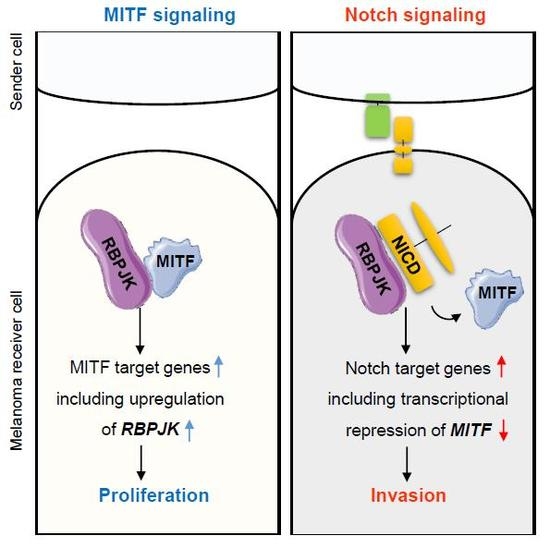
Negative Regulatory Loop between Microphthalmia-Associated Transcription Factor (MITF) and Notch Signaling
Abstract
1. Introduction
2. Results and Discussion
2.1. Notch signaling Decreases MITF Expression
2.2. MITF Directly Regulates RBPJK Expression
3. Materials and Methods
3.1. Cell Culture
3.2. RNA Purification and qRT-PCR
3.3. Melanoma Co-Culture with CHO-K1 Cells
3.4. Plasmids and Transfection
3.5. Luciferase Reporter Assay
3.6. Western Blot Analyses
3.7. Chromatin Immunoprecipitation
3.8. Invasion Assays
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| DAPI | 4′,6-Diamidino-2-phenylindole |
| DAPT | N-[N-(3,5-Difluorophenacetyl)-L-alanyl]-S-phenylglycine t-butyl ester |
| DMEM | Dulbecco’s modification of Eagle medium |
| DMSO | Dimethyl sulfoxide |
References
- Bowden, G.T. Prevention of non-melanoma skin cancer by targeting ultraviolet-B-light signalling. Nat. Rev. Cancer 2004, 4, 23–35. [Google Scholar] [CrossRef] [PubMed]
- Jhappan, C.; Noonan, F.P.; Merlino, G. Ultraviolet radiation and cutaneous malignant melanoma. Oncogene 2003, 22, 3099–3112. [Google Scholar] [CrossRef]
- Tsao, H.; Atkins, M.B.; Sober, A.J. Management of cutaneous melanoma. N. Engl. J. Med. 2004, 351, 998–1012. [Google Scholar] [CrossRef] [PubMed]
- Gaggioli, C.; Sahai, E. Melanoma invasion-current knowledge and future directions. Pigment Cell Res. 2007, 20, 161–172. [Google Scholar] [CrossRef]
- Dadachova, E.; Casadevall, A. Renaissance of targeting molecules for melanoma. Cancer Biother. Radiopharm. 2006, 21, 545–552. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.; Xie, J.; Miao, J.; Li, R.; Liao, W.; Luo, R. A knockdown of Maml1 that results in melanoma cell senescence promotes an innate and adaptive immune response. Cancer Immunol. Immunother. 2013, 62, 183–190. [Google Scholar] [CrossRef] [PubMed]
- Mozuraitiene, J.; Bielskiene, K.; Atkocius, V.; Labeikyte, D. Molecular alterations in signal pathways of melanoma and new personalized treatment strategies. Target. Notch. Med. 2015, 51, 133–145. [Google Scholar] [CrossRef]
- Bray, S.J. Notch signalling in context. Nat. Rev. Mol. Cell Biol. 2016, 17, 722–735. [Google Scholar] [CrossRef]
- Chiba, S. Notch signaling in stem cell systems. Stem Cells 2006, 24, 2437–2447. [Google Scholar] [CrossRef]
- Louvi, A.; Artavanis-Tsakonas, S. Notch and disease: A growing field. Semin. Cell Dev. Biol. 2012, 23, 473–480. [Google Scholar] [CrossRef]
- Bedogni, B. Notch signaling in melanoma: Interacting pathways and stromal influences that enhance Notch targeting. Pigment Cell Melanoma Res. 2014, 27, 162–168. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.J.; Xiao, M.; Balint, K.; Smalley, K.S.; Brafford, P.; Qiu, R.; Pinnix, C.; Li, X.; Herlyn, M. Notch1 signaling promotes primary melanoma progression by activating mitogen-activated protein kinase/phosphatidylinositol 3-kinase-Akt pathways and up-regulating N-cadherin expression. Cancer Res. 2006, 66, 4182–4190. [Google Scholar] [CrossRef] [PubMed]
- Muller, C.S. Notch signaling and malignant melanoma. Adv. Exp. Med. Biol. 2012, 727, 258–264. [Google Scholar]
