Inhibition of C. albicans Dimorphic Switch by Cobalt(II) Complexes with Ligands Derived from Pyrazoles and Dinitrobenzoate: Synthesis, Characterization and Biological Activity
Abstract
1. Introduction
2. Results and Discussion
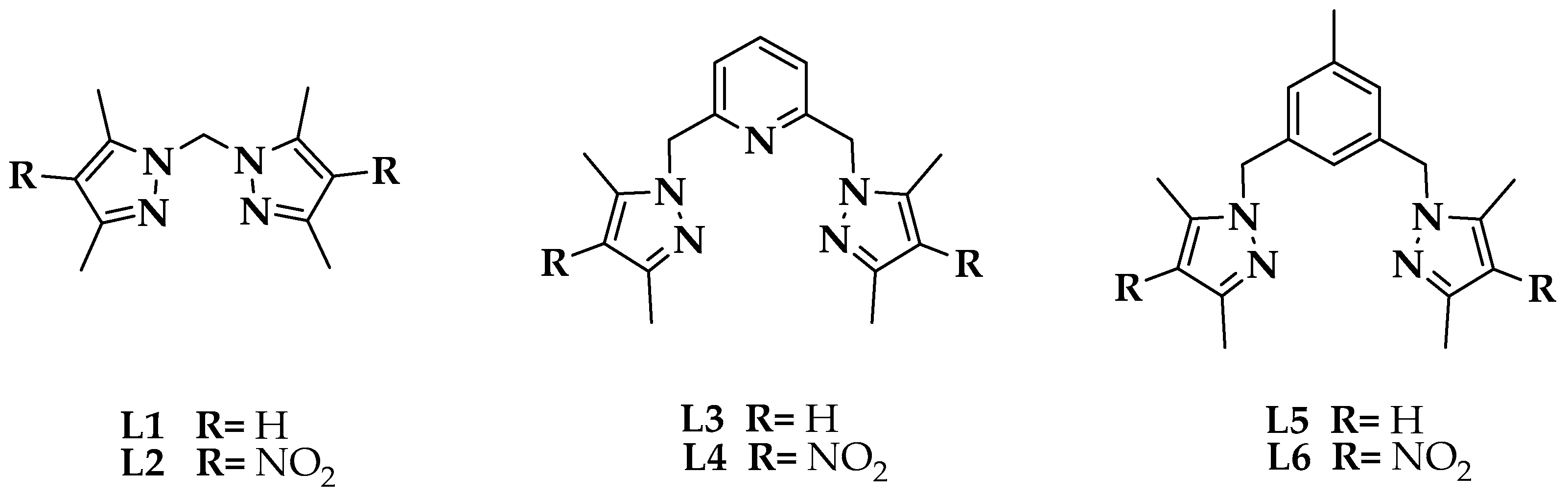
2.1. Synthesis and Characterization of 2,6 bis(4-nitro-3,5-dimethylpyrazol-1-ylmethyl)pyridine L6
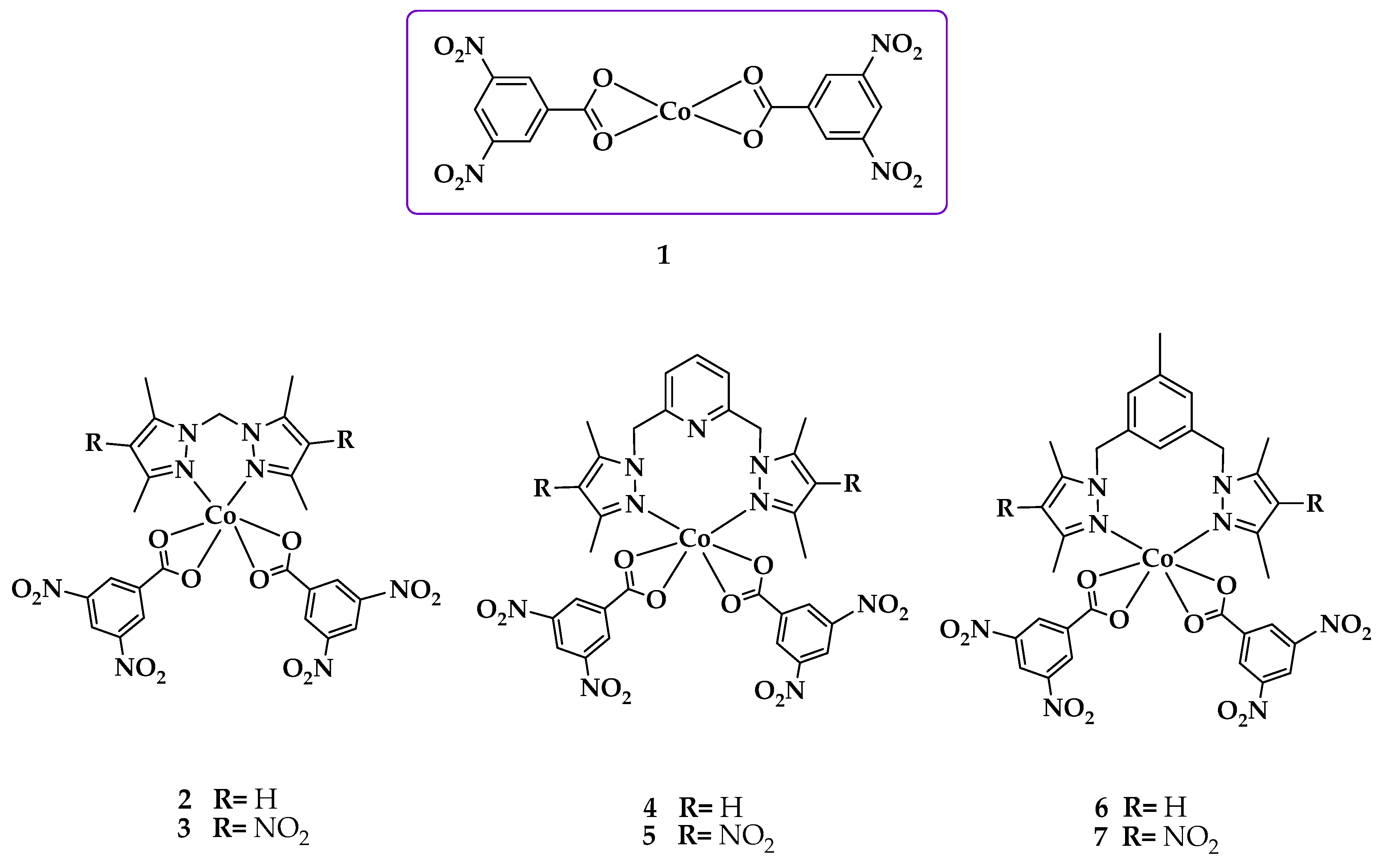
2.2. Synthesis and Characterization of the Cobalt(II) Complexes
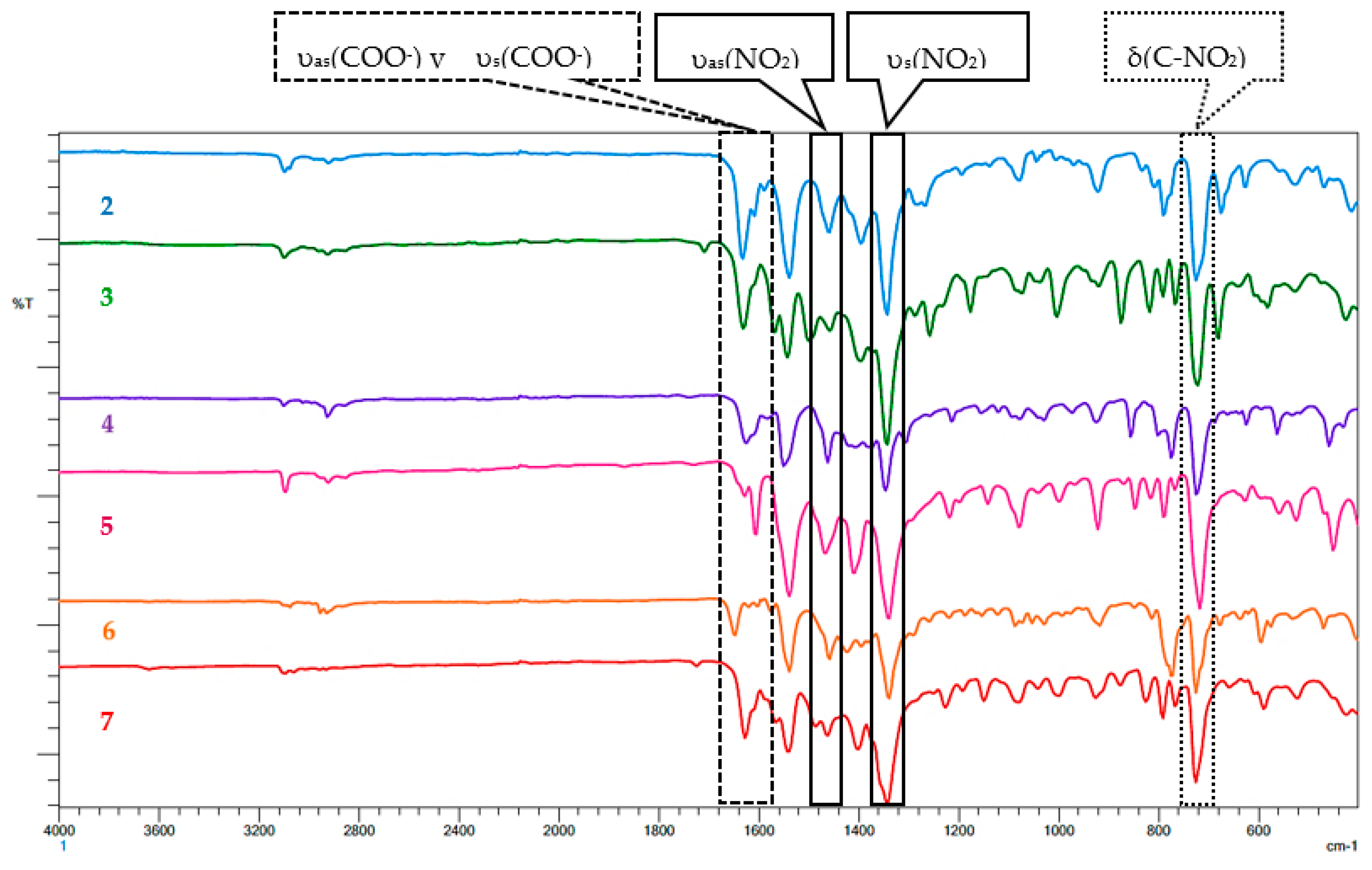
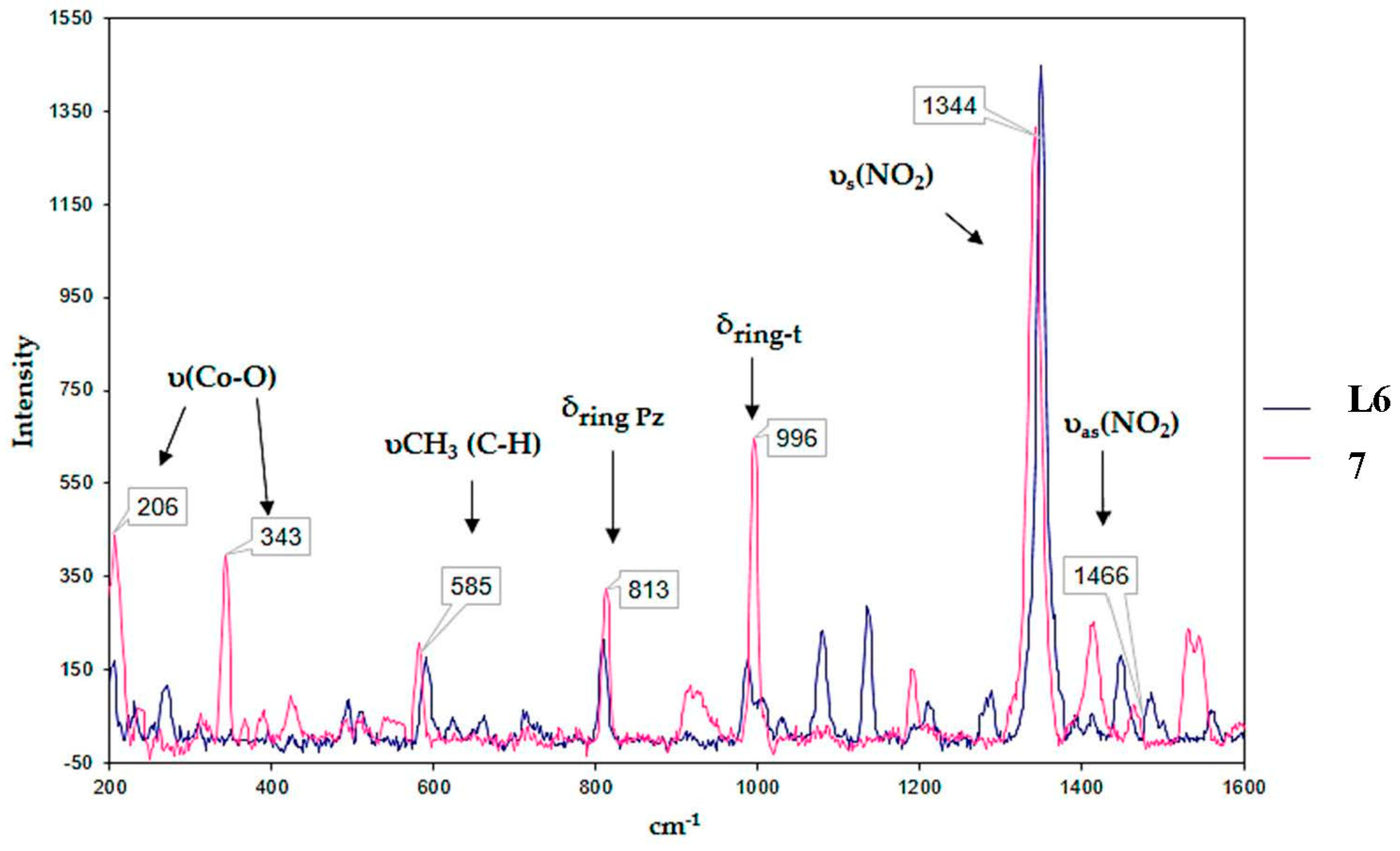
2.2.1. FT-IR and Raman Spectroscopy
2.2.2. UV/Vis Spectroscopy
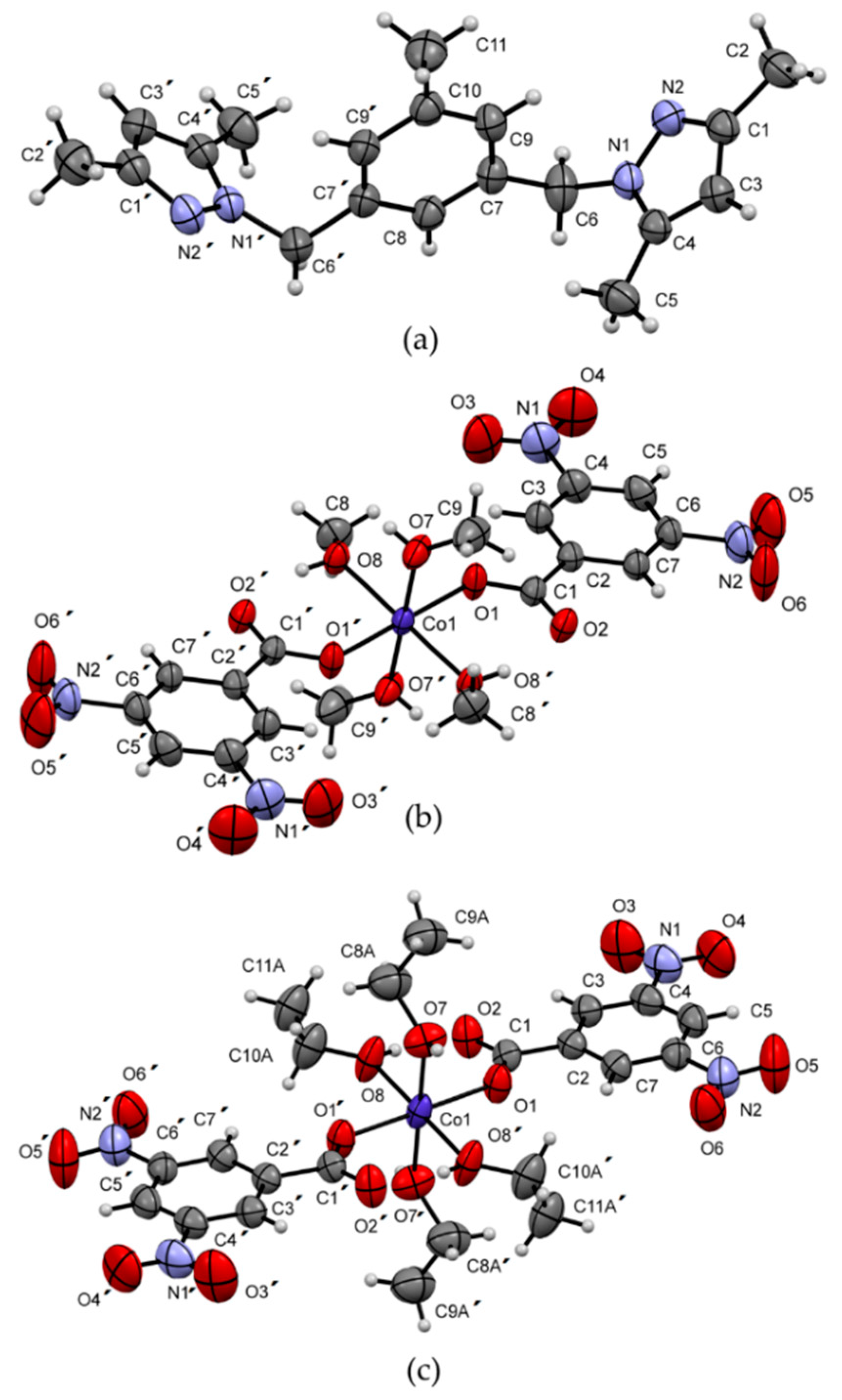
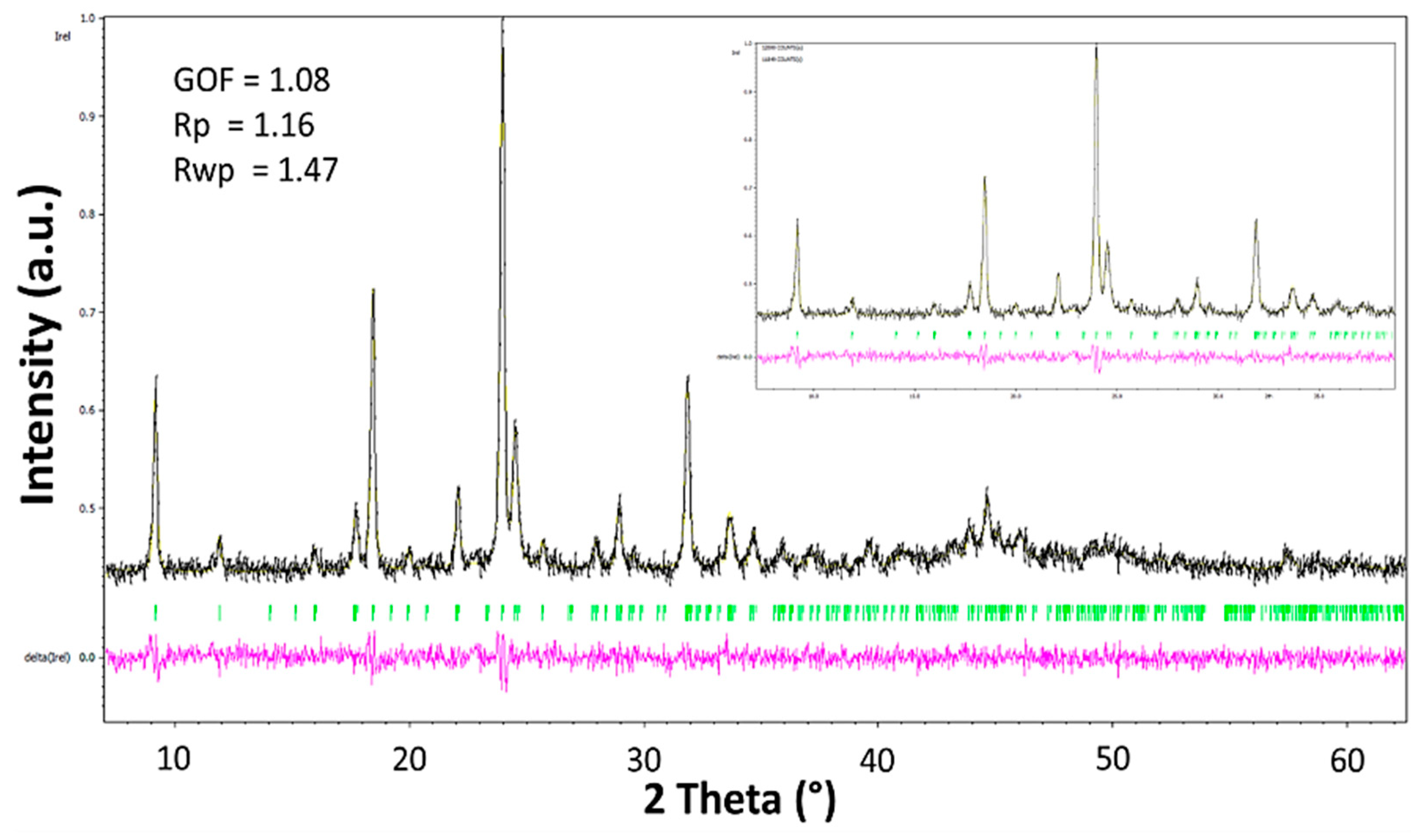
2.2.3. X-Ray Structural Determination of L5 and 1
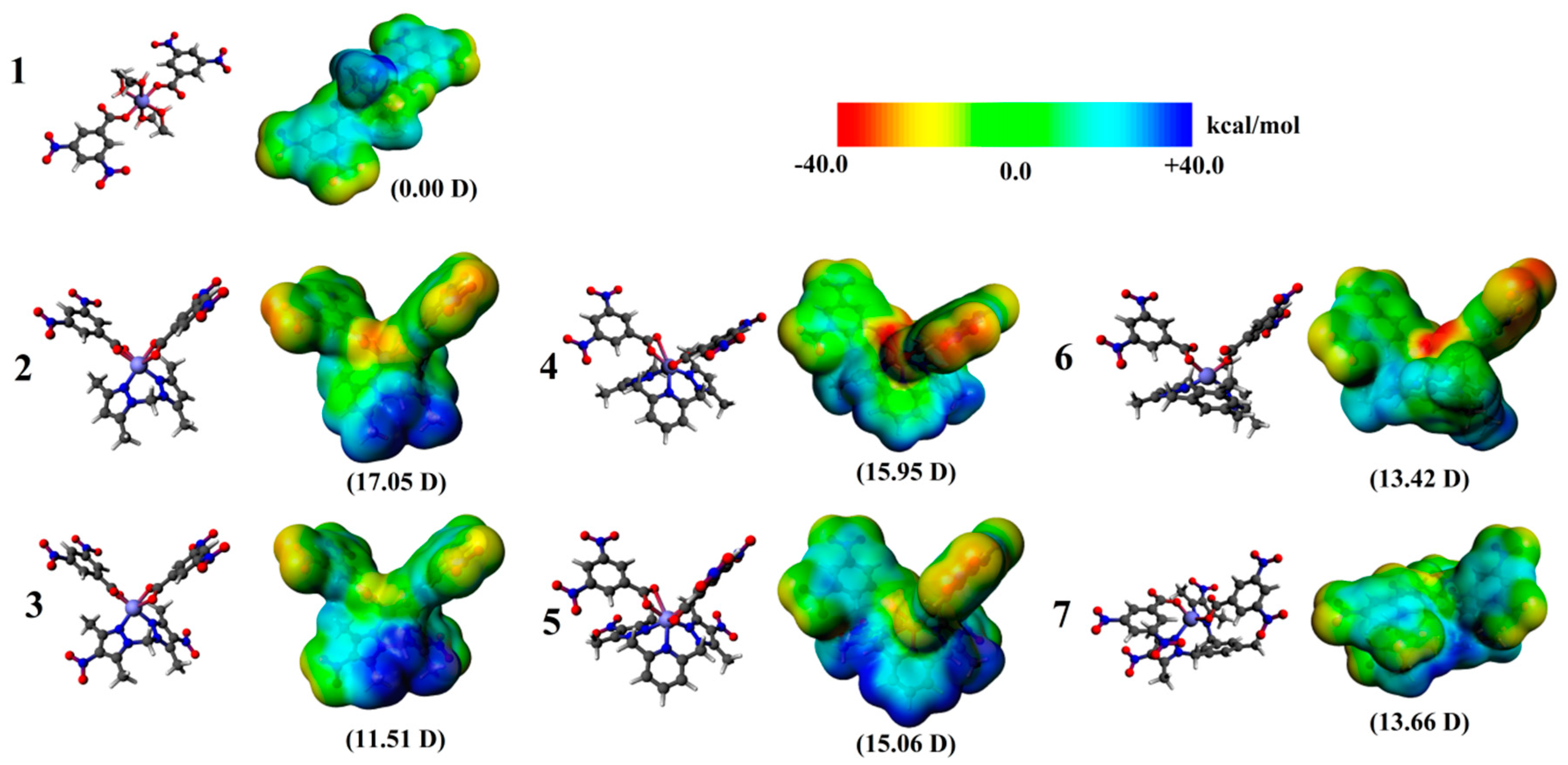
2.3. Quantum-Chemical Calculations
2.4. Biological Activity
