Figure 1.
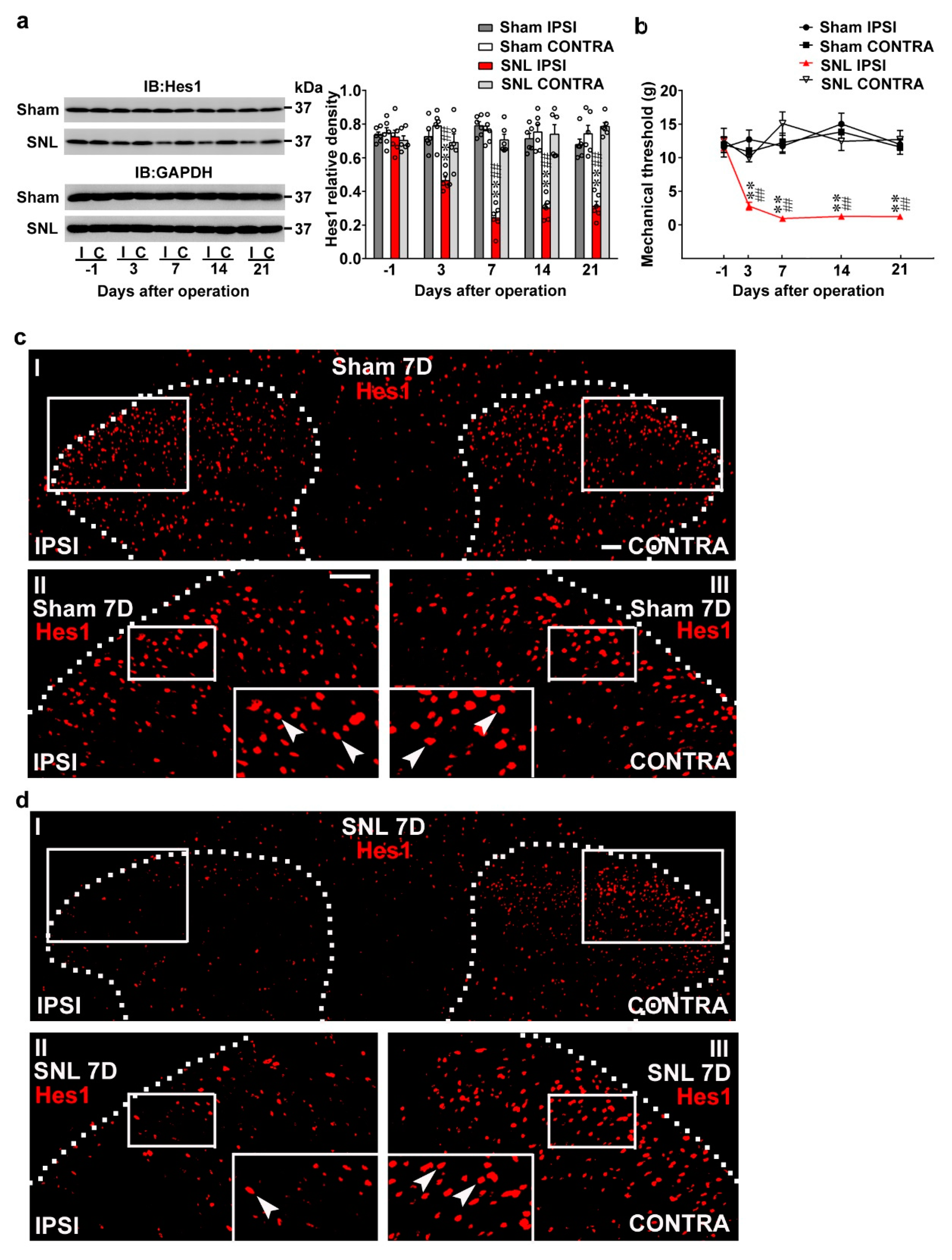
Nerve injury reduced spinal Hes1 expression accompanied with behavioral allodynia. (a) Representative Western blot and statistical analyses (normalized to GAPDH) demonstrating, when compared with the sham operation (Sham), that spinal nerve ligation (SNL) decreased the expression of Hes1 in the ipsilateral (I and IPSI) but not the contralateral (C and CONTRA) dorsal horn at days 3, 7, 14, and 21 after surgery. IB, Immunoblotting. Two-way ANOVA with repeated measures over time, treatment, F (3,20) = 116.6, p < 0.0001; time, F (4,80) = 6.258, p = 0.0002; treatment × time, F(12,80) = 9.003, p < 0.0011. ** p < 0.01 vs. Sham IPSI. ## p < 0.01 vs. SNL day –1. n = 6. (b) Von Frey test demonstrating, when compared with the sham operation, that SNL decreased the tactile withdrawal threshold of the ipsilateral but not the contralateral hind-paw at days 3, 7, 14, and 21 after surgery. Two-way ANOVA with repeated measures over time, treatment, F (3,24) = 59.37, p < 0.0001; time, F(4,96) = 4.514, p = 0.0022; treatment × time, F(12,96) = 6.427, p < 0.0001. ** p < 0.01 vs. Sham IPSI. ## p < 0.01 vs. SNL day –1. n = 7. (c,d) Immunofluorescence images showing, when compared with the sham operation (c), that SNL (d) decreased the number and distribution of Hes1-positive neurons (red) in the ipsilateral dorsal horn (left) of spinal cord slice harvested at day 7 (7D) after surgery. Dashed lines indicate the margin of the dorsal horn. The lower images (II, III) are magnifications of the upper images (I); and the inset images at the bottom of the lower image are further amplified images, with arrows marking the Hes1-positive neurons. Scale bar = 50 μm; Thickness = 30 μm.
Figure 1.
Nerve injury reduced spinal Hes1 expression accompanied with behavioral allodynia. (a) Representative Western blot and statistical analyses (normalized to GAPDH) demonstrating, when compared with the sham operation (Sham), that spinal nerve ligation (SNL) decreased the expression of Hes1 in the ipsilateral (I and IPSI) but not the contralateral (C and CONTRA) dorsal horn at days 3, 7, 14, and 21 after surgery. IB, Immunoblotting. Two-way ANOVA with repeated measures over time, treatment, F (3,20) = 116.6, p < 0.0001; time, F (4,80) = 6.258, p = 0.0002; treatment × time, F(12,80) = 9.003, p < 0.0011. ** p < 0.01 vs. Sham IPSI. ## p < 0.01 vs. SNL day –1. n = 6. (b) Von Frey test demonstrating, when compared with the sham operation, that SNL decreased the tactile withdrawal threshold of the ipsilateral but not the contralateral hind-paw at days 3, 7, 14, and 21 after surgery. Two-way ANOVA with repeated measures over time, treatment, F (3,24) = 59.37, p < 0.0001; time, F(4,96) = 4.514, p = 0.0022; treatment × time, F(12,96) = 6.427, p < 0.0001. ** p < 0.01 vs. Sham IPSI. ## p < 0.01 vs. SNL day –1. n = 7. (c,d) Immunofluorescence images showing, when compared with the sham operation (c), that SNL (d) decreased the number and distribution of Hes1-positive neurons (red) in the ipsilateral dorsal horn (left) of spinal cord slice harvested at day 7 (7D) after surgery. Dashed lines indicate the margin of the dorsal horn. The lower images (II, III) are magnifications of the upper images (I); and the inset images at the bottom of the lower image are further amplified images, with arrows marking the Hes1-positive neurons. Scale bar = 50 μm; Thickness = 30 μm.