- Zhang, K.; Wong, P.; Zhang, L.; Jacobs, B.; Borden, E.C.; Aster, J.C.; Bedogni, B. A Notch1-neuregulin1 autocrine signaling loop contributes to melanoma growth. Oncogene 2012, 31, 4609–4618. [Google Scholar] [CrossRef] [PubMed]
- Golan, T.; Messer, A.R.; Amitai-Lange, A.; Melamed, Z.; Ohana, R.; Bell, R.E.; Kapitansky, O.; Lerman, G.; Greenberger, S.; Khaled, M.; et al. Interactions of Melanoma Cells with Distal Keratinocytes Trigger Metastasis via Notch Signaling Inhibition of MITF. Mol. Cell 2015, 59, 664–676. [Google Scholar] [CrossRef] [PubMed]
- Hori, K.; Sen, A.; Artavanis-Tsakonas, S. Notch signaling at a glance. J. Cell Sci. 2013, 126, 2135–2140. [Google Scholar] [CrossRef]
- Kovall, R.A.; Gebelein, B.; Sprinzak, D.; Kopan, R. The Canonical Notch Signaling Pathway: Structural and Biochemical Insights into Shape, Sugar, and Force. Dev. Cell 2017, 41, 228–241. [Google Scholar] [CrossRef] [PubMed]
- Borggrefe, T.; Oswald, F. The Notch signaling pathway: Transcriptional regulation at Notch target genes. Cell. Mol. Life Sci. 2009, 66, 1631–1646. [Google Scholar] [CrossRef]
- Levy, C.; Khaled, M.; Fisher, D.E. MITF: Master regulator of melanocyte development and melanoma oncogene. Trends Mol. Med. 2006, 12, 406–414. [Google Scholar] [CrossRef]
- Hartman, M.L.; Czyz, M. MITF in melanoma: Mechanisms behind its expression and activity. Cell. Mol. Life Sci. 2015, 72, 1249–1260. [Google Scholar] [CrossRef]
- Kawakami, A.; Fisher, D.E. The master role of microphthalmia-associated transcription factor in melanocyte and melanoma biology. Lab. Investig. 2017, 97, 649–656. [Google Scholar] [CrossRef] [PubMed]
- Eccles, M.R.; He, S.; Ahn, A.; Slobbe, L.J.; Jeffs, A.R.; Yoon, H.S.; Baguley, B.C. MITF and PAX3 Play Distinct Roles in Melanoma Cell Migration; Outline of a “Genetic Switch” Theory Involving MITF and PAX3 in Proliferative and Invasive Phenotypes of Melanoma. Front. Oncol. 2013, 3, 229. [Google Scholar] [CrossRef]
- Hoek, K.S.; Eichhoff, O.M.; Schlegel, N.C.; Dobbeling, U.; Kobert, N.; Schaerer, L.; Hemmi, S.; Dummer, R. In vivo switching of human melanoma cells between proliferative and invasive states. Cancer Res. 2008, 68, 650–656. [Google Scholar] [CrossRef] [PubMed]
- Howlin, J.; Cirenajwis, H.; Lettiero, B.; Staaf, J.; Lauss, M.; Saal, L.; Borg, Å.; Gruvberger-Saal, S.; Jönsson, J. Loss of CITED1, an MITF regulator, drives a phenotype switch in vitro and can predict clinical outcome in primary melanoma tumours. PeerJ 2015, 3, e788. [Google Scholar] [CrossRef]
- Carreira, S.; Goodall, J.; Denat, L.; Rodriguez, M.; Nuciforo, P.; Hoek, K.S.; Testori, A.; Larue, L.; Goding, C.R. Mitf regulation of Dia1 controls melanoma proliferation and invasiveness. Genes Dev. 2006, 20, 3426–3439. [Google Scholar] [CrossRef] [PubMed]
- Hoek, K.S.; Schlegel, N.C.; Brafford, P.; Sucker, A.; Ugurel, S.; Kumar, R.; Weber, B.L.; Nathanson, K.L.; Phillips, D.J.; Herlyn, M.; et al. Metastatic potential of melanomas defined by specific gene expression profiles with no BRAF signature. Pigment Cell Res. 2006, 19, 290–302. [Google Scholar] [CrossRef]
- Bharti, K.; Liu, W.; Csermely, T.; Bertuzzi, S.; Arnheiter, H. Alternative promoter use in eye development: The complex role and regulation of the transcription factor MITF. Development 2008, 135, 1169–1178. [Google Scholar] [CrossRef] [PubMed]
