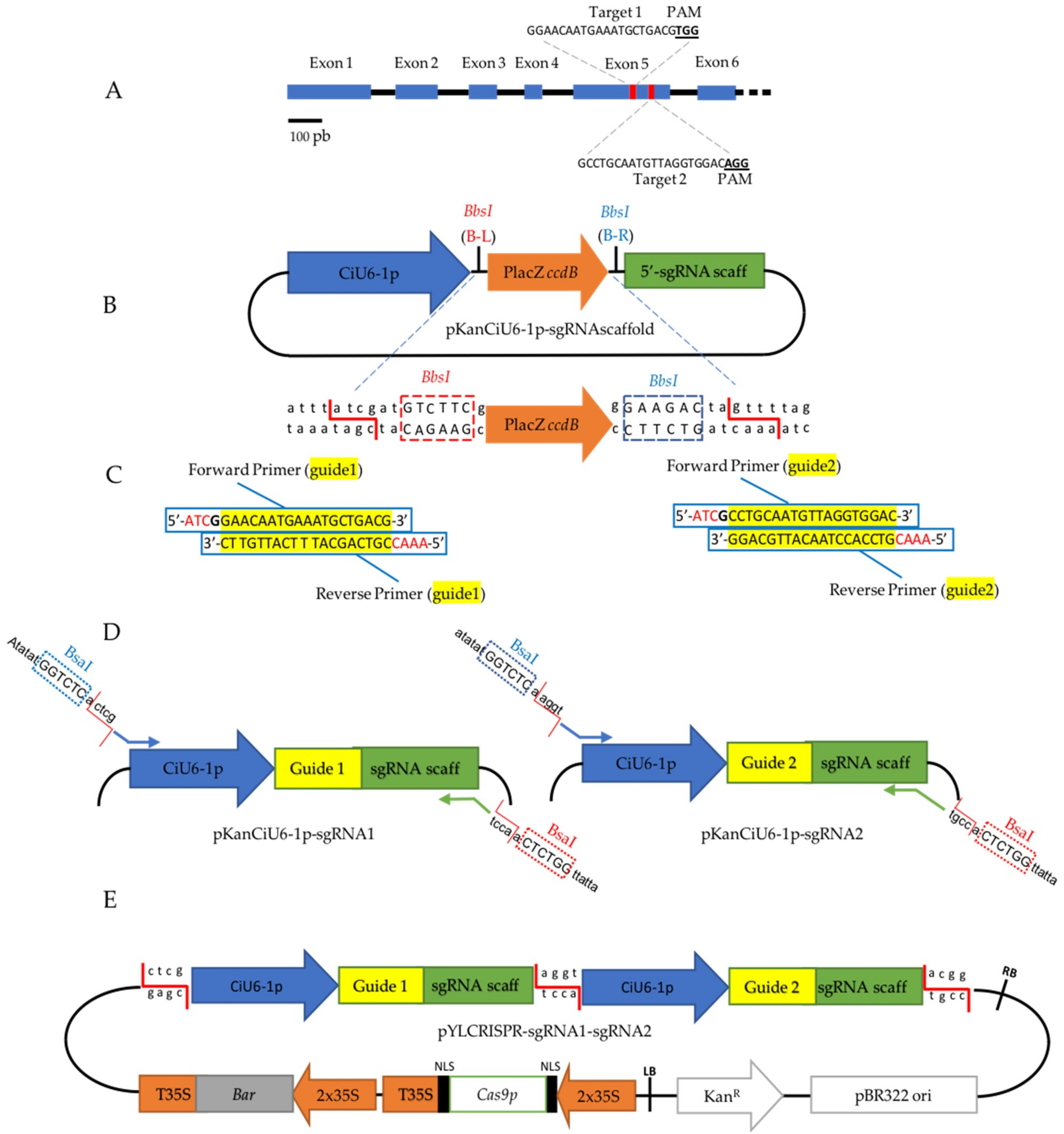
2.2. Target Selection and Plasmid Construction for the CRISPR/Cas9 System
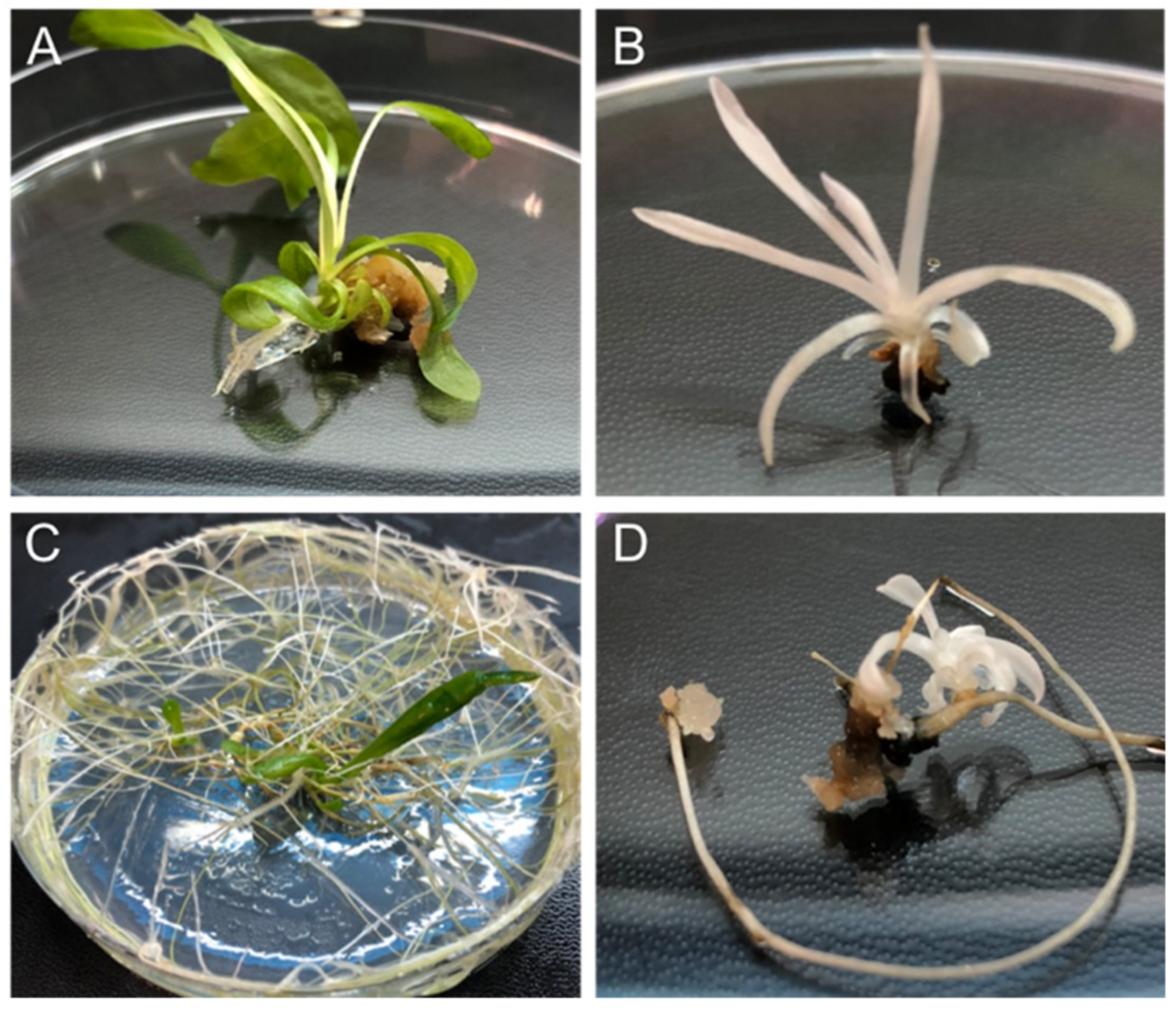
2.3. Regeneration of CRISPR-Edited Plants From Protoplasts and From Hairy Roots Lines
2.4. Identification of Mutation Events in Genome Edited Plants
2.5. Transgene Detection in Albino Plants
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Sequence Identification and gRNA Design
4.3. Vector Construction
4.4. A. rhizogenes Transformation
4.5. Hairy Root Induction and Maintenance
4.6. Protoplast Isolation, Transformation, and Culture
4.7. DNA Extraction, Detection of Mutation, and CAS9 Amplification
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
- 1)
- 2)
- Accession numbers: U6-1: MK455772, U6-2: MK455773, U6-3: MK455774, U6-4: MK455775, U6-5: MK455776), U6-6: MK455777, U6-7: MK455778, U6-8: MK455779, CiPDS: MK455771, HaPDS: AHA36971.1.
