Comparison of Radiosensitization by HDAC Inhibitors CUDC-101 and SAHA in Pancreatic Cancer Cells
Abstract
1. Introduction
2. Results
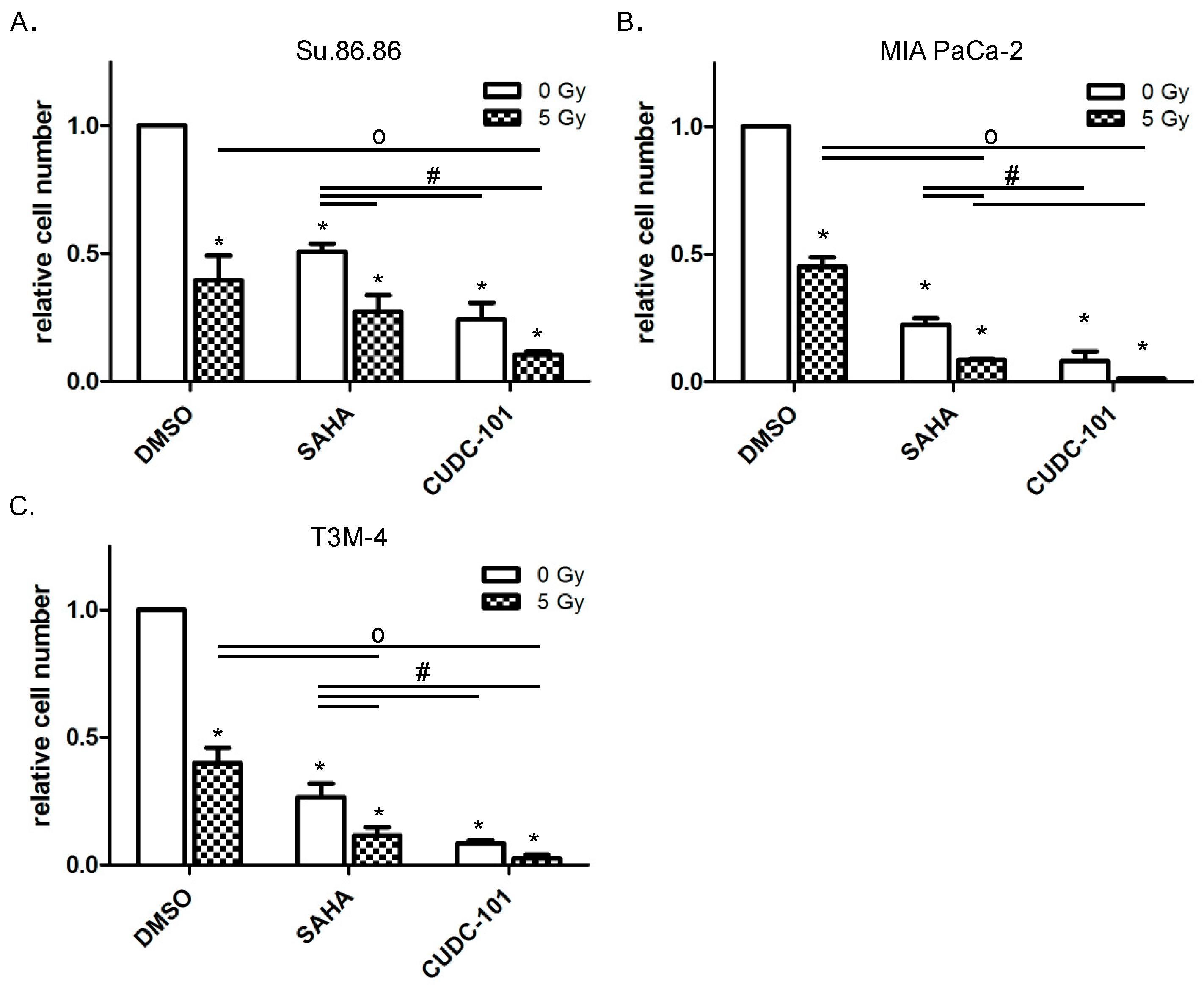
2.1. HDAC Inhibitors CUDC-101 and SAHA Increase the Radiation Sensitivity of Pancreatic Cancer Cells
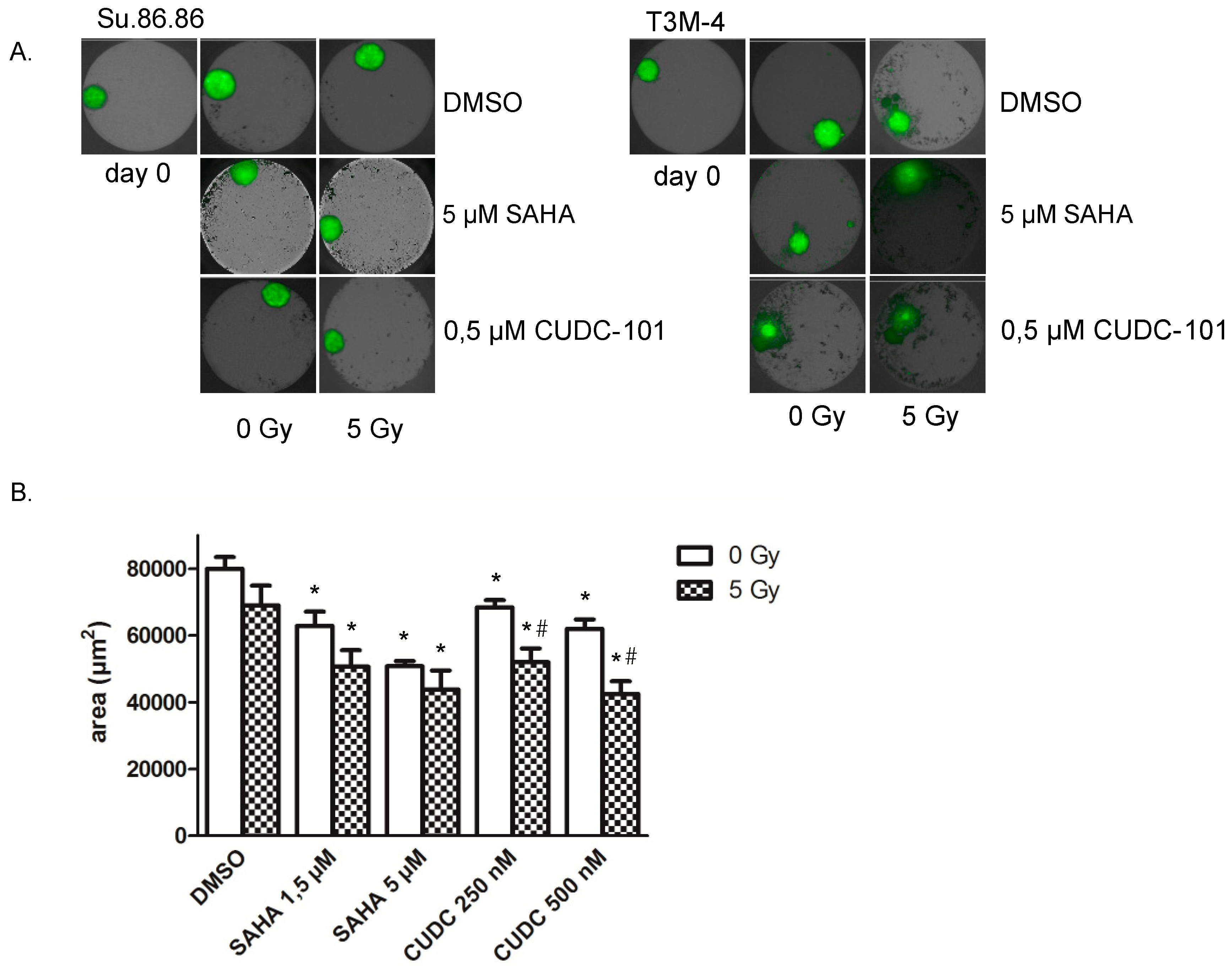
2.2. 3D Microtissue Growth Is Delayed More Efficiently by Combined HDAC Inhibitor and Radiation Treatment
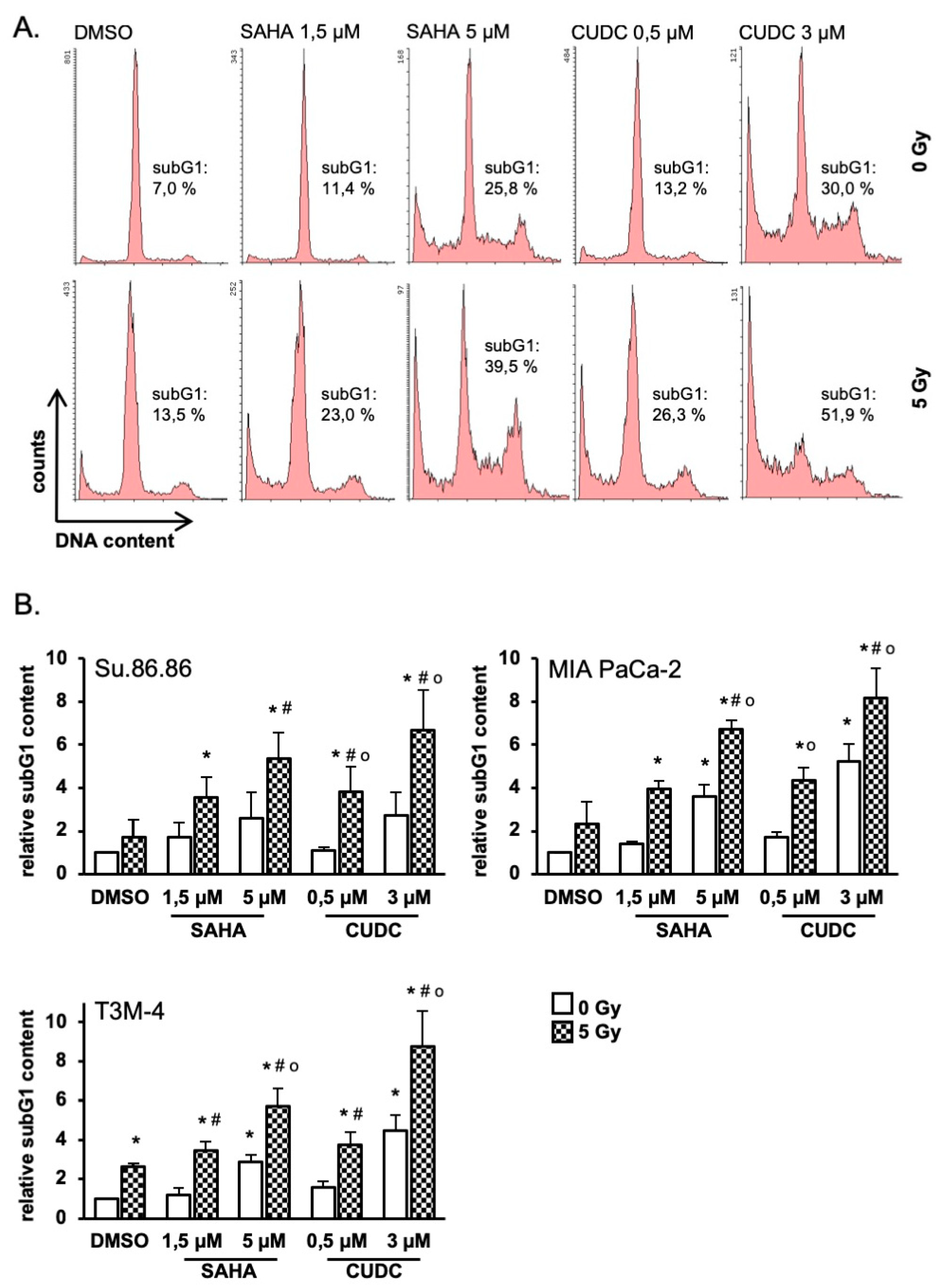
2.3. HDAC Inhibitors Promote Radiation-Induced Apoptosis
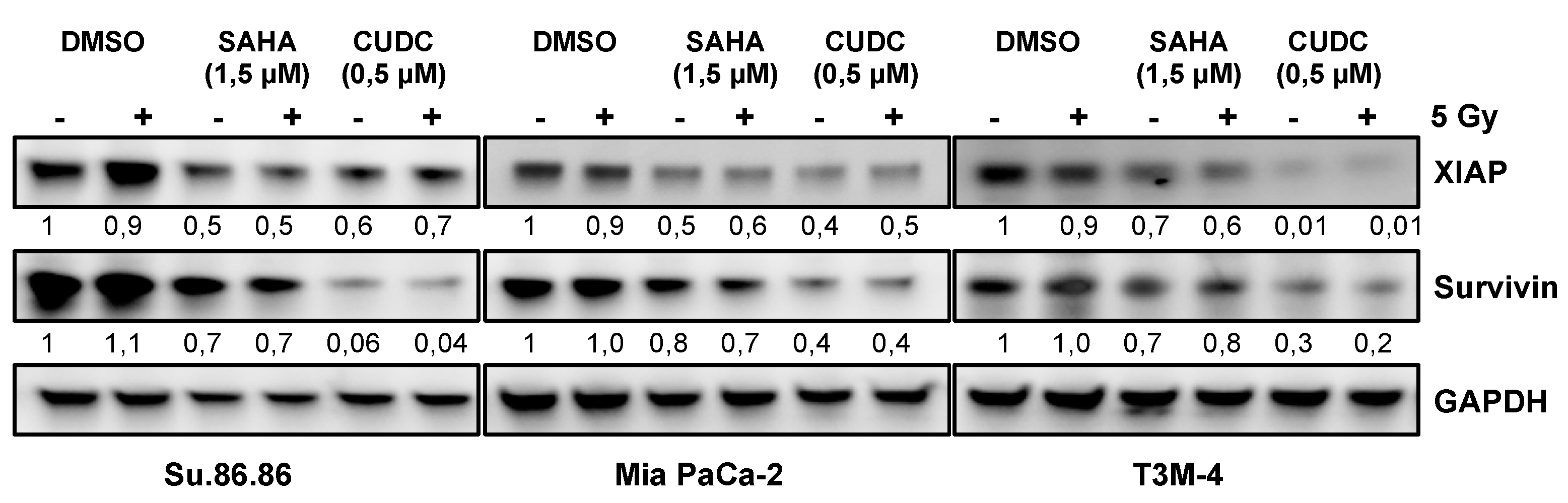
2.4. Prosurvival Factors XIAP and Survivin Are Reduced after Combined CUDC-101 and Radiation Treatment
3. Discussion
3.1. SAHA and CUDC-1 Both Increase Radiosensitivity of Pancreatic Cancer Cells
3.2. IR Increases the Apoptotic Response to SAHA and CUDC-101 Treatment
3.3. Reduced XIAP and Survivin May Account for Reduced Survival
4. Material and Methods
4.1. Cell Lines and Irradiation
4.2. HDAC Inhibitors
4.3. Immunoblot Analysis
4.4. Cell Proliferation
4.5. Cell Viability
4.6. Clonogenic Survival
4.7. 3D Microtissue Analyses
4.8. SubG1 Analysis
4.9. γ-H2AX Foci
4.10. Human Apoptosis Protein Array
4.11. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Ryan, D.P.; Hong, T.S.; Bardeesy, N. Pancreatic adenocarcinoma. N. Engl. J. Med. 2014, 371, 1039–1049. [Google Scholar] [CrossRef] [PubMed]
- Vincent, A.; Herman, J.; Schulick, R.; Hruban, R.H.; Goggins, M. Pancreatic cancer. Lancet 2011, 378, 607–620. [Google Scholar] [CrossRef]
- Andren-Sandberg, A. Pancreatic cancer: Chemotherapy and radiotherapy. N. Am. J. Med. Sci. 2011, 3, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Gong, J.; Tuli, R.; Shinde, A.; Hendifar, A.E. Meta-analyses of treatment standards for pancreatic cancer. Mol. Clin. Oncol. 2016, 4, 315–325. [Google Scholar] [CrossRef] [PubMed]
