Integrated Approach Reveals Role of Mitochondrial Germ-Line Mutation F18L in Respiratory Chain, Oxidative Alterations, Drug Sensitivity, and Patient Prognosis in Glioblastoma
Abstract
1. Introduction
2. Results
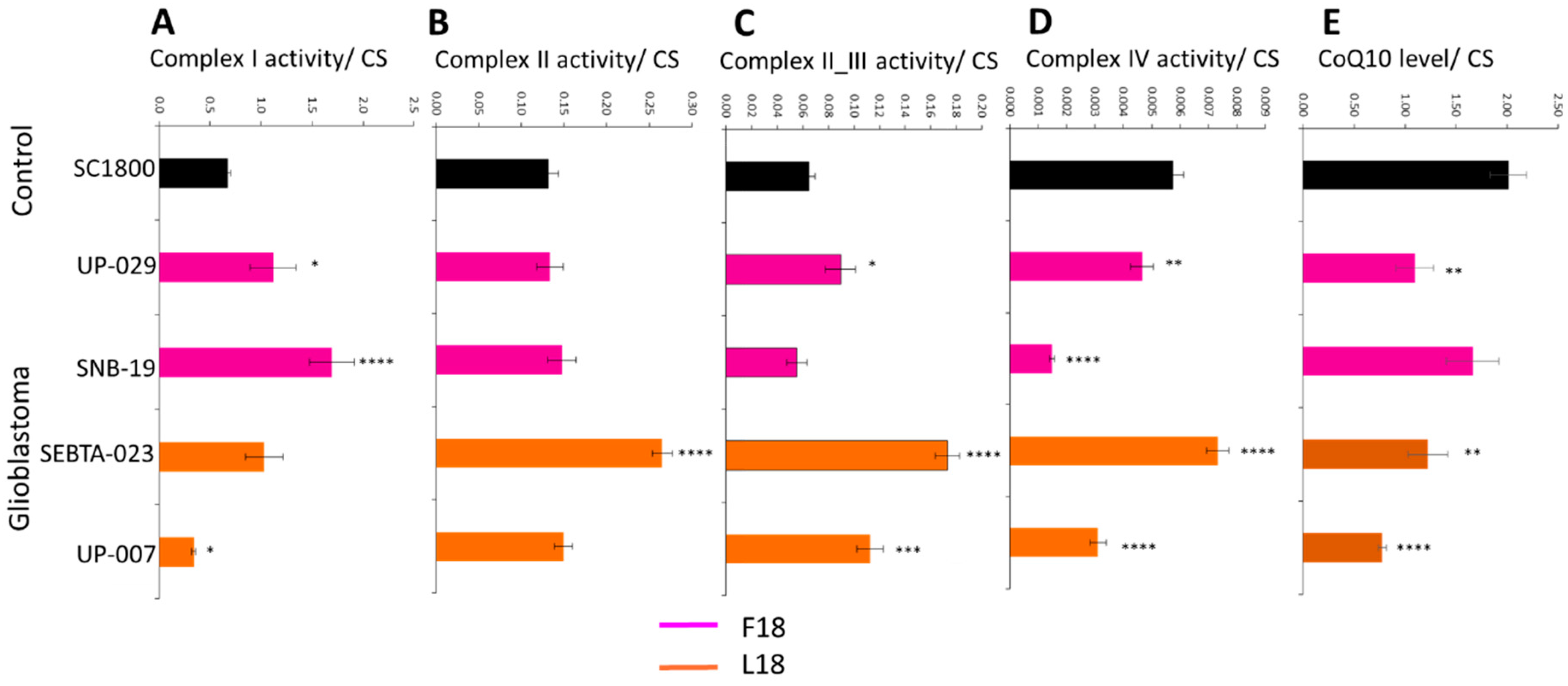
2.1. MRC Complex Activities and CoQ10 Levels Differ in Glioblastoma Cells Relative to Control
2.2. Individual Analysis of MRC Complexes and CoQ10 in Glioblastoma Cells Reveals Biological Variability
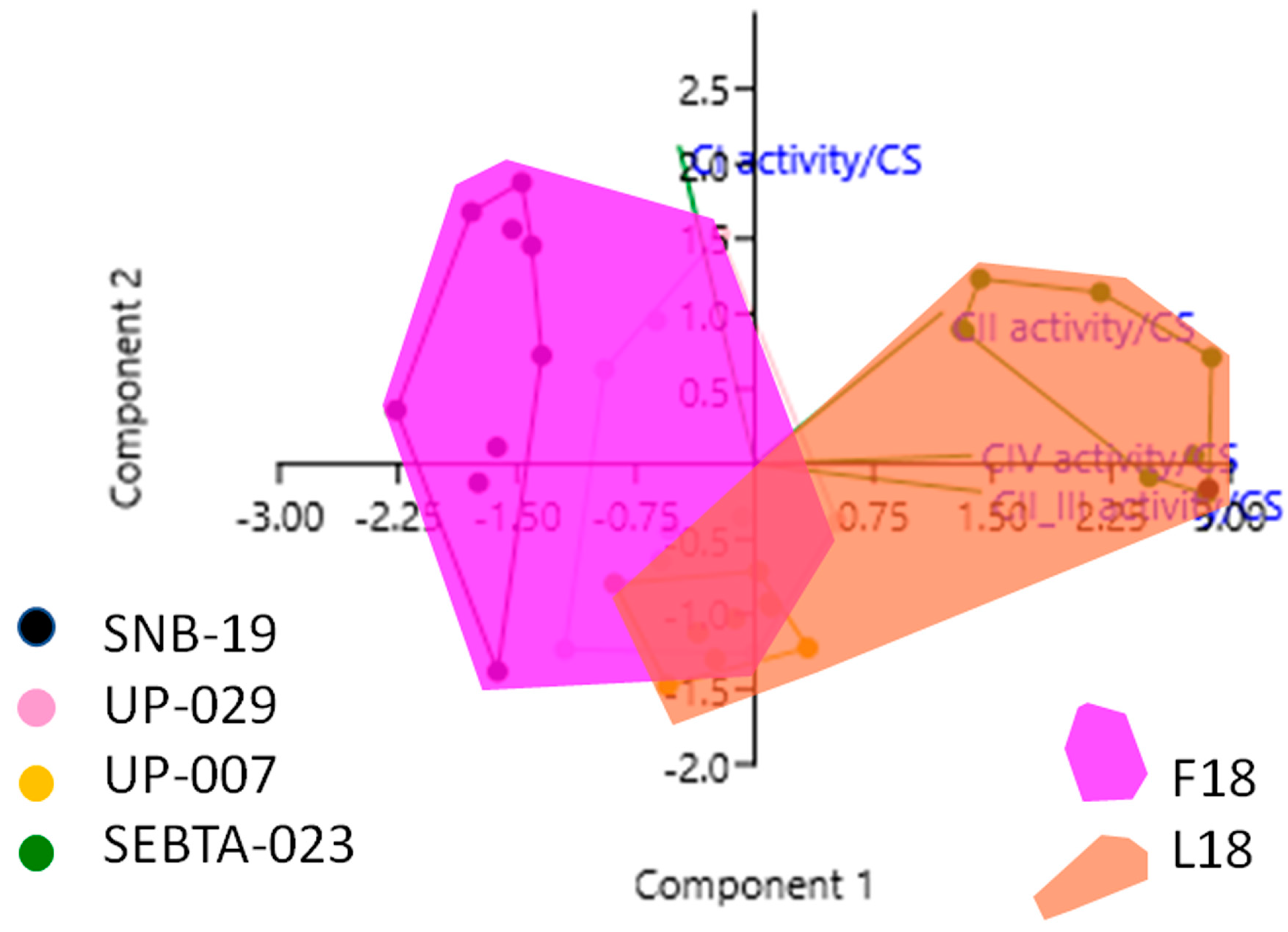
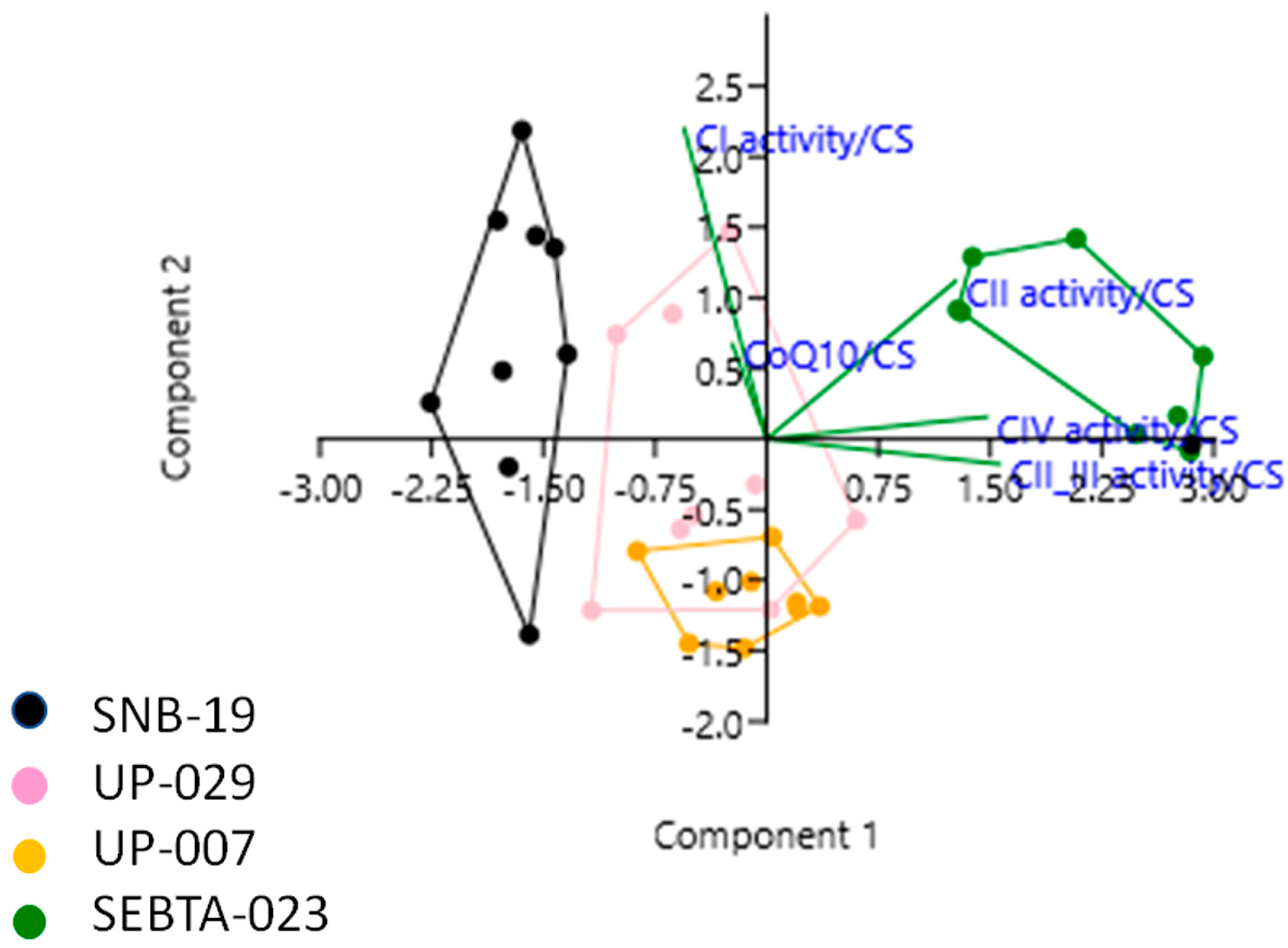
2.3. Simultaneous Analysis of Mitochondrial Parameters Reveals Novel Associations
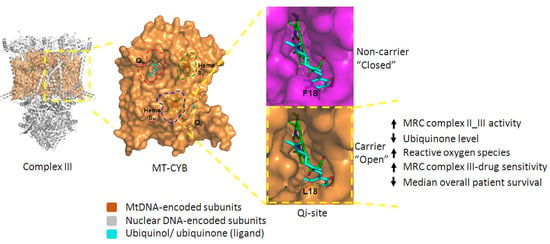
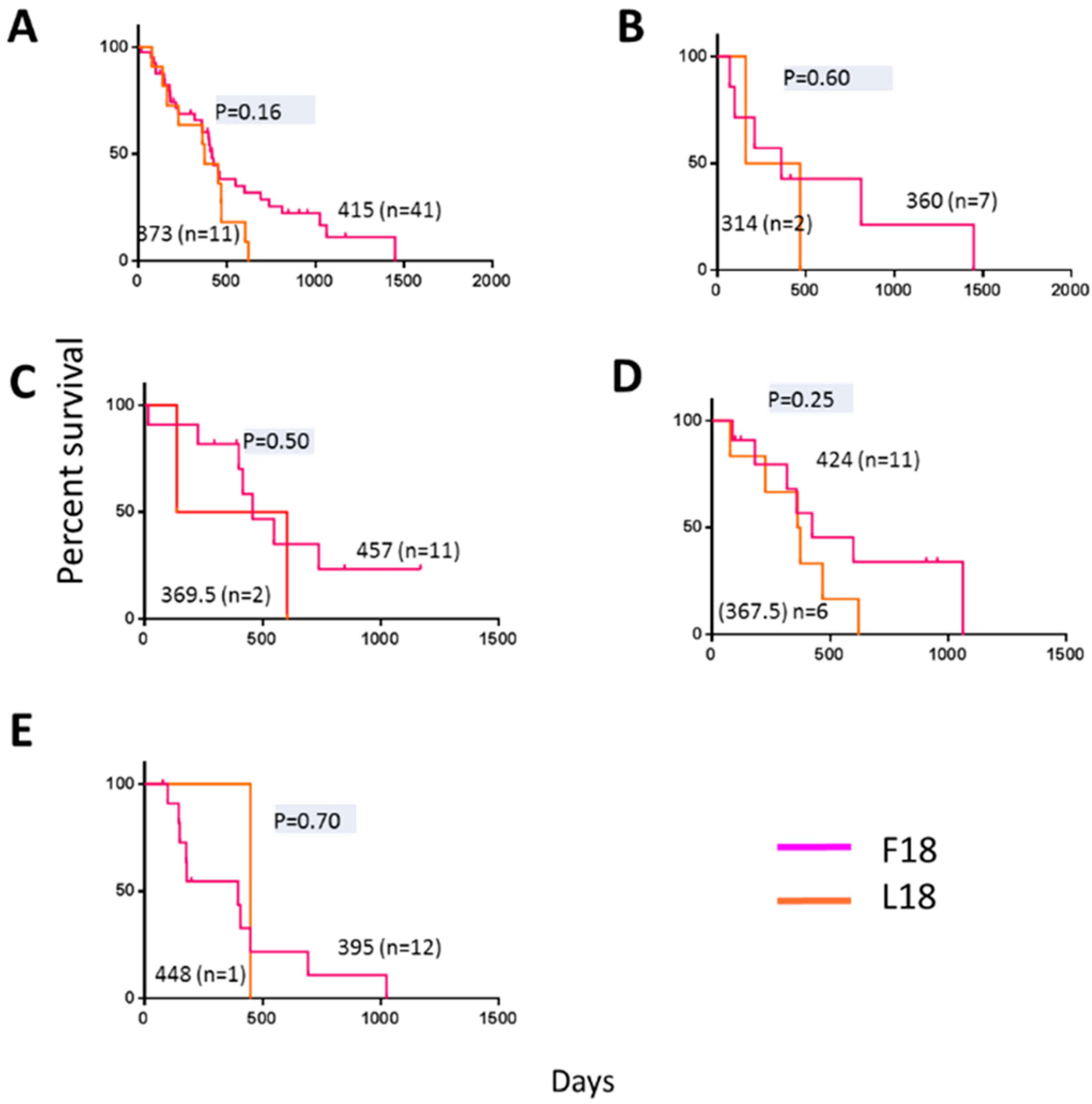
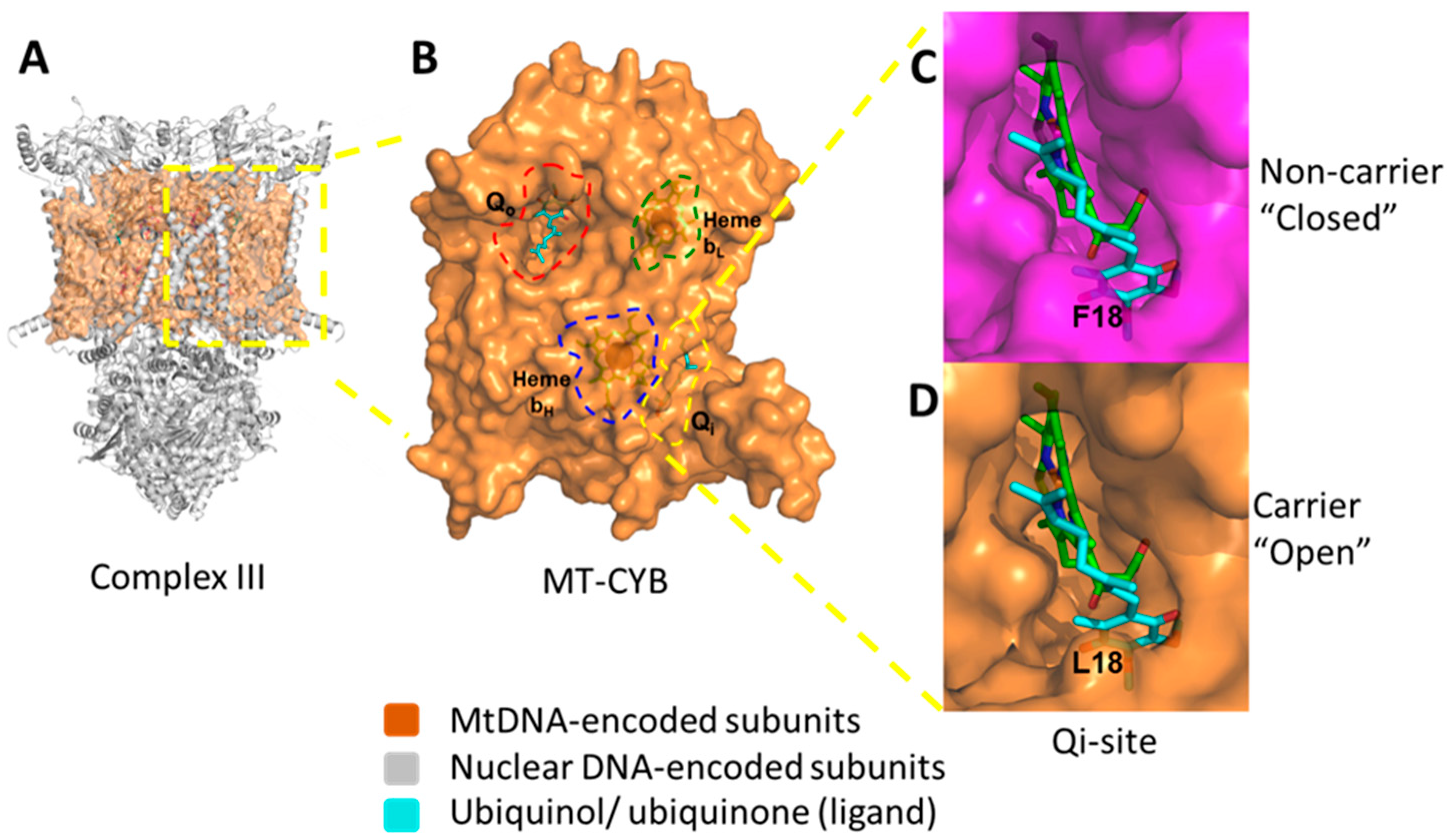
2.4. F18L-Carriers Have Poorer Prognosis
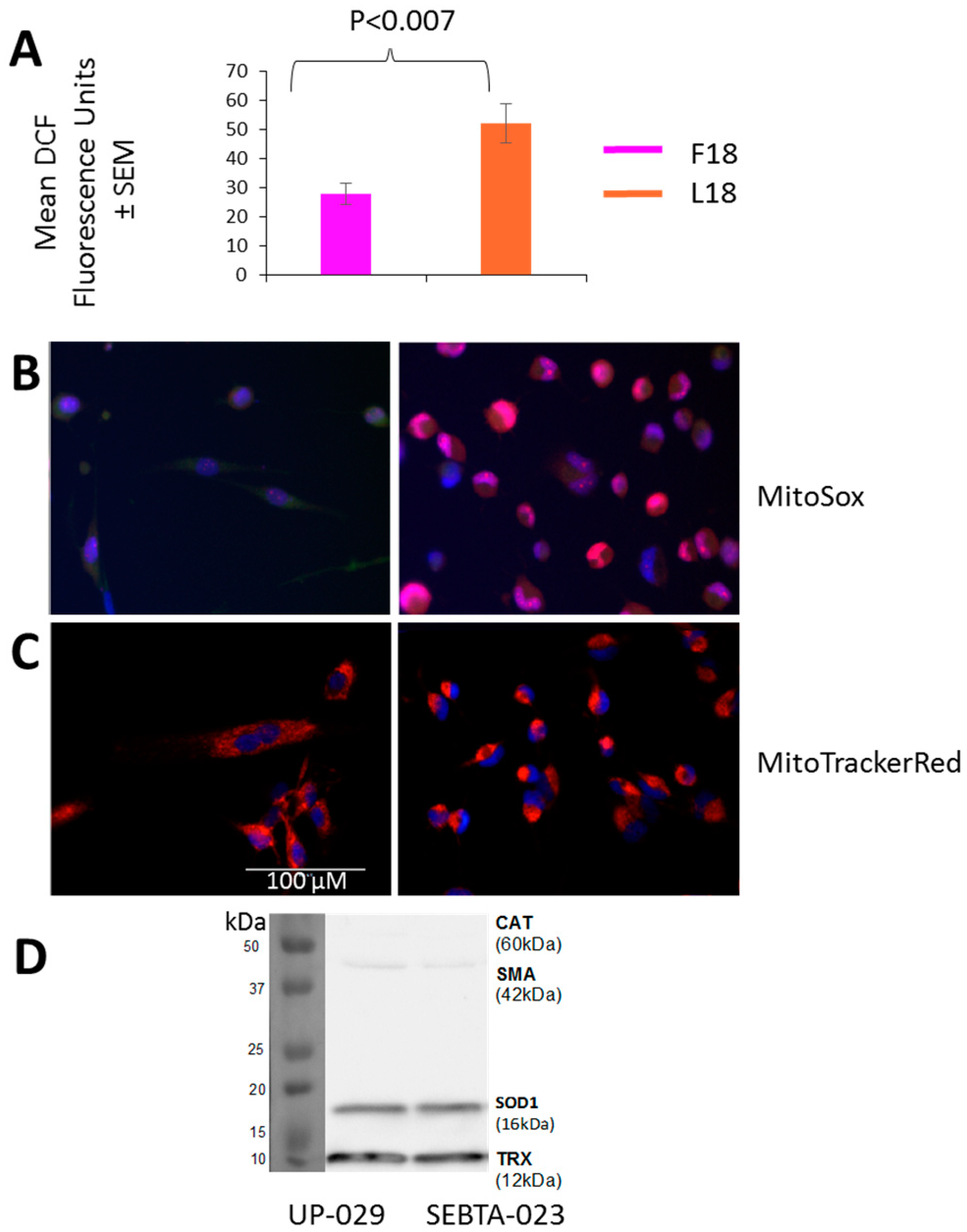
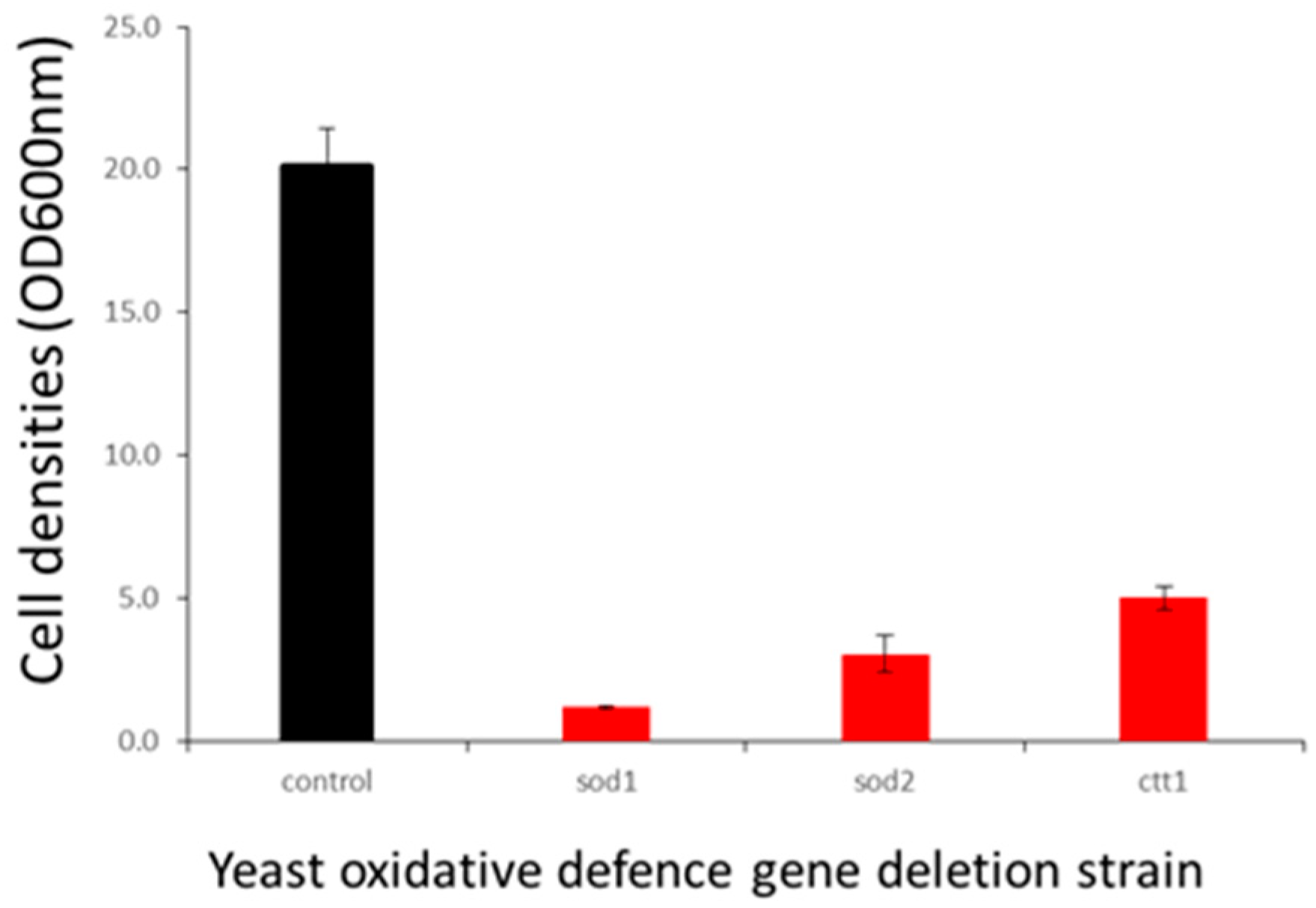
2.5. F18L-Carriers Have Elevated Oxidative Stress
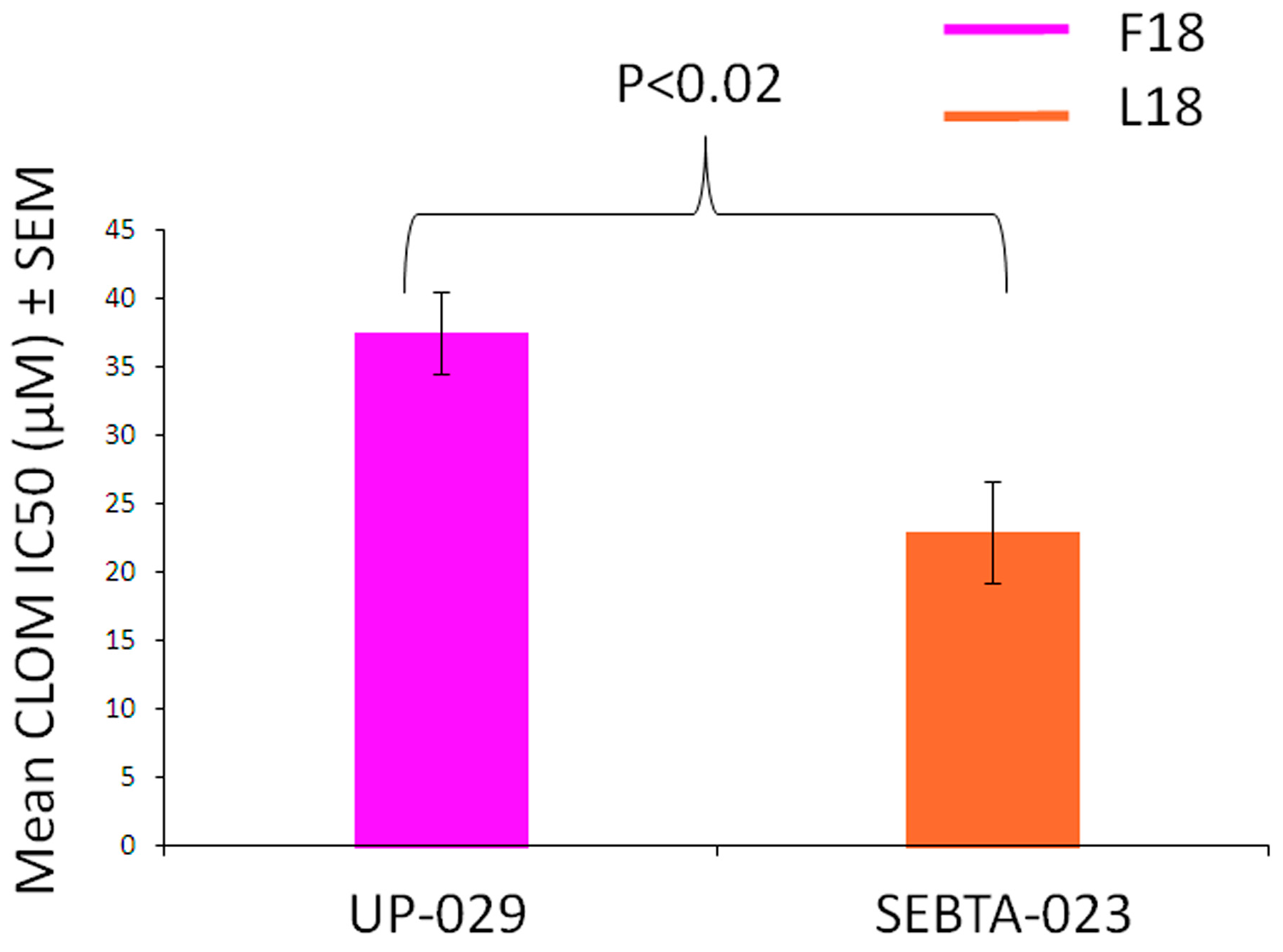
2.6. F18L-Carriers Are More Sensitive to Clomipramine
3. Discussion
4. Materials and Methods
4.1. Patient-Derived Glioblastoma Cell Cultures
4.2. Patient-Derived Glioblastoma Tissues
4.3. F18L Mutation Identification
4.4. Measurement of MRC Enzyme and Citrate Synthase Activity and CoQ10 Levels
4.5. Intracellular ROS Detection
4.6. Live Cell Imaging of Mitochondrial ROS and Networks
4.7. Immunoblotting
4.8. Analysis of Cell Proliferation with and without Clomipramine
4.9. Statistical Analysis
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Ostrom, Q.T.; Bauchet, L.; Davis, F.G.; Deltour, I.; Fisher, J.L.; Langer, C.E.; Pekmezci, M.; Schwartzbaum, J.A.; Turner, M.C.; Walsh, K.M.; et al. The epidemiology of glioma in adults: A “state of the science” review. Neuro Oncol. 2014, 16, 896–913. [Google Scholar] [CrossRef]