![Ijms 20 04177 g001 Ijms 20 04177 g001]()
Figure 2.
Focal knockdown of spinal Hes1 expression elicits no motor deficits but provokes behavioral allodynia. (a) Representative Western blot and statistical analyses (normalized to GAPDH) showing the Hes1 levels in the dorsal horn dissected from naïve animals (Naïve) were not affected by implantation of an intrathecal catheter alone (it) or spinal administration with polyethylenimine (PEI, 10 μL; daily for 4 days) or missense siRNA (MS siRNA, 5 μg, 10 μL; daily for 4 days) but was dose-dependently decreased by intrathecal administration with Hes1 siRNA (Hes1 siRNA; 1, 3, and 5 μg; 10 μL; daily for 4 days) at day 4 after the start of treatments. IB, Immunoblotting. One-way ANOVA, post-hoc Tukey test, F (6,35) = 47.9, p < 0.0001. ** p < 0.01 vs. Naïve. n = 6. (b) Rota-rod test demonstrating no statistical difference in the motor performance among naïve animals as well as naïve animals administered with polyethylenimine, missense siRNA, or Hes1 siRNA (5 μg, 10 μL). The gray bar at the bottom indicates the duration of the reagents’ administration. Two-way ANOVA with repeated measures over time, treatment, F (3,24) = 1.170, p = 0.3419; time, F (4,96) = 0.2094, p = 0.9327; treatment × time, F(12,96) = 0.8289, p = 0.6205. n = 7. (c) Results of the von Frey test demonstrating that administration to naïve rats with Hes1 siRNA (5 μg, 10 μL), but not polyethylenimine, or missense siRNA decreased the withdrawal threshold of the hind paw at days 2, 3, and 4 after the injection. The gray bar at the bottom indicates the duration of the reagents’ administration. Two-way ANOVA with repeated measures over time, treatment, F (3,24) = 40.16, p < 0.0001; time, F (4,96) = 2.481, p = 0.0489; treatment × time, F (12,96) = 2.226, p = 0.0160. ** p < 0.01 vs. Naïve. n = 7.
Figure 2.
Focal knockdown of spinal Hes1 expression elicits no motor deficits but provokes behavioral allodynia. (a) Representative Western blot and statistical analyses (normalized to GAPDH) showing the Hes1 levels in the dorsal horn dissected from naïve animals (Naïve) were not affected by implantation of an intrathecal catheter alone (it) or spinal administration with polyethylenimine (PEI, 10 μL; daily for 4 days) or missense siRNA (MS siRNA, 5 μg, 10 μL; daily for 4 days) but was dose-dependently decreased by intrathecal administration with Hes1 siRNA (Hes1 siRNA; 1, 3, and 5 μg; 10 μL; daily for 4 days) at day 4 after the start of treatments. IB, Immunoblotting. One-way ANOVA, post-hoc Tukey test, F (6,35) = 47.9, p < 0.0001. ** p < 0.01 vs. Naïve. n = 6. (b) Rota-rod test demonstrating no statistical difference in the motor performance among naïve animals as well as naïve animals administered with polyethylenimine, missense siRNA, or Hes1 siRNA (5 μg, 10 μL). The gray bar at the bottom indicates the duration of the reagents’ administration. Two-way ANOVA with repeated measures over time, treatment, F (3,24) = 1.170, p = 0.3419; time, F (4,96) = 0.2094, p = 0.9327; treatment × time, F(12,96) = 0.8289, p = 0.6205. n = 7. (c) Results of the von Frey test demonstrating that administration to naïve rats with Hes1 siRNA (5 μg, 10 μL), but not polyethylenimine, or missense siRNA decreased the withdrawal threshold of the hind paw at days 2, 3, and 4 after the injection. The gray bar at the bottom indicates the duration of the reagents’ administration. Two-way ANOVA with repeated measures over time, treatment, F (3,24) = 40.16, p < 0.0001; time, F (4,96) = 2.481, p = 0.0489; treatment × time, F (12,96) = 2.226, p = 0.0160. ** p < 0.01 vs. Naïve. n = 7.
![Ijms 20 04177 g002 Ijms 20 04177 g002]()
Figure 3.
Lentiviral vector-mediated Hes1 expression reversed neuropathic allodynia. (a) Experimental timeline of spinal nerve ligation (SNL), lentiviral vector (LV) transfection, as well as behavioral test and biochemical analyses. Seven days after operation, lentivirus vector was spinally injected to animals, and the behavioral test or biochemical analyses were carried out 14 days after transfection. (b) Representative western blot and statistical analyses were performed (normalized to GAPDH). Transfecting naïve animals (Naïve) with the Hes1-encoding vector (Hes1 LV), but not with the control vector (Control LV) or implantation of the intrathecal catheter itself (it), increased the abundance of Hes1 in the dorsal horn sample. IB, immunoblotting. One-way ANOVA, post-hoc Tukey test, F(3,20) = 5.516, p = 0.0063. ** p < 0.01 vs. Naïve. n = 6. (c) Results of the von Frey test demonstrating that transfecting SNL animals with Hes1-encoding vector (SNL 21D + Hes1 LV), but not the control vector (SNL 21D + Control LV), ameliorated the SNL-decreased withdrawal threshold in the ipsilateral hind paw tested at day 21 after operation. One-way ANOVA, post-hoc Tukey test, F (3,24) = 51.08, p < 0.0001. ** p < 0.01 vs. Sham 21D. ## p < 0.01 vs. SNL 21D. n = 7. (d) Representative western blot and statistical analyses (normalized to GAPDH) showing that transfection of SNL animals with Hes1-encoding vector (SNL 21D + Hes1 LV) but not the control vector (SNL 21D + Control LV) reversed the SNL-decreased Hes1 expression in the ipsilateral dorsal horn sample dissected at day 21 after operation. One-way ANOVA, post-hoc Tukey test, F (3,20) = 48.96, p < 0.0001. ** p < 0.01 vs. Sham 21D. ## p < 0.01 vs. SNL 21D. n = 6.
Figure 3.