- Bolden, J.E.; Peart, M.J.; Johnstone, R.W. Anticancer activities of histone deacetylase inhibitors. Nat. Rev. Drug Discov. 2006, 5, 769–784. [Google Scholar] [CrossRef] [PubMed]
- Mottamal, M.; Zheng, S.; Huang, T.L.; Wang, G. Histone deacetylase inhibitors in clinical studies as templates for new anticancer agents. Molecules 2015, 20, 3898–3941. [Google Scholar] [CrossRef]
- Delcuve, G.P.; Khan, D.H.; Davie, J.R. Roles of histone deacetylases in epigenetic regulation: Emerging paradigms from studies with inhibitors. Clin. Epigenetics 2012, 4, 5. [Google Scholar] [CrossRef]
- Seto, E.; Yoshida, M. Erasers of histone acetylation: The histone deacetylase enzymes. Cold Spring Harb Perspect. Biol. 2014, 6, a018713. [Google Scholar] [CrossRef]
- Suraweera, A.; O’Byrne, K.J.; Richard, D.J. Combination Therapy With Histone Deacetylase Inhibitors (HDACi) for the Treatment of Cancer: Achieving the Full Therapeutic Potential of HDACi. Front. Oncol. 2018, 8, 92. [Google Scholar] [CrossRef]
- Hessmann, E.; Johnsen, S.A.; Siveke, J.T.; Ellenrieder, V. Epigenetic treatment of pancreatic cancer: Is there a therapeutic perspective on the horizon? Gut 2017, 66, 168–179. [Google Scholar] [CrossRef]
- Namdar, M.; Perez, G.; Ngo, L.; Marks, P.A. Selective inhibition of histone deacetylase 6 (HDAC6) induces DNA damage and sensitizes transformed cells to anticancer agents. Proc. Natl. Acad. Sci. USA 2010, 107, 20003–20008. [Google Scholar] [CrossRef] [PubMed]
- Stiborova, M.; Eckschlager, T.; Poljakova, J.; Hrabeta, J.; Adam, V.; Kizek, R.; Frei, E. The synergistic effects of DNA-targeted chemotherapeutics and histone deacetylase inhibitors as therapeutic strategies for cancer treatment. Curr. Med. Chem. 2012, 19, 4218–4238. [Google Scholar] [CrossRef] [PubMed]