Appendix B
| Name | Sequences (5′-3′) |
|---|---|
| Forward primer (guide1) | ATCGGAACAATGAAATGCTGACG |
| Reverse primer (guide1) | AAACCGTCAGCATTTCATTGTTC |
| Forward primer (guide2) | ATCGCCTGCAATGTTAGGTGGAC |
| Reverse primer (guide2) | AAACGTCCACCTAACATTGCAGG |
| GG1-F | ATATATGGTCTCACTCGAAAGAACCAACCTGTTTTCATAGC |
| GG1-R | ATTATTGGTCTCAACCTAAAAAAAGCACCGACTCGGTG |
| GG2-R | ATATATGGTCTCAAGGTAAAGAACCAACCTGTTTTCATAGC |
| GG2-F | ATTATTGGTCTCAACCGAAAAAAAGCACCGACTCGGTG |
| E5-F | TTTCTTGAAGTTGGAGCTTACC |
| E5-R | AAACCTCAGTAGTAACTCGATCTG |
| C9-F | AAGCACGTTGCTCAGATCCT |
| C9-R | CCGTTCGTCTCGATAAGAGG |
References
- Bortesi, L.; Fischer, R. The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol. Adv. 2015, 33, 41–52. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.G.; Cha, J.; Chandrasegaran, S. Hybrid restriction enzymes: Zinc finger fusions to Fok I cleavage domain. Proc. Natl. Acad. Sci. USA 1996, 93, 1156–1160. [Google Scholar] [CrossRef] [PubMed]
- Christian, M.; Cermak, T.; Doyle, E.L.; Schmidt, C.; Zhang, F.; Hummel, A.; Bogdanove, A.J.; Voytas, D.F. Targeting DNA Double-Strand Breaks with TAL Effector Nucleases. Genetics 2010, 186, 757–761. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.X.; Wang, W.; Le, H.T.T.; Vu, G.T.H. The Power of CRISPR-Cas9-Induced Genome Editing to Speed Up Plant Breeding. Int. J. Genom. 2016, 2016, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Deltcheva, E.; Chylinski, K.; Sharma, C.M.; Gonzales, K.; Chao, Y.; Pirzada, Z.A.; Eckert, M.R.; Vogel, J.; Charpentier, E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 2011, 471, 602–609. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Zhang, Z.; Hua, K.; Gao, X.; Mao, Y.; Botella, J.R.; Zhu, J.K. A Highly Efficient Cell Division-Specific CRISPR/Cas9 System Generates Homozygous Mutants for Multiple Genes in Arabidopsis. Int. J. Sci. 2018, 19, 3925. [Google Scholar] [CrossRef] [PubMed]
- Belhaj, K.; Chaparro-Garcia, A.; Kamoun, S.; Nekrasov, V. Plant genome editing made easy: Targeted mutagenesis in model and crop plants using the CRISPR/Cas system. Plant Methods 2013, 9, 39. [Google Scholar] [CrossRef] [PubMed]
- Engler, C.; Gruetzner, R.; Kandzia, R.; Marillonnet, S. Golden Gate Shuffling: A One-Pot DNA Shuffling Method Based on Type IIs Restriction Enzymes. PLoS ONE 2009, 4, e5553. [Google Scholar] [CrossRef] [PubMed]
- Xing, H.L.; Dong, L.; Wang, Z.P.; Zhang, H.Y.; Han, C.H.; Liu, B.; Wang, X.C.; Chen, Q.J. A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 2014, 14, 327. [Google Scholar] [CrossRef] [PubMed]
- Li, J.F.; Norville, J.E.; Aach, J.; McCormack, M.; Zhang, D.; Bush, J.; Church, G.M.; Sheen, J. Multiplex and homologous recombination—Mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat. Biotechnol. 2013, 31, 688–691. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z.; Zhang, K.; Chen, K.L.; Gao, C.X. Targeted mutagenesis in Zea mays using TALENs and the CRISPR/Cas system. J. Genet. Genom. 2014, 41, 63–68. [Google Scholar] [CrossRef] [PubMed]