3. Materials and Methods
3.1. General Information
3.2. Synthesis of 3,5-bis(3,5-dimethyl-4-nitropyrazol-1-ylmethyl)toluene L6
3.3. Synthesis of the Cobalt(II) Complexes (1–7)
3.3.1. Synthesis of bis(dinitrobenzoate-O,O′) of Co(II) ([Co(dnb)2]) (1)
3.3.2. Synthesis of Dinitrobenzoate [bis(3,5-dimethylpyrazol-1-yl)methane] of Co(II) ([Co(L1)(dnb)2]) (2)
3.3.3. Synthesis of Dinitrobenzoate[bis(3,5-dimethyl-4-nitro-pyrazol-1-yl)methane] of Co(II) ([Co(L2)(dnb)2]) (3)
3.3.4. Synthesis of Dinitrobenzoate[2,6-bis(3,5-dimethylpyrazol-1-ylmethyl)pyridine] of Co(II) ([Co(L3)(dnb)2]) (4)
3.3.5. Synthesis of Dinitrobenzoate[2,6-bis(3,5-dimethyl-4-nitro-pyrazol-1-ylmethyl)pyridine] of Co(II) ([Co(L4)(dnb)2]) (5)
3.3.6. Synthesis of Dinitrobenzoate[3,5-bis(3,5-dimethylpyrazol-1-ylmethyl)toluene] of Co(II) ([Co(L5)(dnb)2]) (6)