- Feng, W.; Cai, D.; Zhang, B.; Lou, G.; Zou, X. Combination of HDAC inhibitor TSA and silibinin induces cell cycle arrest and apoptosis by targeting survivin and cyclinB1/Cdk1 in pancreatic cancer cells. Biomed. Pharm. 2015, 74, 257–264. [Google Scholar] [CrossRef] [PubMed]
- Iwahashi, S.; Ishibashi, H.; Utsunomiya, T.; Morine, Y.; Ochir, T.L.; Hanaoka, J.; Mori, H.; Ikemoto, T.; Imura, S.; Shimada, M. Effect of histone deacetylase inhibitor in combination with 5-fluorouracil on pancreas cancer and cholangiocarcinoma cell lines. J. Med. Invest. 2011, 58, 106–109. [Google Scholar] [CrossRef] [PubMed]
- Iwahashi, S.; Shimada, M.; Utsunomiya, T.; Morine, Y.; Imura, S.; Ikemoto, T.; Mori, H.; Hanaoka, J.; Sugimoto, K.; Saito, Y. Histone deacetylase inhibitor augments anti-tumor effect of gemcitabine and pegylated interferon-α on pancreatic cancer cells. Int. J. Clin. Oncol. 2011, 16, 671–678. [Google Scholar] [CrossRef] [PubMed]
- Groselj, B.; Sharma, N.L.; Hamdy, F.C.; Kerr, M.; Kiltie, A.E. Histone deacetylase inhibitors as radiosensitisers: Effects on DNA damage signalling and repair. Br. J. Cancer 2013, 108, 748–754. [Google Scholar] [CrossRef] [PubMed]
- Butler, L.M.; Agus, D.B.; Scher, H.I.; Higgins, B.; Rose, A.; Cordon-Cardo, C.; Thaler, H.T.; Rifkind, R.A.; Marks, P.A.; Richon, V.M. Suberoylanilide hydroxamic acid, an inhibitor of histone deacetylase, suppresses the growth of prostate cancer cells in vitro and in vivo. Cancer Res. 2000, 60, 5165–5170. [Google Scholar]
- Lauffer, B.E.; Mintzer, R.; Fong, R.; Mukund, S.; Tam, C.; Zilberleyb, I.; Flicke, B.; Ritscher, A.; Fedorowicz, G.; Vallero, R.; et al. Histone deacetylase (HDAC) inhibitor kinetic rate constants correlate with cellular histone acetylation but not transcription and cell viability. J. Biol. Chem. 2013, 288, 26926–26943. [Google Scholar] [CrossRef]