- Dubrow, R.; Darefsky, A.S.; Jacobs, D.I.; Park, L.S.; Rose, M.G.; Laurans, M.S.; King, J.T. Time trends in glioblastoma multiforme survival: The role of temozolomide. Neuro Oncol. 2013, 15, 1750–1761. [Google Scholar] [CrossRef]
- Phillips, H.S.; Kharbanda, S.; Chen, R.; Forrest, W.F.; Soriano, R.H.; Wu, T.D.; Misra, A.; Nigro, J.M.; Colman, H.; Soroceanu, L.; et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell 2006, 9, 157–173. [Google Scholar] [CrossRef] [PubMed]
- Sturm, D.; Witt, H.; Hovestadt, V.; Khuong-Quang, D.A.; Jones, D.T.; Konermann, C.; Pfaff, E.; Tonjes, M.; Sill, M.; Bender, S.; et al. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell 2012, 22, 425–437. [Google Scholar] [CrossRef] [PubMed]
- Verhaak, R.G.; Hoadley, K.A.; Purdom, E.; Wang, V.; Qi, Y.; Wilkerson, M.D.; Miller, C.R.; Ding, L.; Golub, T.; Mesirov, J.P.; et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 2010, 17, 98–110. [Google Scholar] [CrossRef] [PubMed]
- Deighton, R.F.; Le Bihan, T.; Martin, S.F.; Gerth, A.M.J.; McCulloch, M.; Edgar, J.M.; Kerr, L.E.; Whittle, I.R.; McCulloch, J. Interactions among mitochondrial proteins altered in glioblastoma. J. Neurooncol. 2014, 118, 247–256. [Google Scholar] [CrossRef]
- Ordys, B.B.; Launay, S.; Deighton, R.F.; McCulloch, J.; Whittle, I.R. The role of mitochondria in glioma pathophysiology. Mol. Neurobiol. 2010, 42, 64–75. [Google Scholar] [CrossRef]
- Vafai, S.B.; Mootha, V.K. Mitochondrial disorders as windows into an ancient organelle. Nature 2012, 491, 374–383. [Google Scholar] [CrossRef]
- Astuti, D.; Latif, F.; Dallol, A.; Dahia, P.L.; Douglas, F.; George, E.; Skoldberg, F.; Husebye, E.S.; Eng, C.; Maher, E.R. Gene mutations in the succinate dehydrogenase subunit SDHB cause susceptibility to familial pheochromocytoma and to familial paraganglioma. Am. J. Hum. Genet. 2001, 69, 49–54. [Google Scholar] [CrossRef]