- Shan, Q.W.; Wang, Y.P.; Li, J.; Gao, C.C. Genome editing in rice and wheat using the CRISPR/Cas system. Nat. Protoc. 2014, 9, 2395–2410. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Hu, Z.; Chen, R.; Jiang, Q.; Song, G.; Zhang, H.; Xi, Y. Targeted mutagenesis in soybean using the CRISPR-Cas9 system. Sci. Rep. 2015, 5, 10342. [Google Scholar] [CrossRef] [PubMed]
- Cermak, T.; Baltes, N.J.; Cegan, R.; Zhang, Y.; Voytas, D.F. High-frequency, precise modification of the tomato genome. Genome Biol. 2015, 16, 232. [Google Scholar] [CrossRef] [PubMed]
- Tzfira, T.; Li, J.; Lacroix, B.; Citovsky, V. Agrobacterium T-DNA integration: Molecules and models. Trends Genet. 2004, 20, 375–383. [Google Scholar] [CrossRef] [PubMed]
- Tepfer, D. Genetic transformation using Agrobacterium rhizogenes. Physiol. Plant. 1990, 79, 140–146. [Google Scholar] [CrossRef]
- Ron, M.; Kajala, K.; Pauluzzi, G.; Wang, D.; Reynoso, M.A.; Zumstein, K.; Garcha, J.; Winte, S.; Masson, H.; Inagaki, S.; et al. Hairy Root Transformation Using Agrobacterium rhizogenes as a Tool for Exploring Cell Type-Specific Gene Expression and Function Using Tomato as a Model. Plant Physiol. 2014, 166, 455–469. [Google Scholar] [CrossRef] [PubMed]
- Kirchner, T.W.; Niehaus, M.; Debener, T.; Schenk, M.K.; Herde, M. Efficient generation of mutations mediated by CRISPR/Cas9 in the hairy root transformation system of Brassica carinata. PLoS ONE 2017, 12, e0185429. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Tan, H.; Li, Q.; Chen, J.; Gao, S.; Wang, Y.; Chen, W.; Zhang, L. CRISPR/Cas9-mediated efficient targeted mutagenesis of RAS in Salvia miltiorrhiza. Phytochemistry 2018, 148, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Malarz, J.; Stojakowska, A.; Kisiel, W. Long-term cultured hairy roots of chicory—A rich source of hydroxycinnamates and 8-deoxylactucin glucoside. Appl. Biochem. Biotechnol. 2013, 171, 1589–1601. [Google Scholar] [CrossRef] [PubMed]
- Legrand, G.; Delporte, M.; Khelifi, C.; Harant, A.; Vuylsteker, C.; Mörchen, M.; Hance, P.; Hilbert, J.L.; Gagneul, D. Identification and characterization of five BAHD acyltransferases involved in hydroxycinnamoyl ester metabolism in chicory. Front. Plant Sci. 2016, 7, 741. [Google Scholar] [CrossRef] [PubMed]
- Delporte, M.; Bernard, G.; Legrand, G.; Hielscher, B.; Lanoue, A.; Moliné, R.; Rambaud, C.; Mathiron, D.; Besseau, S.; Linka, N.; et al. A BAHD neofunctionalization promotes tetrahydroxycinnamoyl spermine accumulation in pollen coat of the Asteraceae family. J. Exp. Bot. 2018, 69, 5355–5371. [Google Scholar] [CrossRef] [PubMed]
- Bais, H.P.; Suresh, B.; Raghavarao, K.S.M.S.; Ravishankar, G.A. Performance of hairy root cultures of Cichorium intybus L. in bioreactors of different configurations. In Vitro Cell. Dev. Biol. 2002, 38, 573–580. [Google Scholar] [CrossRef]
- Das, S.; Vasudeva, N.; Sharma, S. Cichorium intybus: A concise report on its ethnomedicinal, botanical, and phytopharmacological aspects. Drug Dev. Ther. 2016, 7, 1–12. [Google Scholar] [CrossRef]
- Street, R.A.; Sidana, J.; Prinsloo, G. Cichorium intybus: Traditional Uses, Phytochemistry, Pharmacology, and Toxicology. Evid.-Based Complementary Altern. Med. 2013. [Google Scholar] [CrossRef] [PubMed]