Lentiviral vector-mediated Hes1 expression reversed neuropathic allodynia. (a) Experimental timeline of spinal nerve ligation (SNL), lentiviral vector (LV) transfection, as well as behavioral test and biochemical analyses. Seven days after operation, lentivirus vector was spinally injected to animals, and the behavioral test or biochemical analyses were carried out 14 days after transfection. (b) Representative western blot and statistical analyses were performed (normalized to GAPDH). Transfecting naïve animals (Naïve) with the Hes1-encoding vector (Hes1 LV), but not with the control vector (Control LV) or implantation of the intrathecal catheter itself (it), increased the abundance of Hes1 in the dorsal horn sample. IB, immunoblotting. One-way ANOVA, post-hoc Tukey test, F(3,20) = 5.516, p = 0.0063. ** p < 0.01 vs. Naïve. n = 6. (c) Results of the von Frey test demonstrating that transfecting SNL animals with Hes1-encoding vector (SNL 21D + Hes1 LV), but not the control vector (SNL 21D + Control LV), ameliorated the SNL-decreased withdrawal threshold in the ipsilateral hind paw tested at day 21 after operation. One-way ANOVA, post-hoc Tukey test, F (3,24) = 51.08, p < 0.0001. ** p < 0.01 vs. Sham 21D. ## p < 0.01 vs. SNL 21D. n = 7. (d) Representative western blot and statistical analyses (normalized to GAPDH) showing that transfection of SNL animals with Hes1-encoding vector (SNL 21D + Hes1 LV) but not the control vector (SNL 21D + Control LV) reversed the SNL-decreased Hes1 expression in the ipsilateral dorsal horn sample dissected at day 21 after operation. One-way ANOVA, post-hoc Tukey test, F (3,20) = 48.96, p < 0.0001. ** p < 0.01 vs. Sham 21D. ## p < 0.01 vs. SNL 21D. n = 6.
![Ijms 20 04177 g003 Ijms 20 04177 g003]()
Figure 4.
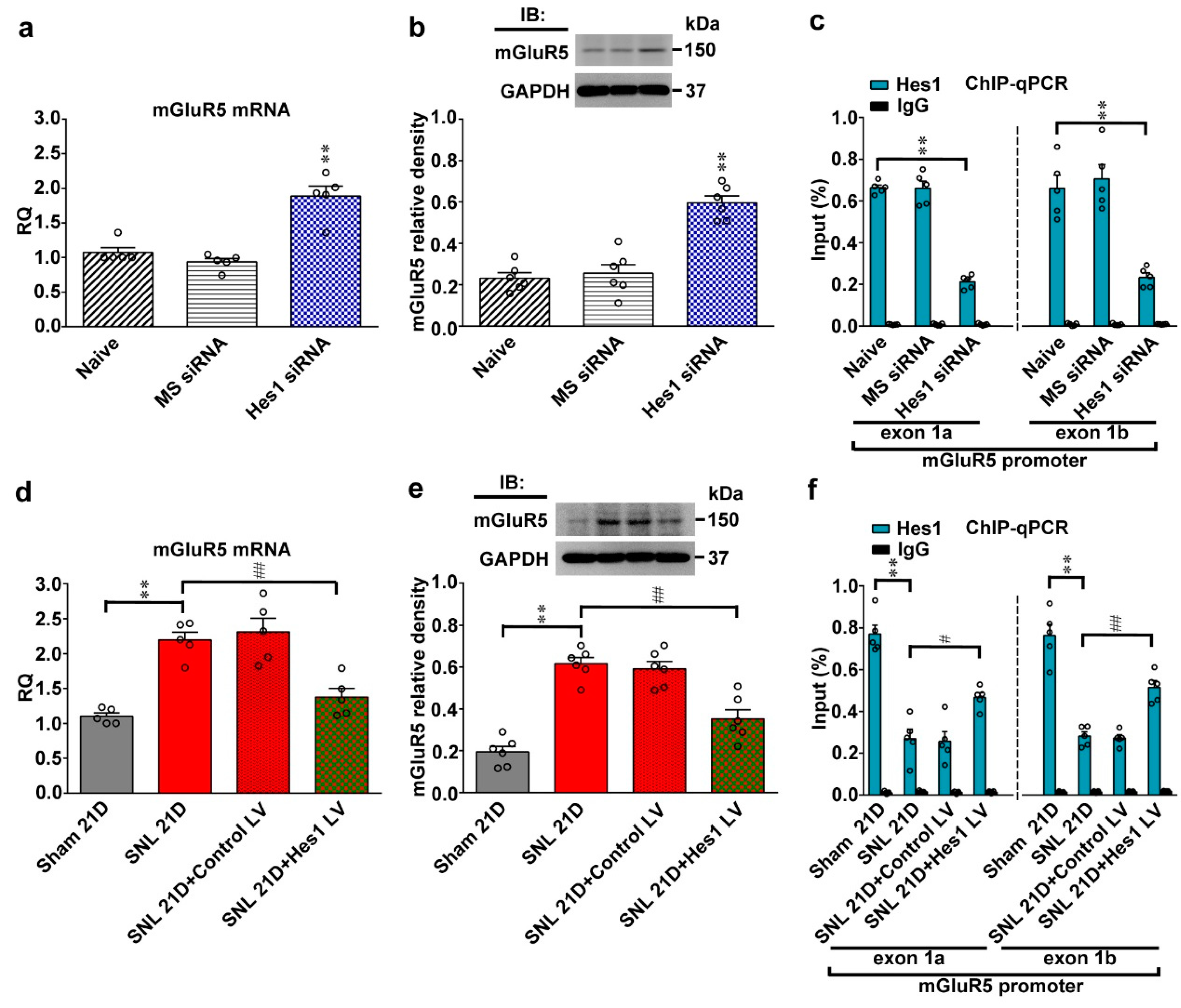
Neuropathic injury diminishes Hes1-repressed spinal mGluR5 transcription. (a,b) RT-PCR and western blot analyses (normalized to GAPDH) demonstrating that the mRNA and protein levels of mGluR5 in dorsal horn samples were increased by administering naïve animals (Naive) with Hes1 siRNA but not with missense siRNA (Hes1 siRNA and MS siRNA, respectively. 5 μg, 10 μL; it, daily for 4 days). IB, immunoblotting. mRNA, one-way ANOVA, post-hoc Tukey test, F (2,12) = 27.91, p < 0.0001. protein, one-way ANOVA, post-hoc Tukey test, F (2,15) = 34.80, p < 0.0001. ** p < 0.01 vs. Naïve. mRNA, n = 5. protein, n = 6. (c) ChIP-qPCR assay of dorsal horn samples demonstrating that administering naïve animals with Hes1 siRNA but not with missense siRNA (Hes1 siRNA and MS siRNA, respectively. 5 μg, 10 μL; it, daily for 4 days) decreased amounts of Hes1 antibody-precipitated exon 1a and exon 1b promoter fragments of mGluR5. ** p < 0.01 vs. Naïve. n = 5. Hes1, exon 1a, one-way ANOVA, post-hoc Tukey test, F (2,12) = 131.7, p < 0.0001. Hes1, exon 1b, one-way ANOVA, post-hoc Tukey test, F (2,12) = 22.70, p < 0.0001. (d,e) RT-PCR and western blot analyses of the ipsilateral dorsal horn samples dissected at day 21 after operation. When compared with the sham operation (Sham 21D), spinal nerve ligation (SNL 21D) increased mRNA and protein levels of mGluR5 that were both reversed by intrathecal administration to SNL animals with Hes1-encoding vector (SNL 21D + Hes1 LV) but not the control vector (SNL 21D + Control LV). mRNA, one-way ANOVA, post-hoc Tukey test, F(3,16) = 20.75, p < 0.0001. protein, one-way ANOVA, post-hoc Tukey test, F (3,20) = 34.70, p < 0.0001. ** p < 0.01 vs. Sham 21D. ## p < 0.01 vs. SNL 21D. mRNA, n = 5. protein, n = 6. (f) ChIP-qPCR assay of dorsal horn samples dissected at day 21 post operation. When compared with the sham operation, SNL decreased the amounts of Hes1 antibody-precipitated exon 1a and exon 1b promoter fragments of mGluR5; effects that were both reversed by transfecting animals with Hes1-encoding vector (SNL 21D + Hes1 LV) but not the control vector (SNL 21D + Control LV). Hes1, exon 1a, one-way ANOVA, post-hoc Tukey test, F (3,16) = 34.72, p < 0.0001. Hes1, exon 1b, one-way ANOVA, post-hoc Tukey test, F (3,16) = 45.74, p < 0.0001. ** p < 0.01 vs. Sham 21D. # p < 0.05, ## p < 0.01 vs. SNL 21D. n = 5.