- Cai, X.; Zhai, H.X.; Wang, J.; Forrester, J.; Qu, H.; Yin, L.; Lai, C.J.; Bao, R.; Qian, C. Discovery of 7-(4-(3-ethynylphenylamino)-7-methoxyquinazolin-6-yloxy)-N-hydroxyheptanamide (CUDc-101) as a potent multi-acting HDAC, EGFR, and HER2 inhibitor for the treatment of cancer. J. Med. Chem. 2010, 53, 2000–2009. [Google Scholar] [CrossRef]
- Lai, C.J.; Bao, R.; Tao, X.; Wang, J.; Atoyan, R.; Qu, H.; Wang, D.G.; Yin, L.; Samson, M.; Forrester, J.; et al. CUDC-101, a multitargeted inhibitor of histone deacetylase, epidermal growth factor receptor, and human epidermal growth factor receptor 2, exerts potent anticancer activity. Cancer Res. 2010, 70, 3647–3656. [Google Scholar] [CrossRef]
- Slingerland, M.; Guchelaar, H.J.; Gelderblom, H. Histone deacetylase inhibitors: An overview of the clinical studies in solid tumors. Anticancer Drugs 2014, 25, 140–149. [Google Scholar] [CrossRef] [PubMed]
- Bots, M.; Johnstone, R.W. Rational combinations using HDAC inhibitors. Clin. Cancer Res. 2009, 15, 3970–3977. [Google Scholar] [CrossRef] [PubMed]
- Shabason, J.E.; Tofilon, P.J.; Camphausen, K. Grand rounds at the National Institutes of Health: HDAC inhibitors as radiation modifiers, from bench to clinic. J. Cell. Mol. Med. 2011, 15, 2735–2744. [Google Scholar] [CrossRef] [PubMed]
- Entin-Meer, M.; Yang, X.; VandenBerg, S.R.; Lamborn, K.R.; Nudelman, A.; Rephaeli, A.; Haas-Kogan, D.A. In vivo efficacy of a novel histone deacetylase inhibitor in combination with radiation for the treatment of gliomas. Neuro. Oncol. 2007, 9, 82–88. [Google Scholar] [CrossRef] [PubMed]