- Baysal, B.E.; Ferrell, R.E.; Willett-Brozick, J.E.; Lawrence, E.C.; Myssiorek, D.; Bosch, A.; van der Mey, A.; Taschner, P.E.; Rubinstein, W.S.; Myers, E.N.; et al. Mutations in SDHD, a mitochondrial complex II gene, in hereditary paraganglioma. Science 2000, 287, 848–851. [Google Scholar] [CrossRef]
- Niemann, S.; Muller, U. Mutations in SDHC cause autosomal dominant paraganglioma, type 3. Nat. Genet. 2000, 26, 268–270. [Google Scholar] [CrossRef] [PubMed]
- Feichtinger, R.G.; Weis, S.; Mayr, J.A.; Zimmermann, F.; Geilberger, R.; Sperl, W.; Kofler, B. Alterations of oxidative phosphorylation complexes in astrocytomas. Glia 2014, 62, 514–525. [Google Scholar] [CrossRef] [PubMed]
- Griguer, C.E.; Cantor, A.B.; Fathallah-Shaykh, H.M.; Gillespie, G.Y.; Gordon, A.S.; Markert, J.M.; Radovanovic, I.; Clement-Schatlo, V.; Shannon, C.N.; Oliva, C.R. Prognostic relevance of cytochrome C oxidase in primary glioblastoma multiforme. PLoS ONE 2013, 8, e61035. [Google Scholar] [CrossRef] [PubMed]
- Oliva, C.R.; Zhang, W.; Langford, C.; Suto, M.J.; Griguer, C.E. Repositioning chlorpromazine for treating chemoresistant glioma through the inhibition of cytochrome c oxidase bearing the COX4–1 regulatory subunit. Oncotarget 2017, 8, 37568–37583. [Google Scholar] [CrossRef] [PubMed]
- Daley, E.; Wilkie, D.; Loesch, A.; Hargreaves, I.P.; Kendall, D.A.; Pilkington, G.J.; Bates, T.E. Chlorimipramine: A novel anticancer agent with a mitochondrial target. Biochem. Biophys. Res. Commun. 2005, 328, 623–632. [Google Scholar] [CrossRef] [PubMed]
- Higgins, S.C.; Pilkington, G.J. The in vitro effects of tricyclic drugs and dexamethasone on cellular respiration of malignant glioma. Anticancer. Res. 2010, 30, 391–397. [Google Scholar] [PubMed]