- Bais, H.P.; Ravishankar, G.A. Cichorium intybus L.—Cultivation, processing, utility, value addition and biotechnology, with an emphasis on current status and future prospects. J. Sci. Food Agric. 2001, 81, 467–484. [Google Scholar] [CrossRef]
- Maroufi, A.; Karimi, M.; Mehdikhanlou, K.; Bockstaele, E.V.; De Loose, M. Regeneration ability and genetic transformation of root type chicory (Cichorium intybus var. sativum). Afr. J. Biotechnol. 2012, 11, 11874–11886. [Google Scholar] [CrossRef]
- Bais, H.P.; Venkatesh, R.T.; Chandrashekar, A.; Ravishankar, G.A. Agrobacterium rhizogenes-mediated transformation of Witloof chicory—In vitro shoot regeneration and induction of flowering. Curr. Sci. 2001, 80, 83–87. [Google Scholar]
- Bogdanović, M.D.; Todorović, S.I.; Banjanac, T.; Dragićević, M.B.; Verstappen, F.W.A.; Bouwmeester, H.J.; Simonović, A.D. Production of guaianolides in Agrobacterium rhizogenes—Transformed chicory regenerants flowering in vitro. Ind. Crops Prod. 2014, 60, 52–59. [Google Scholar] [CrossRef]
- Iaffaldano, B.; Cornish, K.; Zhang, Y. CRISPR/Cas9 genome editing of rubber producing dandelion Taraxacum kok-saghyz using Agrobacterium rhizogenes without selection. Ind. Crops Prod. 2016, 89, 356–362. [Google Scholar] [CrossRef]
- Rambaud, C.; Dubois, J.; Vasseur, J. Male-sterile chicory cybrids obtained by intergeneric protoplast fusion. Theor. Appl. Genet. 1993, 87, 347–352. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Yu, X.; Li, Y.; Sun, Y.; Zhu, Q.; Sun, J. Highly Efficient Targeted Gene Editing in Upland Cotton Using the CRISPR/Cas9 System. Int. J. Mol. Sci. 2018, 19, 3000. [Google Scholar] [CrossRef] [PubMed]
- Waibel, F.; Filipowicz, W. U6 snRNA genes of Arabidopsis are transcribed by RNA polymerase III but contain the same two upstream promoters as RNA polymerase II-transcribed U-snRNA genes. Nucleic Acids Res. 1990, 18, 3451–3458. [Google Scholar] [CrossRef] [PubMed]
- Qin, G.; Gu, H.; Ma, L.; Peng, Y.; Deng, X.W.; Chen, Z.; Qu, L.J. Disruption of phytoene desaturase gene results in albino and dwarf phenotypes in Arabidopsis by impairing chlorophyll, carotenoid, and gibberellin biosynthesis. Cell Res. 2007, 17, 471–482. [Google Scholar] [CrossRef] [PubMed]
- Haeussler, M.; Schönig, K.; Eckert, H.; Eschstruth, A.; Mianné, J.; Renaud, J.B.; Schneider-Maunoury, S.; Shkumatava, A.; Teboul, L.; Kent, J.; et al. Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 2016, 17, 148. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Liu, Y.G. CRISPR/Cas9-Based Multiplex Genome Editing in Monocot and Dicot Plants. Curr. Protoc. Mol. Biol. 2016, 115, 31.6.1–31.6.21. [Google Scholar] [CrossRef] [PubMed]
- Weber, E.; Engler, C.; Gruetzner, R.; Werner, S.; Marillonnet, S. A modular cloning system for standardized assembly of multigene constructs. PLoS ONE 2011, 6, e16765. [Google Scholar] [CrossRef] [PubMed]
- Rambaud, C.; Vasseur, J. Somatic Hybridization in Cichorium intybus L. (Chicory). In Biotechnology in Agriculture and Forestry; Nagata, T., Bajaj, Y.P.S., Eds.; Springer: Berlin, Germany, 2001; Volume 49, pp. 112–123. [Google Scholar]