Figure 4.
Neuropathic injury diminishes Hes1-repressed spinal mGluR5 transcription. (a,b) RT-PCR and western blot analyses (normalized to GAPDH) demonstrating that the mRNA and protein levels of mGluR5 in dorsal horn samples were increased by administering naïve animals (Naive) with Hes1 siRNA but not with missense siRNA (Hes1 siRNA and MS siRNA, respectively. 5 μg, 10 μL; it, daily for 4 days). IB, immunoblotting. mRNA, one-way ANOVA, post-hoc Tukey test, F (2,12) = 27.91, p < 0.0001. protein, one-way ANOVA, post-hoc Tukey test, F (2,15) = 34.80, p < 0.0001. ** p < 0.01 vs. Naïve. mRNA, n = 5. protein, n = 6. (c) ChIP-qPCR assay of dorsal horn samples demonstrating that administering naïve animals with Hes1 siRNA but not with missense siRNA (Hes1 siRNA and MS siRNA, respectively. 5 μg, 10 μL; it, daily for 4 days) decreased amounts of Hes1 antibody-precipitated exon 1a and exon 1b promoter fragments of mGluR5. ** p < 0.01 vs. Naïve. n = 5. Hes1, exon 1a, one-way ANOVA, post-hoc Tukey test, F (2,12) = 131.7, p < 0.0001. Hes1, exon 1b, one-way ANOVA, post-hoc Tukey test, F (2,12) = 22.70, p < 0.0001. (d,e) RT-PCR and western blot analyses of the ipsilateral dorsal horn samples dissected at day 21 after operation. When compared with the sham operation (Sham 21D), spinal nerve ligation (SNL 21D) increased mRNA and protein levels of mGluR5 that were both reversed by intrathecal administration to SNL animals with Hes1-encoding vector (SNL 21D + Hes1 LV) but not the control vector (SNL 21D + Control LV). mRNA, one-way ANOVA, post-hoc Tukey test, F(3,16) = 20.75, p < 0.0001. protein, one-way ANOVA, post-hoc Tukey test, F (3,20) = 34.70, p < 0.0001. ** p < 0.01 vs. Sham 21D. ## p < 0.01 vs. SNL 21D. mRNA, n = 5. protein, n = 6. (f) ChIP-qPCR assay of dorsal horn samples dissected at day 21 post operation. When compared with the sham operation, SNL decreased the amounts of Hes1 antibody-precipitated exon 1a and exon 1b promoter fragments of mGluR5; effects that were both reversed by transfecting animals with Hes1-encoding vector (SNL 21D + Hes1 LV) but not the control vector (SNL 21D + Control LV). Hes1, exon 1a, one-way ANOVA, post-hoc Tukey test, F (3,16) = 34.72, p < 0.0001. Hes1, exon 1b, one-way ANOVA, post-hoc Tukey test, F (3,16) = 45.74, p < 0.0001. ** p < 0.01 vs. Sham 21D. # p < 0.05, ## p < 0.01 vs. SNL 21D. n = 5.
![Ijms 20 04177 g004 Ijms 20 04177 g004]()
Figure 5.
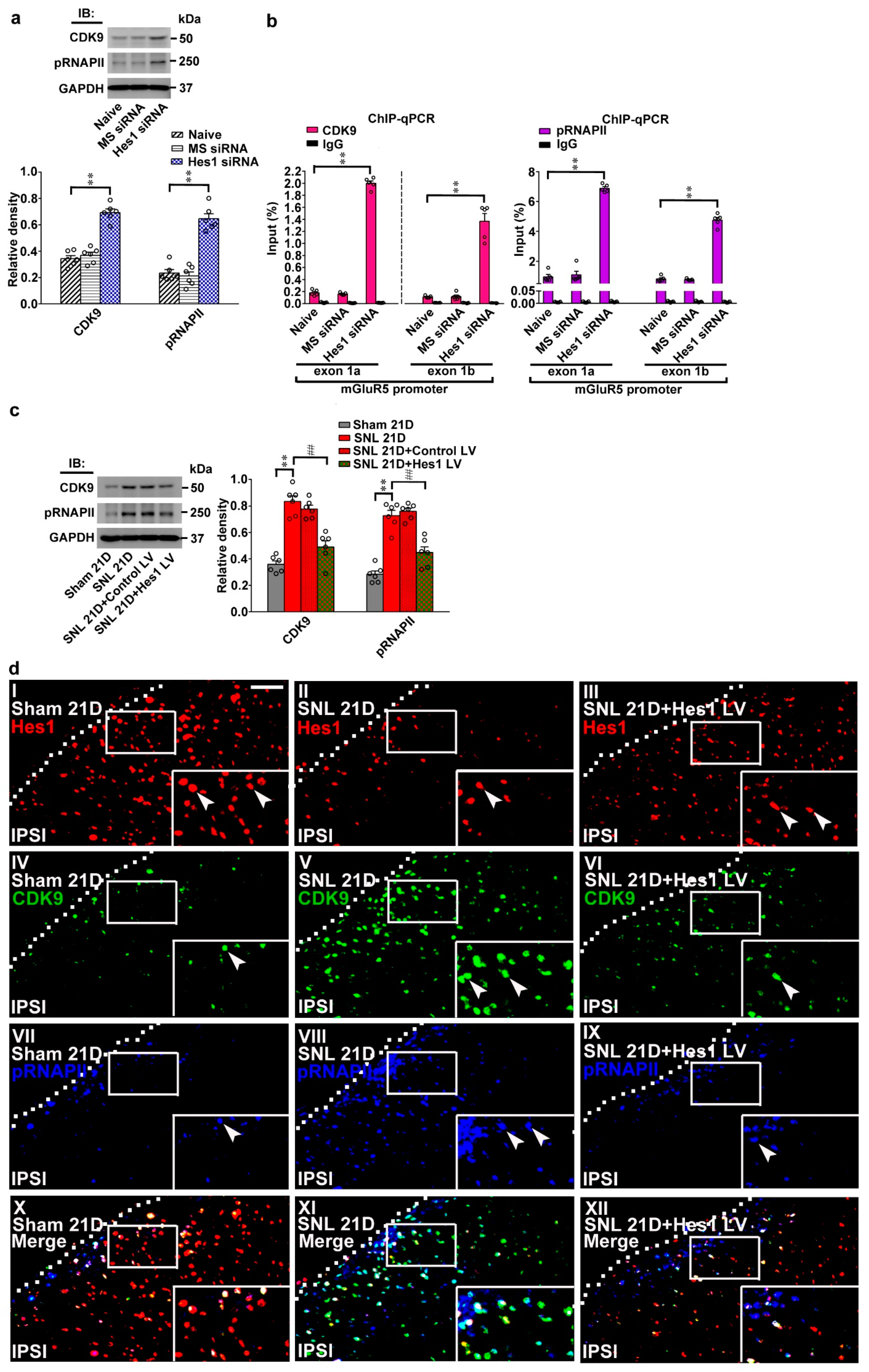