- Levkovitz, Y.; Gil-Ad, I.; Zeldich, E.; Dayag, M.; Weizman, A. Differential induction of apoptosis by antidepressants in glioma and neuroblastoma cell lines: Evidence for p-c-Jun, cytochrome c, and caspase-3 involvement. J. Mol. Neurosci. 2005, 27, 29–42. [Google Scholar] [CrossRef]
- Pilkington, G.J.; Parker, K.; Murray, S.A. Approaches to mitochondrially mediated cancer therapy. Semin. Cancer Biol. 2008, 18, 226–235. [Google Scholar] [CrossRef] [PubMed]
- Shchors, K.; Massaras, A.; Hanahan, D. Dual Targeting of the Autophagic Regulatory Circuitry in Gliomas with Repurposed Drugs Elicits Cell-Lethal Autophagy and Therapeutic Benefit. Cancer Cell 2015, 28, 456–471. [Google Scholar] [CrossRef] [PubMed]
- Lloyd, R.E.; Keatley, K.; Littlewood, D.T.; Meunier, B.; Holt, W.V.; An, Q.; Higgins, S.C.; Polyzoidis, S.; Stephenson, K.F.; Ashkan, K.; et al. Identification and functional prediction of mitochondrial complex III and IV mutations associated with glioblastoma. Neuro. Oncol. 2015, 17, 942–952. [Google Scholar] [CrossRef] [PubMed]
- Yeung, K.Y.; Dickinson, A.; Donoghue, J.F.; Polekhina, G.; White, S.J.; Grammatopoulos, D.K.; McKenzie, M.; Johns, T.G.; St John, J.C. The identification of mitochondrial DNA variants in glioblastoma multiforme. Acta Neuro. Commun. 2014, 2, 1. [Google Scholar] [CrossRef] [PubMed]
- Oliva, C.R.; Nozell, S.E.; Diers, A.; McClugage, S.G.; Sarkaria, J.N.; Markert, J.M.; Darley-Usmar, V.M.; Bailey, S.M.; Gillespie, G.Y.; Landar, A.; et al. Acquisition of temozolomide chemoresistance in gliomas leads to remodeling of mitochondrial electron transport chain. J. Biol. Chem. 2010, 285, 39759–39767. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Vazquez, E.J.; Moghaddas, S.; Hoppel, C.L.; Lesnefsky, E.J. Production of reactive oxygen species by mitochondria: Central role of complex III. J. Biol. Chem. 2003, 278, 36027–36031. [Google Scholar] [CrossRef] [PubMed]