- Gerelchuluun, A.; Maeda, J.; Manabe, E.; Brents, C.A.; Sakae, T.; Fujimori, A.; Chen, D.J.; Tsuboi, K.; Kato, T.A. Histone Deacetylase Inhibitor Induced Radiation Sensitization Effects on Human Cancer Cells after Photon and Hadron Radiation Exposure. Int. J. Mol. Sci. 2018, 19, 496. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Jing, S.; Li, Y.; Gao, Y.; Yu, S.; Li, Z.; Zhao, Y.; Piao, J.; Ma, S.; Chen, X. The effects of SAHA on radiosensitivity in pancreatic cancer cells by inducing apoptosis and targeting RAD51. Biomed. Pharm. 2017, 89, 705–710. [Google Scholar] [CrossRef] [PubMed]
- Emanuele, S.; Lauricella, M.; Tesoriere, G. Histone deacetylase inhibitors: Apoptotic effects and clinical implications (Review). Int. J. Oncol. 2008, 33, 637–646. [Google Scholar] [CrossRef]
- Ruckert, F.; Samm, N.; Lehner, A.K.; Saeger, H.D.; Grutzmann, R.; Pilarsky, C. Simultaneous gene silencing of Bcl-2, XIAP and Survivin re-sensitizes pancreatic cancer cells towards apoptosis. BMC Cancer 2010, 10. [Google Scholar] [CrossRef]
- Rathore, R.; McCallum, J.E.; Varghese, E.; Florea, A.M.; Busselberg, D. Overcoming chemotherapy drug resistance by targeting inhibitors of apoptosis proteins (IAPs). Apoptosis 2017, 22, 898–919. [Google Scholar] [CrossRef]
- Ohnishi, K.; Scuric, Z.; Schiestl, R.H.; Okamoto, N.; Takahashi, A.; Ohnishi, T. siRNA targeting NBS1 or XIAP increases radiation sensitivity of human cancer cells independent of TP53 status. Radiat Res. 2006, 166, 454–462. [Google Scholar] [CrossRef]
- Wang, D.G.; Sun, Y.B.; Ye, F.; Li, W.; Kharbuja, P.; Gao, L.; Zhang, D.Y.; Suo, J. Anti-tumor activity of the X-linked inhibitor of apoptosis (XIAP) inhibitor embelin in gastric cancer cells. Mol. Cell. Biochem. 2014, 386, 143–152. [Google Scholar] [CrossRef] [PubMed]