- Ma, X.; Chen, L.; Zhu, Q.; Chen, Y.; Liu, Y.G. Rapid Decoding of Sequence-Specific Nuclease-Induced Heterozygous and Biallelic Mutations by Direct Sequencing of PCR Products. Mol. Plant 2015, 8, 1285–1287. [Google Scholar] [CrossRef] [PubMed]
- Shan, S.; Mavrodiev, E.; Li, R.; Zhang, Z.; Hauser, B.A.; Soltis, P.S.; Soltis, D.E.; Yang, B. Application of CRISPR/Cas9 to Tragopogon (Asteraceae), an evolutionary model for the study of polyploidy. Mol. Ecol. Resour. 2018, 18, 1427–1443. [Google Scholar] [CrossRef] [PubMed]
- Bertier, L.D.; Ron, M.; Huo, H.; Bradford, K.J.; Britt, A.B.; Michelmore, R.W. High-Resolution Analysis of the Efficiency, Heritability, and Editing Outcomes of CRISPR/Cas9-Induced Modifications of NCED4 in Lettuce (Lactuca sativa). G3 2018, 8, 1513–1521. [Google Scholar] [CrossRef] [PubMed]
- Hill, K.; Schaller, G.E. Enhancing plant regeneration in tissue culture A molecular approach through manipulation of cytokinin sensitivity. Plant Signal. Behav. 2013, 8, e25709. [Google Scholar] [CrossRef]
- Shan, Q.; Wang, Y.; Li, J.; Zhang, Y.; Chen, K.; Ling, Z.; Zhang, K.; Liu, J.; Xi, J.J.; Qiu, J.L.; et al. Targeted genome modification of crop plants using a CRISPR-Cas system. Nat. Biotechnol. 2013, 31, 684–686. [Google Scholar] [CrossRef] [PubMed]
- Upadhyay, S.K.; Kumar, J.; Alok, A.; Tuli, R. RNA-Guided Genome Editing for Target Gene Mutations in Wheat. G3 2013, 3, 2233–2238. [Google Scholar] [CrossRef] [PubMed]
- Fan, D.; Liu, T.; Li, C.; Jiao, B.; Li, S.; Hou, Y.; Luo, K. Efficient CRISPR/Cas9-mediated Targeted Mutagenesis in Populus in the First Generation. Sci. Rep. 2015, 5, 12217. [Google Scholar] [CrossRef] [PubMed]
- Nishitani, C.; Hirai, N.; Komori, S.; Wada, M.; Okada, K.; Osakabe, K.; Yamamoto, T.; Osakabe, Y. Efficient Genome Editing in Apple Using a CRISPR/Cas9 system. Sci. Rep. 2016, 6, 31481. [Google Scholar] [CrossRef] [PubMed]
- Tian, S.; Jiang, L.; Gao, Q.; Zhang, J.; Zong, M.; Zhang, H.; Ren, Y.; Guo, S.; Gong, G.; Liu, F.; et al. Efficient CRISPR/Cas9-based gene knockout in watermelon. Plant Cell Rep. 2017, 36, 399–406. [Google Scholar] [CrossRef] [PubMed]
- Odipio, J.; Alicai, T.; Ingelbrecht, I.; Nusinow, D.; Bart, R.; Taylor, N.J. Efficient CRISPR/Cas9 Genome Editing of Phytoene desaturase in Cassava. Front. Plant Sci. 2017, 8, 1780. [Google Scholar] [CrossRef] [PubMed]
- Naim, F.; Dugdale, B.; Kleidon, J.; Brinin, A.; Shand, K.; Waterhouse, P.; Dale, J. Gene editing the phytoene desaturase alleles of Cavendish banana using CRISPR/Cas9. Transgenic Res. 2018, 27, 451–460. [Google Scholar] [CrossRef] [PubMed]
- Kaur, N.; Alok, A.; Shivani; Kaur, N.; Pandey, P.; Awasthi, P.; Tiwari, S. CRISPR/Cas9-mediated efficient editing in phytoene desaturase (PDS) demonstrates precise manipulation in banana cv. Rasthali genome. Funct. Integr. Genom. 2018, 18, 89–99. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Padh, H.; Shrivastava, N. Hairy root cultures: A suitable biological system for studying secondary metabolic pathways in plants. Eng. Life Sci. 2013, 13, 62–75. [Google Scholar] [CrossRef]