- Hargreaves, I.P. Coenzyme Q10 as a therapy for mitochondrial disease. Int. J. Biochem. Cell. Biol. 2014, 49, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Al-Mehdi, A.B.; Pastukh, V.M.; Swiger, B.M.; Reed, D.J.; Patel, M.R.; Bardwell, G.C.; Pastukh, V.V.; Alexeyev, M.F.; Gillespie, M.N. Perinuclear mitochondrial clustering creates an oxidant-rich nuclear domain required for hypoxia-induced transcription. Sci. Signal. 2012, 5, ra47. [Google Scholar] [CrossRef] [PubMed]
- Del Bufalo, D.; Trisciuoglio, D.; Biroccio, A.; Marcocci, L.; Buglioni, S.; Candiloro, A.; Scarsella, M.; Leonetti, C.; Zupi, G. Bcl-2 overexpression decreases BCNU sensitivity of a human glioblastoma line through enhancement of catalase activity. J. Cell. Biochem. 2001, 83, 473–483. [Google Scholar] [CrossRef] [PubMed]
- Dudek, H.; Farbiszewski, R.; Rydzewska, M.; Michno, T.; Kozlowski, A. Evaluation of antioxidant enzymes activity and concentration of non-enzymatic antioxidants in human brain tumours. Wiad. Lek. 2004, 57, 16–19. [Google Scholar] [PubMed]
- Kohsaka, S.; Takahashi, K.; Wang, L.; Tanino, M.; Kimura, T.; Nishihara, H.; Tanaka, S. Inhibition of GSH synthesis potentiates temozolomide-induced bystander effect in glioblastoma. Cancer Lett. 2013, 331, 68–75. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.C.; Kim, D.W.; Jung, K.Y.; Park, I.C.; Park, M.J.; Kim, M.S.; Woo, S.H.; Rhee, C.H.; Yoo, H.; Lee, S.H.; et al. Increased expression of antioxidant enzymes in radioresistant variant from U251 human glioblastoma cell line. Int. J. Mol. Med. 2004, 13, 883–887. [Google Scholar] [CrossRef] [PubMed]