Neuropathic injury impedes Hes1-suppressed CDK9 recruitment and RNAPII phosphorylation on mGluR5 promoters. (a) Representative western blot and statistical analyses (normalized to GAPDH) of dorsal horn samples demonstrating the levels of CDK9 and phosphorylated RNAPII (pRNAPII) were increased by spinally administering naïve animals (Naïve) with Hes1-siRNA but not with missense siRNA (Hes1 siRNA and MS siRNA, respectively, 5 μg, 10 μL; daily for 4 days). IB, immunoblotting. CDK9, one-way ANOVA, post-hoc Tukey test, F (2,15) = 64.17, p < 0.0001. pRNAPII, one-way ANOVA, post-hoc Tukey test, F (2,15) = 59.83, p < 0.0001. ** p < 0.01 vs. Naïve. n = 6. (b) ChIP-qPCR assay of dorsal horn samples demonstrating that administering naïve animals with Hes1 siRNA, but not missense siRNA (Hes1 siRNA and MS siRNA, respectively. 5 μg, 10 μL; daily for 4 days), increased amounts of both CDK9 and pRNAPII antibody-precipitated exon 1a and exon 1b promoter fragments of mGluR5. CDK9, exon 1a, one-way ANOVA, post-hoc Tukey test, F (2,12) = 1494, p < 0.0001. CDK9, exon 1b, one-way ANOVA, post-hoc Tukey test, F(2,12) = 88.97, p < 0.0001. pRNAPII, exon 1a, one-way ANOVA, post-hoc Tukey test, F (2,12) = 358.4, p < 0.0001. pRNAPII, exon 1b, one-way ANOVA, post-hoc Tukey test, F (2,12) = 373.5, p < 0.0001. ** p < 0.01 vs. Naïve. n = 5. (c) Representative western blot and statistical analyses of dorsal horn samples dissected at day 21 after operation. When compared with the sham operation (Sham 21D), spinal nerve ligation (SNL 21D) increased spinal CDK9 and pRNAPII levels; that were both reversed by administering SNL animals with Hes1-encoding vector (SNL 21D + Hes1 LV) but not the control vector (SNL 21D + Control LV). CDK9, one-way ANOVA, post-hoc Tukey test, F (3,20) = 38.06, p < 0.0001. pRNAPII, one-way ANOVA, post-hoc Tukey test, F (3,20) = 43.90, p < 0.0001. ** p < 0.01 vs. Sham 21D. ## p < 0.01 vs. SNL 21D. n = 6. (d) Immunofluorescence images of spinal slices dissected at day 21 post operation. Compared with the sham operation (left), SNL (middle) decreased the number and distribution of the Hes1-positive (red) neurons but increased that of the CDK9-postive (green) and pRNAPII-postive (blue) neurons in the dorsal horn; these effects were all reversed by transfecting SNL animals with Hes1-encoding vector (right). Dashed lines indicate the margin of the dorsal horn. The inset images at the bottom are amplifications of the upper marked area, with arrows indicating the immunopositive neurons. Scale bar = 50 μm; Thickness = 30 μm. (e) ChIP-qPCR assay of dorsal horn samples dissected at day 21 post operation. When compared with the sham operation, SNL increased the amounts of CDK9 and pRNAPII antibodies-precipitated exon 1a and exon 1b promoter fragments of mGluR5; effects were both reversed by transfecting SNL animals with Hes1-encoding vector but not the control vector. CDK9, exon 1a, one-way ANOVA, post-hoc Tukey test, F (3,16) = 1812, p < 0.0001. CDK9, exon 1b, one-way ANOVA, post-hoc Tukey test, F (3,16) = 675.6, p < 0.0001. pRNAPII, exon 1a, one-way ANOVA, post-hoc Tukey test, F (3,16) = 638.7 p < 0.0001. pRNAPII, exon 1b, one-way ANOVA, post-hoc Tukey test, F (3,16) = 103.0 p < 0.0001. ** p < 0.01 vs. Sham 21D. # p < 0.05, ## p < 0.01 vs. SNL 21D. n = 5.
Figure 5.