3.3.7. Synthesis of Dinitrobenzoate[3,5-bis(3,5-dimethyl-4-nitro-pyrazol-1-ylmethyl)toluene] of Co(II) ([Co(L6)(dnb)2]) (7)
3.4. Biological Studies
3.4.1. In Vitro Anti T. cruzi Activity
3.4.2. In Vitro Antifungal Susceptibility Testing
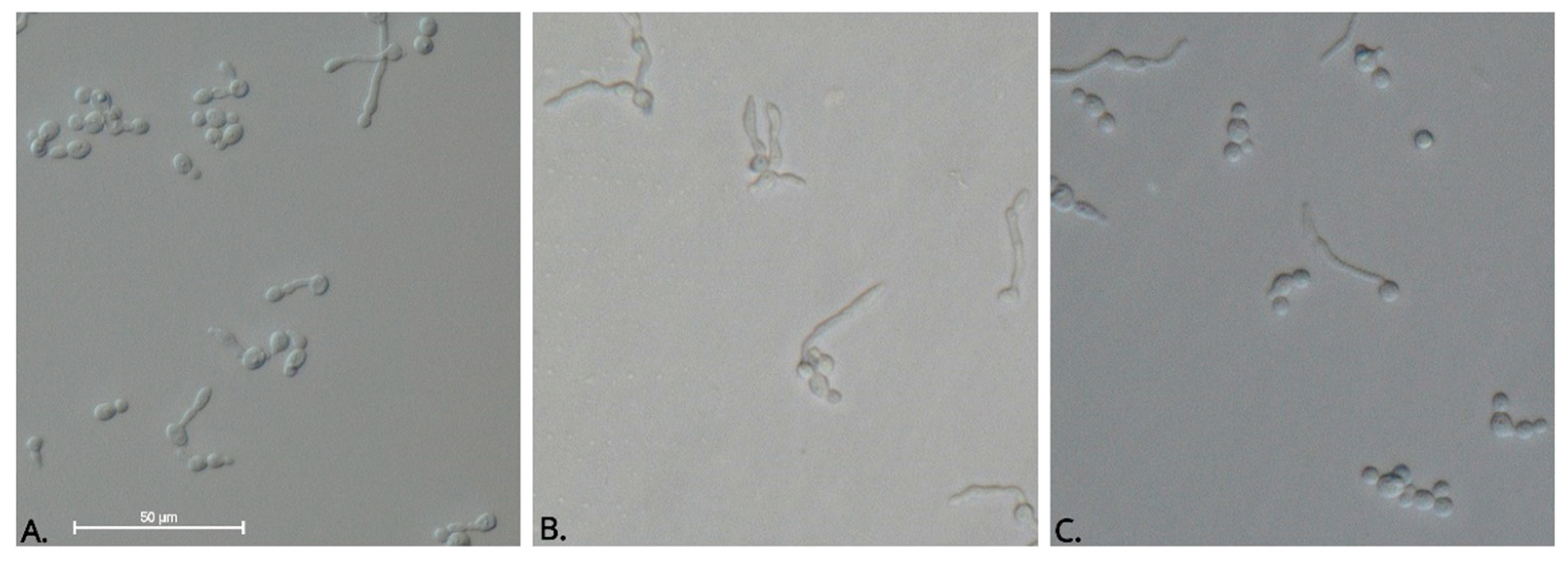
3.4.3. Germ Tube Inhibition Assay
3.4.4. Cytotoxicity in Mammalian Cells
3.4.5. Statistical Analysis
In Vitro anti-T. cruzi Activity
Cytotoxicity in Mammalian Cells
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Arendrup, M.C.; Patterson, T.F. Multidrug-Resistant Candida: Epidemiology, Molecular Mechanisms, and Treatment. J. Infect. Dis. 2017, 216, 445–451. [Google Scholar] [CrossRef] [PubMed]
- Kljun, J.; Scott, A.J.; Lanišnik Rižner, T.; Keiser, J.; Turel, I. Synthesis and biological evaluation of organoruthenium complexes with azole antifungal agents. First crystal structure of a tioconazole metal complex. Organometallics 2014, 33, 1594–1601. [Google Scholar] [CrossRef]