- Gontier, L.; Blassiau, C.; Mörchen, M.; Cadalen, T.; Poiret, M.; Hendriks, T.; Quillet, M.C. High-density genetic maps for loci involved in nuclear male sterility (NMS1) and sporophytic self-incompatibility (S-locus) in chicory (Cichorium intybus L., Asteraceae). Theor. Appl. Genet. 2013, 126, 2103–2121. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.S.; Hsu, C.T.; Yang, L.H.; Lee, L.Y.; Fu, J.Y.; Cheng, Q.W.; Wu, F.H.; Hsiao, H.C.W.; Zhang, Y.; Zhang, R.; et al. Application of protoplast technology to CRISPR/Cas9 mutagenesis: From single-cell mutation detection to mutant plant regeneration. Plant Biotechnol. J. 2018, 16, 1295–1310. [Google Scholar] [CrossRef] [PubMed]
- Woo, J.W.; Kim, J.; Kwon, S.I.; Corvalan, C.; Cho, S.W.; Kim, H.; Kim, S.G.; Kim, S.T.; Choe, S.; Kim, J.S. DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat. Biotechnol. 2015, 33, 1162–1165. [Google Scholar] [CrossRef] [PubMed]
- Heller, R. Researches on the mineral nutrition of plant development. Ann. Sci. Nat. Bot. Biol. Vg. 1953, 14, 1–223. [Google Scholar]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Nagel, R.; Elliott, A.; Masel, A.; Birch, R.G.; Manners, M.J. Electroporation of binary Ti plasmid vector into Agrobacterium tumefaciens, and Agrobacterium rhizogenes. FEMS Microbiol. Lett. 1990, 67, 325–328. [Google Scholar] [CrossRef]
- Yoo, S.D.; Cho, Y.H.; Sheen, J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2007, 2, 1565–1572. [Google Scholar] [CrossRef] [PubMed]


| Target 1 | Target 2 | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Transformation type | N° of lines | N° of albino lines | Mutation frequency (%) | Biallelic mutation | Mono allelic mutation | No mutation | Mutation efficiency | Biallelic mutation | Mono allelic mutation | No mutation | Mutation efficiency | ||||
| HM1 | HZ2 | % | HM1 | HZ2 | % | ||||||||||
| Protoplast | 198 | 9 | 4.5 | 2 | 3 | 55 | 1 | 3 | 66.6% | 5 | 4 | 100 | 0 | 0 | 100% |
| Hairy root | 32 | 10 | 31.3 | 1 | 5 | 60 | 3 | 1 | 90.0% | 1 | 7 | 80 | 1 | 1 | 90% |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bernard, G.; Gagneul, D.; Alves Dos Santos, H.; Etienne, A.; Hilbert, J.-L.; Rambaud, C. Efficient Genome Editing Using CRISPR/Cas9 Technology in Chicory. Int. J. Mol. Sci. 2019, 20, 1155. https://doi.org/10.3390/ijms20051155
Bernard G, Gagneul D, Alves Dos Santos H, Etienne A, Hilbert J-L, Rambaud C. Efficient Genome Editing Using CRISPR/Cas9 Technology in Chicory. International Journal of Molecular Sciences. 2019; 20(5):1155. https://doi.org/10.3390/ijms20051155
Chicago/Turabian StyleBernard, Guillaume, David Gagneul, Harmony Alves Dos Santos, Audrey Etienne, Jean-Louis Hilbert, and Caroline Rambaud. 2019. "Efficient Genome Editing Using CRISPR/Cas9 Technology in Chicory" International Journal of Molecular Sciences 20, no. 5: 1155. https://doi.org/10.3390/ijms20051155
APA StyleBernard, G., Gagneul, D., Alves Dos Santos, H., Etienne, A., Hilbert, J.-L., & Rambaud, C. (2019). Efficient Genome Editing Using CRISPR/Cas9 Technology in Chicory. International Journal of Molecular Sciences, 20(5), 1155. https://doi.org/10.3390/ijms20051155