- Yi, X.P.; Han, T.; Li, Y.X.; Long, X.Y.; Li, W.Z. Simultaneous silencing of XIAP and survivin causes partial mesenchymal-epithelial transition of human pancreatic cancer cells via the PTEN/PI3K/Akt pathway. Mol. Med. Rep. 2015, 12, 601–608. [Google Scholar] [CrossRef] [PubMed]
- Heider, T.; Mutschelknaus, L.; Radulovic, V.; Winkler, K.; Kimmel, J.; Anastasov, N.; Atkinson, M.J.; Moertl, S. Radiation induced transcriptional and post-transcriptional regulation of the hsa-miR-23a~27a~24-2 cluster suppresses apoptosis by stabilizing XIAP. Biochim. Biophys. Acta Gene Regul. Mech. 2017, 1860, 1127–1137. [Google Scholar] [CrossRef] [PubMed]
- Yoon, D.H.; Shin, J.S.; Jin, D.H.; Hong, S.W.; Jung, K.A.; Kim, S.M.; Hong, Y.S.; Kim, K.P.; Lee, J.L.; Suh, C.; et al. The survivin suppressant YM155 potentiates chemosensitivity to gemcitabine in the human pancreatic cancer cell line MiaPaCa-2. Anticancer Res. 2012, 32, 1681–1688. [Google Scholar] [PubMed]
- Moussata, D.; Amara, S.; Siddeek, B.; Decaussin, M.; Hehlgans, S.; Paul-Bellon, R.; Mornex, F.; Gerard, J.P.; Romestaing, P.; Rodel, F.; et al. XIAP as a radioresistance factor and prognostic marker for radiotherapy in human rectal adenocarcinoma. Am. J. Pathol. 2012, 181, 1271–1278. [Google Scholar] [CrossRef] [PubMed]
- Rodel, F.; Sprenger, T.; Kaina, B.; Liersch, T.; Rodel, C.; Fulda, S.; Hehlgans, S. Survivin as a prognostic/predictive marker and molecular target in cancer therapy. Curr. Med. Chem. 2012, 19, 3679–3688. [Google Scholar] [CrossRef] [PubMed]
- Schneider, G.; Henrich, A.; Greiner, G.; Wolf, V.; Lovas, A.; Wieczorek, M.; Wagner, T.; Reichardt, S.; von Werder, A.; Schmid, R.M.; et al. Cross talk between stimulated NF-kappaB and the tumor suppressor p53. Oncogene 2010, 29, 2795–2806. [Google Scholar] [CrossRef] [PubMed]