- Dadar, M.; Tiwari, R.; Karthik, K.; Chakraborty, S.; Shahali, Y.; Dhama, K. Candida albicans—Biology, molecular characterization, pathogenicity, and advances in diagnosis and control—An update. Microb. Pathog. 2018, 117, 128–138. [Google Scholar] [CrossRef] [PubMed]
- Bonney, K.M. Chagas disease in the 21st Century: A public health success or an emerging threat? Parasite 2014, 21, 11. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Delgado, R.A.; Navarro, M.; Lazardi, K.; Atencio, R.; Capparelli, M.; Vargas, F.; Urbina, J.A.; Bouillez, A.; Noels, A.F.; Masi, D. Toward a Novel Metal Based Chemotherapy against Tropical Diseases 4. Synthesis and Characterization of New Metal-Clotrimazole Complexes and Evaluation of Their Activity against Trypanosoma Cruzi. Inorganica Chim. Acta 1998, 275, 528–540. [Google Scholar] [CrossRef]
- Bello-Vieda, N.J.; Pastrana, H.F.; Garavito, M.F.; Ávila, A.G.; Celis, A.M.; Muñoz-Castro, A.; Restrepo, S.; Hurtado, J.J. Antibacterial activities of azole complexes combined with silver nanoparticles. Molecules 2018, 23, 361. [Google Scholar] [CrossRef]
- Urbina, J.A. Ergosterol biosynthesis and drug development for Chagas disease. Mem. Inst. Oswaldo Cruz 2009, 104, 311–318. [Google Scholar] [CrossRef] [PubMed]
- Murcia, R.; Leal, S.; Roa, M.; Nagles, E.; Muñoz-Castro, A.; Hurtado, J. Development of Antibacterial and Antifungal Triazole Chromium(III) and Cobalt(II) Complexes: Synthesis and Biological Activity Evaluations. Molecules 2018, 23, 2013. [Google Scholar] [CrossRef]