- Shwetha, S.D.; Shastry, A.H.; Arivazhagan, A.; Santosh, V. Manganese superoxide dismutase (MnSOD) is a malignant astrocytoma specific biomarker and associated with adverse prognosis in p53 expressing glioblastoma. Pathol. Res. Pract. 2016, 212, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.; Laleve, A.; Vallieres, C.; McGeehan, J.E.; Lloyd, R.E.; Meunier, B. Human Mitochondrial Cytochrome b Variants Studied in Yeast: Not All Are Silent Polymorphisms. Hum. Mutat. 2016, 37, 933–941. [Google Scholar] [CrossRef] [PubMed]
- Libby, C.J.; Tran, A.N.; Scott, S.E.; Griguer, C.; Hjelmeland, A.B. The pro-tumorigenic effects of metabolic alterations in glioblastoma including brain tumor initiating cells. Biochim. Biophys. Acta Rev. Cancer 2018, 1869, 175–188. [Google Scholar] [CrossRef] [PubMed]
- Larman, T.C.; DePalma, S.R.; Hadjipanayis, A.G.; Cancer Genome Atlas Research, N.; Protopopov, A.; Zhang, J.; Gabriel, S.B.; Chin, L.; Seidman, C.E.; Kucherlapati, R.; et al. Spectrum of somatic mitochondrial mutations in five cancers. Proc. Natl. Acad. Sci. USA 2012, 109, 14087–14091. [Google Scholar] [CrossRef] [PubMed]
- DeHaan, C.; Habibi-Nazhad, B.; Yan, E.; Salloum, N.; Parliament, M.; Allalunis-Turner, J. Mutation in mitochondrial complex I ND6 subunit is associated with defective response to hypoxia in human glioma cells. Mol. Cancer 2004, 3, 19. [Google Scholar] [CrossRef] [PubMed]
- Oliva, C.R.; Markert, T.; Gillespie, G.Y.; Griguer, C.E. Nuclear-encoded cytochrome c oxidase subunit 4 regulates BMI1 expression and determines proliferative capacity of high-grade gliomas. Oncotarget 2015, 6, 4330–4344. [Google Scholar] [CrossRef] [PubMed]
- Miranda, S.; Foncea, R.; Guerrero, J.; Leighton, F. Oxidative stress and upregulation of mitochondrial biogenesis genes in mitochondrial DNA-depleted HeLa cells. Biochem. Biophys. Res. Commun. 1999, 258, 44–49. [Google Scholar] [CrossRef] [PubMed]