Neuropathic injury impedes Hes1-suppressed CDK9 recruitment and RNAPII phosphorylation on mGluR5 promoters. (a) Representative western blot and statistical analyses (normalized to GAPDH) of dorsal horn samples demonstrating the levels of CDK9 and phosphorylated RNAPII (pRNAPII) were increased by spinally administering naïve animals (Naïve) with Hes1-siRNA but not with missense siRNA (Hes1 siRNA and MS siRNA, respectively, 5 μg, 10 μL; daily for 4 days). IB, immunoblotting. CDK9, one-way ANOVA, post-hoc Tukey test, F (2,15) = 64.17, p < 0.0001. pRNAPII, one-way ANOVA, post-hoc Tukey test, F (2,15) = 59.83, p < 0.0001. ** p < 0.01 vs. Naïve. n = 6. (b) ChIP-qPCR assay of dorsal horn samples demonstrating that administering naïve animals with Hes1 siRNA, but not missense siRNA (Hes1 siRNA and MS siRNA, respectively. 5 μg, 10 μL; daily for 4 days), increased amounts of both CDK9 and pRNAPII antibody-precipitated exon 1a and exon 1b promoter fragments of mGluR5. CDK9, exon 1a, one-way ANOVA, post-hoc Tukey test, F (2,12) = 1494, p < 0.0001. CDK9, exon 1b, one-way ANOVA, post-hoc Tukey test, F(2,12) = 88.97, p < 0.0001. pRNAPII, exon 1a, one-way ANOVA, post-hoc Tukey test, F (2,12) = 358.4, p < 0.0001. pRNAPII, exon 1b, one-way ANOVA, post-hoc Tukey test, F (2,12) = 373.5, p < 0.0001. ** p < 0.01 vs. Naïve. n = 5. (c) Representative western blot and statistical analyses of dorsal horn samples dissected at day 21 after operation. When compared with the sham operation (Sham 21D), spinal nerve ligation (SNL 21D) increased spinal CDK9 and pRNAPII levels; that were both reversed by administering SNL animals with Hes1-encoding vector (SNL 21D + Hes1 LV) but not the control vector (SNL 21D + Control LV). CDK9, one-way ANOVA, post-hoc Tukey test, F (3,20) = 38.06, p < 0.0001. pRNAPII, one-way ANOVA, post-hoc Tukey test, F (3,20) = 43.90, p < 0.0001. ** p < 0.01 vs. Sham 21D. ## p < 0.01 vs. SNL 21D. n = 6. (d) Immunofluorescence images of spinal slices dissected at day 21 post operation. Compared with the sham operation (left), SNL (middle) decreased the number and distribution of the Hes1-positive (red) neurons but increased that of the CDK9-postive (green) and pRNAPII-postive (blue) neurons in the dorsal horn; these effects were all reversed by transfecting SNL animals with Hes1-encoding vector (right). Dashed lines indicate the margin of the dorsal horn. The inset images at the bottom are amplifications of the upper marked area, with arrows indicating the immunopositive neurons. Scale bar = 50 μm; Thickness = 30 μm. (e) ChIP-qPCR assay of dorsal horn samples dissected at day 21 post operation. When compared with the sham operation, SNL increased the amounts of CDK9 and pRNAPII antibodies-precipitated exon 1a and exon 1b promoter fragments of mGluR5; effects were both reversed by transfecting SNL animals with Hes1-encoding vector but not the control vector. CDK9, exon 1a, one-way ANOVA, post-hoc Tukey test, F (3,16) = 1812, p < 0.0001. CDK9, exon 1b, one-way ANOVA, post-hoc Tukey test, F (3,16) = 675.6, p < 0.0001. pRNAPII, exon 1a, one-way ANOVA, post-hoc Tukey test, F (3,16) = 638.7 p < 0.0001. pRNAPII, exon 1b, one-way ANOVA, post-hoc Tukey test, F (3,16) = 103.0 p < 0.0001. ** p < 0.01 vs. Sham 21D. # p < 0.05, ## p < 0.01 vs. SNL 21D. n = 5.
![Ijms 20 04177 g005a Ijms 20 04177 g005a]()
![Ijms 20 04177 g005b Ijms 20 04177 g005b]()
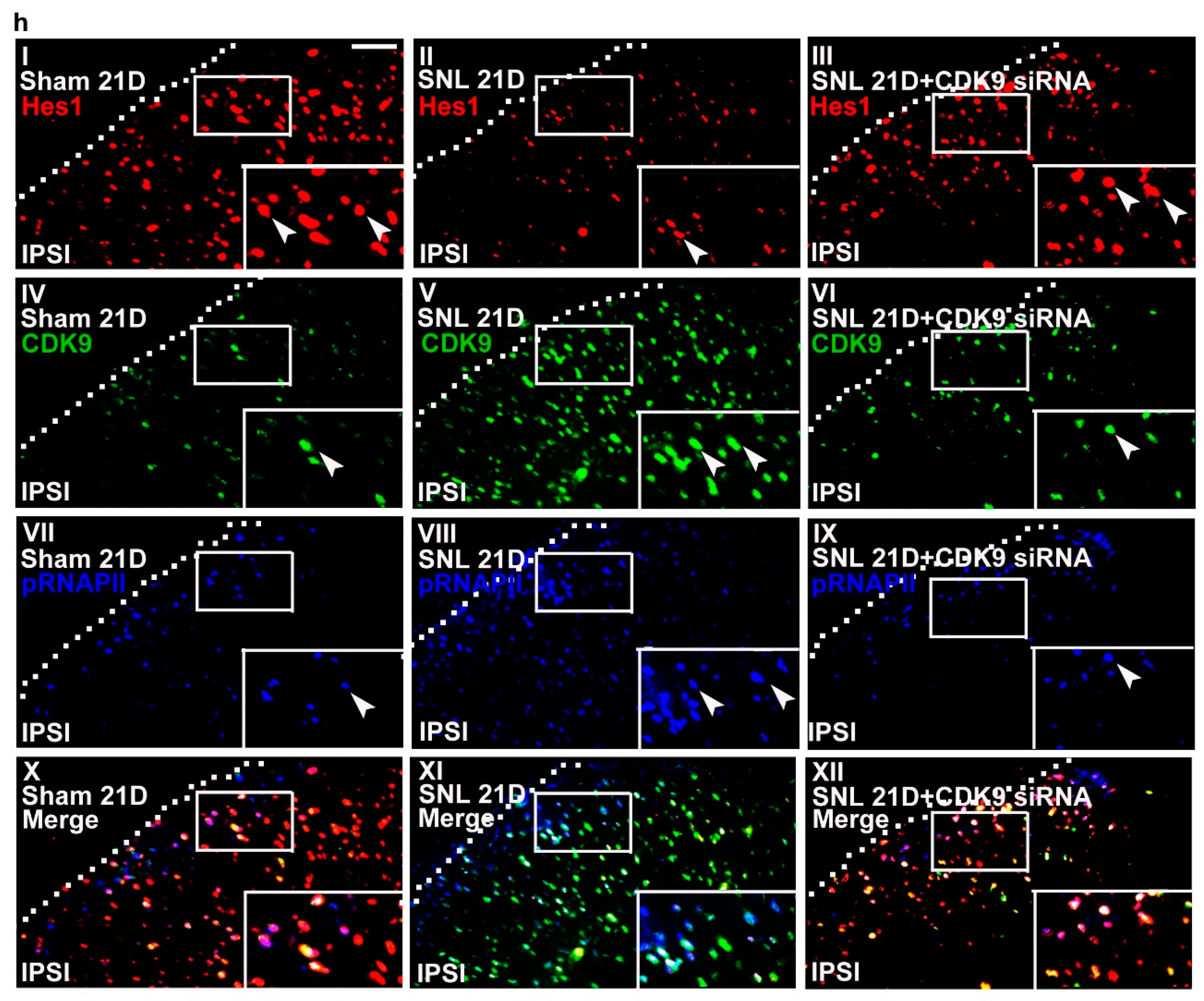
Figure 6.