- Levy, C.; Khaled, M.; Iliopoulos, D.; Janas, M.M.; Schubert, S.; Pinner, S.; Chen, P.H.; Li, S.; Fletcher, A.L.; Yokoyama, S.; et al. Intronic miR-211 assumes the tumor suppressive function of its host gene in melanoma. Mol. Cell 2010, 40, 841–849. [Google Scholar] [CrossRef]
- Tshori, S.; Gilon, D.; Beeri, R.; Nechushtan, H.; Kaluzhny, D.; Pikarsky, E.; Razin, E. Transcription factor MITF regulates cardiac growth and hypertrophy. J. Clin. Investig. 2006, 116, 2673–2681. [Google Scholar] [CrossRef]
- Rachmin, I.; Amsalem, E.; Golomb, E.; Beeri, R.; Gilon, D.; Fang, P.; Nechushtan, H.; Kay, G.; Guo, M.; Yiqing, L.; et al. FHL2 switches MITF from activator to repressor of Erbin expression during cardiac hypertrophy. Int. J. Cardiol. 2015, 195, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.L.; Liu, J.C. Role of Notch signaling in the mammalian heart. Braz. J. Med. Biol. Res. 2014, 47, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Tabach, Y.; Golan, T.; Hernandez-Hernandez, A.; Messer, A.R.; Fukuda, T.; Kouznetsova, A.; Liu, J.-G.; Lilienthal, I.; Levy, C.; Ruvkun, G. Human disease locus discovery and mapping to molecular pathways through phylogenetic profiling. Mol. Syst. Biol. 2013, 9, 692. [Google Scholar] [CrossRef]
- Chang, P.J.; Boonsiri, J.; Wang, S.S.; Chen, L.Y.; Miller, G. Binding of RBP-Jkappa (CSL) protein to the promoter of the Kaposi’s sarcoma-associated herpesvirus ORF47 (gL) gene is a critical but not sufficient determinant of transactivation by ORF50 protein. Virology 2010, 398, 38–48. [Google Scholar] [CrossRef] [PubMed]
- Sprinzak, D.; Lakhanpal, A.; Lebon, L.; Santat, L.A.; Fontes, M.E.; Anderson, G.A.; Anderson, G.A.; Garcia-Ojalvo, J.; Elowitz, M.B. Cis-interactions between Notch and Delta generate mutually exclusive signalling states. Nature 2010, 465, 86–90. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Carey, M.; Workman, J.L. The role of chromatin during transcription. Cell 2007, 128, 707–719. [Google Scholar] [CrossRef]
- Lanz, T.A.; Wood, K.M.; Richter, K.E.; Nolan, C.E.; Becker, S.L.; Pozdnyakov, N.; Martin, B.-A.; Du, P.; Oborski, C.E.; Wood, D.E.; et al. Pharmacodynamics and pharmacokinetics of the gamma-secretase inhibitor PF-3084014. J. Pharmacol. Exp. Ther. 2010, 334, 269–277. [Google Scholar] [CrossRef]


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Golan, T.; Levy, C. Negative Regulatory Loop between Microphthalmia-Associated Transcription Factor (MITF) and Notch Signaling. Int. J. Mol. Sci. 2019, 20, 576. https://doi.org/10.3390/ijms20030576
Golan T, Levy C. Negative Regulatory Loop between Microphthalmia-Associated Transcription Factor (MITF) and Notch Signaling. International Journal of Molecular Sciences. 2019; 20(3):576. https://doi.org/10.3390/ijms20030576
Chicago/Turabian StyleGolan, Tamar, and Carmit Levy. 2019. "Negative Regulatory Loop between Microphthalmia-Associated Transcription Factor (MITF) and Notch Signaling" International Journal of Molecular Sciences 20, no. 3: 576. https://doi.org/10.3390/ijms20030576
APA StyleGolan, T., & Levy, C. (2019). Negative Regulatory Loop between Microphthalmia-Associated Transcription Factor (MITF) and Notch Signaling. International Journal of Molecular Sciences, 20(3), 576. https://doi.org/10.3390/ijms20030576