- Stojanovic, N.; Hassan, Z.; Wirth, M.; Wenzel, P.; Beyer, M.; Schafer, C.; Brand, P.; Kroemer, A.; Stauber, R.H.; Schmid, R.M.; et al. HDAC1 and HDAC2 integrate the expression of p53 mutants in pancreatic cancer. Oncogene 2017, 36, 1804–1815. [Google Scholar] [CrossRef] [PubMed]
- Schafer, C.; Goder, A.; Beyer, M.; Kiweler, N.; Mahendrarajah, N.; Rauch, A.; Nikolova, T.; Stojanovic, N.; Wieczorek, M.; Reich, T.R.; et al. Class I histone deacetylases regulate p53/NF-κB crosstalk in cancer cells. Cell Signal 2017, 29, 218–225. [Google Scholar] [CrossRef] [PubMed]
- Park, S.J.; Kim, S.M.; Moon, J.H.; Kim, J.H.; Shin, J.S.; Hong, S.W.; Shin, Y.J.; Lee, D.H.; Lee, E.Y.; Hwang, I.Y.; et al. SAHA, an HDAC inhibitor, overcomes erlotinib resistance in human pancreatic cancer cells by modulating E-cadherin. Tumour Biol. 2016, 37, 4323–4330. [Google Scholar] [CrossRef]
- Drucker, B.J.; Marincola, F.M.; Siao, D.Y.; Donlon, T.A.; Bangs, C.D.; Holder, W.D., Jr. A new human pancreatic carcinoma cell line developed for adoptive immunotherapy studies with lymphokine-activated killer cells in nude mice. In Vitro Cell. Dev. Biol. 1988, 24, 1179–1187. [Google Scholar] [CrossRef] [PubMed]
- Okabe, T.; Yamaguchi, N.; Ohsawa, N. Establishment and characterization of a carcinoembryonic antigen (CEA)-producing cell line from a human carcinoma of the exocrine pancreas. Cancer 1983, 51, 662–668. [Google Scholar] [CrossRef]
- Yunis, A.A.; Arimura, G.K.; Russin, D.J. Human pancreatic carcinoma (MIA PaCa-2) in continuous culture: Sensitivity to asparaginase. Int. J. Cancer 1977, 19, 128–135. [Google Scholar] [CrossRef] [PubMed]
- Anastasov, N.; Hofig, I.; Mall, S.; Krackhardt, A.M.; Thirion, C. Optimized Lentiviral Transduction Protocols by Use of a Poloxamer Enhancer, Spinoculation, and scFv-Antibody Fusions to VSV-G. Methods Mol. Biol. 2016, 1448, 49–61. [Google Scholar] [PubMed]