- Janiszewska, M.; Suva, M.L.; Riggi, N.; Houtkooper, R.H.; Auwerx, J.; Clement-Schatlo, V.; Radovanovic, I.; Rheinbay, E.; Provero, P.; Stamenkovic, I. Imp2 controls oxidative phosphorylation and is crucial for preserving glioblastoma cancer stem cells. Genes. Dev. 2012, 26, 1926–1944. [Google Scholar] [CrossRef] [PubMed]
- Montero, R.; Grazina, M.; Lopez-Gallardo, E.; Montoya, J.; Briones, P.; Navarro-Sastre, A.; Land, J.M.; Hargreaves, I.P.; Artuch, R. Coenzyme Q10 deficiency in mitochondrial DNA depletion syndromes. Mitochondrion 2013, 13, 337–341. [Google Scholar] [CrossRef]
- Enriquez, J.A.; Lenaz, G. Coenzyme q and the respiratory chain: Coenzyme q pool and mitochondrial supercomplexes. Mol. Syndromol. 2014, 5, 119–140. [Google Scholar] [CrossRef] [PubMed]
- Wallace, D.C. The mitochondrial genome in human adaptive radiation and disease: On the road to therapeutics and performance enhancement. Gene 2005, 354, 169–180. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, J.F.; Sabelnykova, V.Y.; Weischenfeldt, J.; Simon, R.; Aguiar, J.A.; Alkallas, R.; Heisler, L.E.; Zhang, J.; Watson, J.D.; Chua, M.L.K.; et al. Mitochondrial mutations drive prostate cancer aggression. Nat. Commun. 2017, 8, 656. [Google Scholar] [CrossRef] [PubMed]
- Walker, A.J.; Card, T.; Bates, T.E.; Muir, K. Tricyclic antidepressants and the incidence of certain cancers: A study using the GPRD. Br. J. Cancer 2011, 104, 193–197. [Google Scholar] [CrossRef] [PubMed]
- Illumina Resequencing Workflow. Available online: http://supportres.illumina.com/documents/documentation/software_documentation/miseqreporter/miseq-reporter-user-guide-15042295-b.pdf (accessed on 5 July 2019).