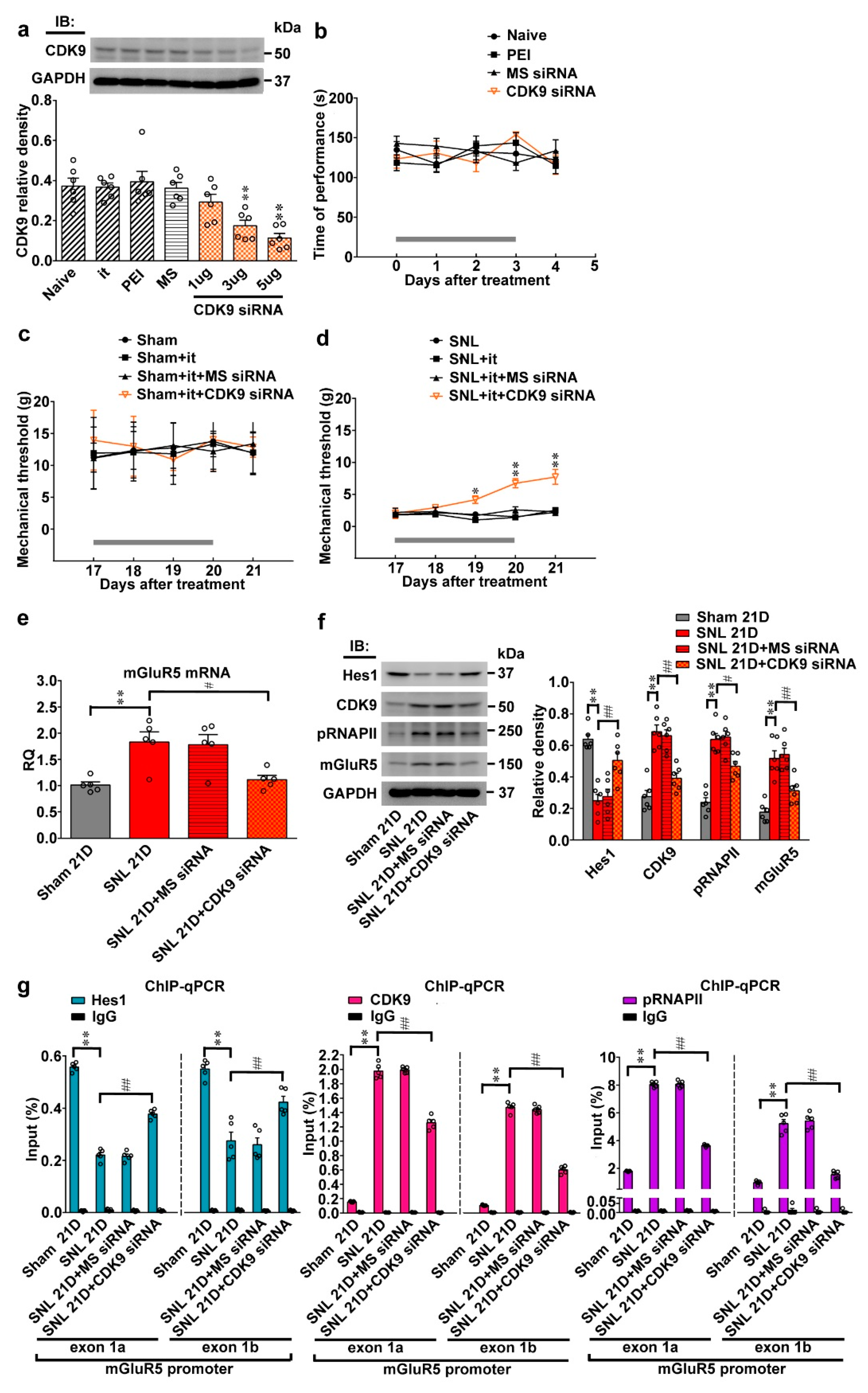
Neuropathic injury induces allodynia with reciprocal changes in spinal Hes1-CDK9 levels. (a) Representative Western blot and statistical analyses (normalized to GAPDH) of the dorsal horn sample dissected from naïve animals (Naïve) at day 4 after the start of treatment. The abundance of CDK9 was not affected by implantation of an intrathecal catheter alone (it) or spinal administration of polyethylenimine (PEI, 10 μL; daily for 4 days) or missense siRNA (MS siRNA, 5 μg, 10 μL; daily for 4 days) but was dose-dependently decreased by intrathecal administration of CDK9 siRNA (CDK9 siRNA; 1, 3, and 5 μg; 10 μL; daily for 4 days). IB, Immunoblotting. One-way ANOVA, post-hoc Tukey test, F (6,35) = 10.16, p < 0.0001. ** p < 0.01 vs. Naïve. n = 6. (b) Rota-rod test demonstrating no statistical difference in the motor performance among naïve animals as well as naïve animals administered with polyethylenimine, missense siRNA, or CDK9 siRNA (5 μg, 10 μL). The gray bar at the bottom indicates the duration of the reagents’ administration. n = 7. Two-way ANOVA with repeated measures over time, treatment, F (3,24) = 0.4792, p = 0.6997; time, F (4,96) = 0.2094, p = 0.9351; treatment × time, F (12,96) = 1.123, p = 0.3510. (c,d) Von Frey test demonstrating that while it exhibited no effect on the sham-operated group, spinal administration of CDK9 siRNA (5 μg, 10 μL), but not missense siRNA or polyethylenimine, increased the withdrawal threshold of the ipsilateral hind paw of SNL rats at days 19, 20, and 21 after operation. The gray bar at the bottom indicates the duration of the reagents’ administration. * p < 0.05, ** p < 0.01 vs. SNL. n = 7. Sham, Two-way ANOVA with repeated measures over time, treatment, F (3,24) = 0.2648, p = 0.8501; time, F (4,96) = 0.4465, p = 0.7747; treatment × time, F(12,96) = 0.3796, p = 0.9679. SNL, Two-way ANOVA with repeated measures over time, treatment, F (3,24) = 32.28, p < 0.0001; time, F(4,96) = 7.267, p < 0.0001; treatment × time, F(12,96) = 5.291, p < 0.0001. (e,f) RT-PCR and western blot analyses (normalized to GAPDH) of ipsilateral dorsal horn samples dissected at day 21 post operation. When compared with the sham operation (Sham 21D), SNL (SNL 21D) increased the abundance of mGluR5 mRNA as well as CDK9, phosphorylated RNAPII (pRNAPII), and mGluR5 protein but decreased that of Hes1 protein. These effects were all reversed by administering SNL animals with CDK9 siRNA (SNL 21D + CDK9 siRNA) but not the missense siRNA (SNL 21D + MS siRNA). mGluR5 mRNA, one-way ANOVA, post-hoc Tukey test, F (3,16) = 8.639, p = 0.0012. Hes1, one-way ANOVA, post-hoc Tukey test, F (3,20) = 19.59, p < 0.0001. CDK9, one-way ANOVA, post-hoc Tukey test, F (3,20) = 27.98, p < 0.0001. pRNAPII, one-way ANOVA, post-hoc Tukey test, F (3,20) = 32,00, p < 0.0001. mGluR5 protein, one-way ANOVA, post-hoc Tukey test, F (3,20) = 21.31, p < 0.0001. ** p < 0.01 vs. Sham 21D. ## p < 0.01 vs. SNL 21D. mRNA, n = 5. protein, n = 6. (g) ChIP-qPCR assay of dorsal horn samples dissected at day 21 post operation. Compared to the sham operation, SNL decreased the levels of Hes1 antibody-precipitated but increased that of CDK9 and pRNAPII antibodies-precipitated exon 1a and exon 1b promoter fragments of mGluR5. These effects were all reversed by administering SNL animals with CDK9-targeting antisense siRNA, but not with the missense siRNA. Hes1, exon 1a, one-way ANOVA, post-hoc Tukey test, F (3,16) = 380.8, p < 0.0001. Hes1, exon 1b, one-way ANOVA, post-hoc Tukey test, F (3,16) = 29.54, p < 0.0001. CDK9, exon 1a, one-way ANOVA, post-hoc Tukey test, F(3,16) = 727.1, p < 0.0001. CDK9, exon 1b, one-way ANOVA, post-hoc Tukey test, F (3,16) = 856.4, p < 0.0001. pRNAPII, exon 1a, one-way ANOVA, post-hoc Tukey test, F (3,16) = 1888 p < 0.0001. pRNAPII, exon 1b, one-way ANOVA, post-hoc Tukey test, F(3,16) = 129.1 p < 0.0001. ** p < 0.01 vs. Sham 21D. # p < 0.05, ## p < 0.01 vs. SNL 21D. n = 5. (h) Immunofluorescence images of spinal slices dissected at day 21 post operation. When compared with the sham operation (left), SNL (middle) decreased the number and distribution of Hes1-positive (red) but increased that of CDK9- (green) and pRNAPII-positive (blue) neurons in the dorsal horn ipsilateral to operation; these effects were all reversed by administering SNL animals with CDK9 siRNA (SNL 21D + CDK9 siRNA) (right). Dashed lines indicate the margin of the dorsal horn. The images at the bottom are amplifications of the marked area, with arrows indicating the immunopositive neurons. Scale bar = 50 μm; Thickness = 50 μm.