- Anastasov, N.; Hofig, I.; Radulovic, V.; Strobel, S.; Salomon, M.; Lichtenberg, J.; Rothenaigner, I.; Hadian, K.; Kelm, J.M.; Thirion, C.; et al. A 3D-microtissue-based phenotypic screening of radiation resistant tumor cells with synchronized chemotherapeutic treatment. BMC Cancer 2015, 15, 466. [Google Scholar] [CrossRef]
- Kraemer, A.; Anastasov, N.; Angermeier, M.; Winkler, K.; Atkinson, M.J.; Moertl, S. MicroRNA-mediated processes are essential for the cellular radiation response. Radiat. Res. 2011, 176, 575–586. [Google Scholar] [CrossRef] [PubMed]


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moertl, S.; Payer, S.; Kell, R.; Winkler, K.; Anastasov, N.; Atkinson, M.J. Comparison of Radiosensitization by HDAC Inhibitors CUDC-101 and SAHA in Pancreatic Cancer Cells. Int. J. Mol. Sci. 2019, 20, 3259. https://doi.org/10.3390/ijms20133259
Moertl S, Payer S, Kell R, Winkler K, Anastasov N, Atkinson MJ. Comparison of Radiosensitization by HDAC Inhibitors CUDC-101 and SAHA in Pancreatic Cancer Cells. International Journal of Molecular Sciences. 2019; 20(13):3259. https://doi.org/10.3390/ijms20133259
Chicago/Turabian StyleMoertl, Simone, Sarah Payer, Rosemarie Kell, Klaudia Winkler, Natasa Anastasov, and Michael J. Atkinson. 2019. "Comparison of Radiosensitization by HDAC Inhibitors CUDC-101 and SAHA in Pancreatic Cancer Cells" International Journal of Molecular Sciences 20, no. 13: 3259. https://doi.org/10.3390/ijms20133259
APA StyleMoertl, S., Payer, S., Kell, R., Winkler, K., Anastasov, N., & Atkinson, M. J. (2019). Comparison of Radiosensitization by HDAC Inhibitors CUDC-101 and SAHA in Pancreatic Cancer Cells. International Journal of Molecular Sciences, 20(13), 3259. https://doi.org/10.3390/ijms20133259