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Ruan, J.; Durbin, R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008, 18, 1851–1858. [Google Scholar] [CrossRef] [PubMed]
- DePristo, M.A.; Banks, E.; Poplin, R.; Garimella, K.V.; Maguire, J.R.; Hartl, C.; Philippakis, A.A.; del Angel, G.; Rivas, M.A.; Hanna, M.; et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011, 43, 491–498. [Google Scholar] [CrossRef]
- Milne, I.; Stephen, G.; Bayer, M.; Cock, P.J.; Pritchard, L.; Cardle, L.; Shaw, P.D.; Marshall, D. Using Tablet for visual exploration of second-generation sequencing data. Brief. Bioinform. 2013, 14, 193–202. [Google Scholar] [CrossRef]
- Lowe, T.M.; Chan, P.P. tRNAscan-SE On-line: Integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 2016, 44, W54–W57. [Google Scholar] [CrossRef]
- Laslett, D.; Canback, B. ARWEN: A program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics 2008, 24, 172–175. [Google Scholar] [CrossRef]
- TCGA BWA with Mark Duplicates and Cocleaning Workflow. Available online: https://docs.gdc.cancer.gov/Data/Bioinformatics_Pipelines/DNA_Seq_Variant_Calling_Pipeline (accessed on 5 July 2019).
- Samtools. Available online: http://samtools.sourceforge.net/ (accessed on 5 July 2019).
- Rimmer, A.; Phan, H.; Mathieson, I.; Iqbal, Z.; Twigg, S.R.F.; WGS500 Consortium; Wilkie, A.O.M.; McVean, G.; Lunter, G. Integrating mapping-, assembly- and haplotype-based approaches for calling variants in clinical sequencing applications. Nat. Genet. 2014, 46, 912–918. [Google Scholar] [CrossRef]
- Birch-Machin, M.A.; Turnbull, D.M. Assaying mitochondrial respiratory complex activity in mitochondria isolated from human cells and tissues. Methods Cell Biol. 2001, 65, 97–117. [Google Scholar]
- King, A.L.; Mantena, S.K.; Andringa, K.K.; Millender-Swain, T.; Dunham-Snary, K.J.; Oliva, C.R.; Griguer, C.E.; Bailey, S.M. The methyl donor S-adenosylmethionine prevents liver hypoxia and dysregulation of mitochondrial bioenergetic function in a rat model of alcohol-induced fatty liver disease. Redox. Biol. 2016, 9, 188–197. [Google Scholar] [CrossRef]
- Wharton, D.C.; Tzagoloff, A. Cytochrome oxidase from beef heart mitochondria. Methods Enzymol. 1967, 10, 245–250. [Google Scholar]
- Larsen, S.; Nielsen, J.; Hansen, C.N.; Nielsen, L.B.; Wibrand, F.; Stride, N.; Schroder, H.D.; Boushel, R.; Helge, J.W.; Dela, F.; et al. Biomarkers of mitochondrial content in skeletal muscle of healthy young human subjects. J. Physiol. 2012, 590, 3349–3360. [Google Scholar] [CrossRef] [PubMed]
- Hargreaves, P.; Rahman, S.; Guthrie, P.; Taanman, J.W.; Leonard, J.V.; Land, J.M.; Heales, S.J. Diagnostic value of succinate ubiquinone reductase activity in the identification of patients with mitochondrial DNA depletion. J. Inherit. Metab. Dis. 2002, 25, 7–16. [Google Scholar] [CrossRef] [PubMed]
- Duncan, A.J.; Heales, S.J.; Mills, K.; Eaton, S.; Land, J.M.; Hargreaves, I.P. Determination of coenzyme Q10 status in blood mononuclear cells, skeletal muscle, and plasma by HPLC with di-propoxy-coenzyme Q10 as an internal standard. Clin. Chem. 2005, 51, 2380–2382. [Google Scholar] [CrossRef] [PubMed]
- Selak, M.A.; de Chadarevian, J.P.; Melvin, J.J.; Grover, W.D.; Salganicoff, L.; Kaye, E.M. Mitochondrial activity in Pompe’s disease. Pediatr. Neurol. 2000, 23, 54–57. [Google Scholar] [CrossRef]
- Hammer, Ø.; Harper, D.A.T.; Ryan, P.D. PAST: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2000, 4, 9. [Google Scholar]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Keatley, K.; Stromei-Cleroux, S.; Wiltshire, T.; Rajala, N.; Burton, G.; Holt, W.V.; Littlewood, D.T.J.; Briscoe, A.G.; Jung, J.; Ashkan, K.; et al. Integrated Approach Reveals Role of Mitochondrial Germ-Line Mutation F18L in Respiratory Chain, Oxidative Alterations, Drug Sensitivity, and Patient Prognosis in Glioblastoma. Int. J. Mol. Sci. 2019, 20, 3364. https://doi.org/10.3390/ijms20133364
Keatley K, Stromei-Cleroux S, Wiltshire T, Rajala N, Burton G, Holt WV, Littlewood DTJ, Briscoe AG, Jung J, Ashkan K, et al. Integrated Approach Reveals Role of Mitochondrial Germ-Line Mutation F18L in Respiratory Chain, Oxidative Alterations, Drug Sensitivity, and Patient Prognosis in Glioblastoma. International Journal of Molecular Sciences. 2019; 20(13):3364. https://doi.org/10.3390/ijms20133364
Chicago/Turabian StyleKeatley, Kathleen, Samuel Stromei-Cleroux, Tammy Wiltshire, Nina Rajala, Gary Burton, William V. Holt, D. Timothy J. Littlewood, Andrew G. Briscoe, Josephine Jung, Keyoumars Ashkan, and et al. 2019. "Integrated Approach Reveals Role of Mitochondrial Germ-Line Mutation F18L in Respiratory Chain, Oxidative Alterations, Drug Sensitivity, and Patient Prognosis in Glioblastoma" International Journal of Molecular Sciences 20, no. 13: 3364. https://doi.org/10.3390/ijms20133364
APA StyleKeatley, K., Stromei-Cleroux, S., Wiltshire, T., Rajala, N., Burton, G., Holt, W. V., Littlewood, D. T. J., Briscoe, A. G., Jung, J., Ashkan, K., Heales, S. J., Pilkington, G. J., Meunier, B., McGeehan, J. E., Hargreaves, I. P., & McGeehan, R. E. (2019). Integrated Approach Reveals Role of Mitochondrial Germ-Line Mutation F18L in Respiratory Chain, Oxidative Alterations, Drug Sensitivity, and Patient Prognosis in Glioblastoma. International Journal of Molecular Sciences, 20(13), 3364. https://doi.org/10.3390/ijms20133364