Figure 6.
Neuropathic injury induces allodynia with reciprocal changes in spinal Hes1-CDK9 levels. (a) Representative Western blot and statistical analyses (normalized to GAPDH) of the dorsal horn sample dissected from naïve animals (Naïve) at day 4 after the start of treatment. The abundance of CDK9 was not affected by implantation of an intrathecal catheter alone (it) or spinal administration of polyethylenimine (PEI, 10 μL; daily for 4 days) or missense siRNA (MS siRNA, 5 μg, 10 μL; daily for 4 days) but was dose-dependently decreased by intrathecal administration of CDK9 siRNA (CDK9 siRNA; 1, 3, and 5 μg; 10 μL; daily for 4 days). IB, Immunoblotting. One-way ANOVA, post-hoc Tukey test, F (6,35) = 10.16, p < 0.0001. ** p < 0.01 vs. Naïve. n = 6. (b) Rota-rod test demonstrating no statistical difference in the motor performance among naïve animals as well as naïve animals administered with polyethylenimine, missense siRNA, or CDK9 siRNA (5 μg, 10 μL). The gray bar at the bottom indicates the duration of the reagents’ administration. n = 7. Two-way ANOVA with repeated measures over time, treatment, F (3,24) = 0.4792, p = 0.6997; time, F (4,96) = 0.2094, p = 0.9351; treatment × time, F (12,96) = 1.123, p = 0.3510. (c,d) Von Frey test demonstrating that while it exhibited no effect on the sham-operated group, spinal administration of CDK9 siRNA (5 μg, 10 μL), but not missense siRNA or polyethylenimine, increased the withdrawal threshold of the ipsilateral hind paw of SNL rats at days 19, 20, and 21 after operation. The gray bar at the bottom indicates the duration of the reagents’ administration. * p < 0.05, ** p < 0.01 vs. SNL. n = 7. Sham, Two-way ANOVA with repeated measures over time, treatment, F (3,24) = 0.2648, p = 0.8501; time, F (4,96) = 0.4465, p = 0.7747; treatment × time, F(12,96) = 0.3796, p = 0.9679. SNL, Two-way ANOVA with repeated measures over time, treatment, F (3,24) = 32.28, p < 0.0001; time, F(4,96) = 7.267, p < 0.0001; treatment × time, F(12,96) = 5.291, p < 0.0001. (e,f) RT-PCR and western blot analyses (normalized to GAPDH) of ipsilateral dorsal horn samples dissected at day 21 post operation. When compared with the sham operation (Sham 21D), SNL (SNL 21D) increased the abundance of mGluR5 mRNA as well as CDK9, phosphorylated RNAPII (pRNAPII), and mGluR5 protein but decreased that of Hes1 protein. These effects were all reversed by administering SNL animals with CDK9 siRNA (SNL 21D + CDK9 siRNA) but not the missense siRNA (SNL 21D + MS siRNA). mGluR5 mRNA, one-way ANOVA, post-hoc Tukey test, F (3,16) = 8.639, p = 0.0012. Hes1, one-way ANOVA, post-hoc Tukey test, F (3,20) = 19.59, p < 0.0001. CDK9, one-way ANOVA, post-hoc Tukey test, F (3,20) = 27.98, p < 0.0001. pRNAPII, one-way ANOVA, post-hoc Tukey test, F (3,20) = 32,00, p < 0.0001. mGluR5 protein, one-way ANOVA, post-hoc Tukey test, F (3,20) = 21.31, p < 0.0001. ** p < 0.01 vs. Sham 21D. ## p < 0.01 vs. SNL 21D. mRNA, n = 5. protein, n = 6. (g) ChIP-qPCR assay of dorsal horn samples dissected at day 21 post operation. Compared to the sham operation, SNL decreased the levels of Hes1 antibody-precipitated but increased that of CDK9 and pRNAPII antibodies-precipitated exon 1a and exon 1b promoter fragments of mGluR5. These effects were all reversed by administering SNL animals with CDK9-targeting antisense siRNA, but not with the missense siRNA. Hes1, exon 1a, one-way ANOVA, post-hoc Tukey test, F (3,16) = 380.8, p < 0.0001. Hes1, exon 1b, one-way ANOVA, post-hoc Tukey test, F (3,16) = 29.54, p < 0.0001. CDK9, exon 1a, one-way ANOVA, post-hoc Tukey test, F(3,16) = 727.1, p < 0.0001. CDK9, exon 1b, one-way ANOVA, post-hoc Tukey test, F (3,16) = 856.4, p < 0.0001. pRNAPII, exon 1a, one-way ANOVA, post-hoc Tukey test, F (3,16) = 1888 p < 0.0001. pRNAPII, exon 1b, one-way ANOVA, post-hoc Tukey test, F(3,16) = 129.1 p < 0.0001. ** p < 0.01 vs. Sham 21D. # p < 0.05, ## p < 0.01 vs. SNL 21D. n = 5. (h) Immunofluorescence images of spinal slices dissected at day 21 post operation. When compared with the sham operation (left), SNL (middle) decreased the number and distribution of Hes1-positive (red) but increased that of CDK9- (green) and pRNAPII-positive (blue) neurons in the dorsal horn ipsilateral to operation; these effects were all reversed by administering SNL animals with CDK9 siRNA (SNL 21D + CDK9 siRNA) (right). Dashed lines indicate the margin of the dorsal horn. The images at the bottom are amplifications of the marked area, with arrows indicating the immunopositive neurons. Scale bar = 50 μm; Thickness = 50 μm.
![Ijms 20 04177 g006a Ijms 20 04177 g006a]()
![Ijms 20 04177 g006b Ijms 20 04177 g006b]()