- Batista, D.D.G.J.; Da Silva, P.B.; Stivanin, L.; Lachter, D.R.; Silva, R.S.; Felcman, J.; Louro, S.R.W.; Teixeira, L.R.; Soeiro, M.D.N.C. Co(II), Mn(II) and Cu(II) complexes of fluoroquinolones: Synthesis, spectroscopical studies and biological evaluation against Trypanosoma cruzi. Polyhedron 2011, 30, 1718–1725. [Google Scholar] [CrossRef]
- Amalanathan, M.; Rastogi, V.K.; Hubert Joe, I.; Palafox, M.A.; Tomar, R. Density functional theory calculations and vibrational spectral analysis of 3,5-(dinitrobenzoic acid). Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2011, 78, 1437–1444. [Google Scholar] [CrossRef]
- Zhang, C.X.; Zhang, Y.Y.; Sun, Y.Q. Synthesis, structures and magnetic properties of copper (II) and cobalt (II) complexes containing pyridyl-substituted nitronyl nitroxide and 3, 5-dinitrobenzoate. Polyhedron 2010, 29, 1387–1392. [Google Scholar] [CrossRef]
- Fonseca, D.; Páez, C.; Ibarra, L.; García-Huertas, P.; Macías, M.A.; Triana-Chávez, O.; Hurtado, J.J. Metal complex derivatives of bis (pyrazol-1-yl) methane ligands: Synthesis, characterization and anti-Trypanosoma cruzi activity. Transit. Met. Chem. 2018, 44, 135–144. [Google Scholar] [CrossRef]
- Hurtado, J.; Portaluppi, M.; Quijada, R.; Rojas, R.; Valderrama, M. Synthesis, characterization, and reactivity studies in ethylene polymerization of cyclometalated palladium(II) complexes containing terdentate ligands with N,C,N-donors. J. Coord. Chem. 2009, 62, 2772–2781. [Google Scholar] [CrossRef]
- Sundaraganesan, N.; Kavitha, E.; Sebastian, S.; Cornard, J.P.; Martel, M. Experimental FTIR, FT-IR (gas phase), FT-Raman and NMR spectra, hyperpolarizability studies and DFT calculations of 3,5-dimethylpyrazole. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2009, 74, 788–797. [Google Scholar] [CrossRef] [PubMed]
- Tankov, I.; Mitkova, M.; Nikolova, R.; Veli, A. N-Butyl Acetate Synthesis in the Presence of Pyridinium-Based Acidic Ionic Liquids: Influence of the Anion Nature. Catal. Lett. 2017, 147, 2279–2289. [Google Scholar] [CrossRef]
- Bal, S.; Köytepe, S.; Connolly, J.D. Synthesis of carbazole-derived ligands and their metal complexes: Characterization, thermal, catalytic, and electrochemical features. Mon. Fur Chem. 2016, 147, 2061–2071. [Google Scholar] [CrossRef]
- Tamayo, L.V.; Santos, A.F.; Ferreira, I.P.; Santos, V.G.; Lopes, M.T.P.; Beraldo, H. Silver(I) complexes with chromone-derived hydrazones: Investigation on the antimicrobial and cytotoxic effects. BioMetals 2017, 30, 379–392. [Google Scholar] [CrossRef]
- Hernández, L.; Quevedo-Acosta, Y.; Vázquez, K.; Gómez-Treviño, A.; Zarate-Ramos, J.J.; Macías, M.A.; Hurtado, J.J. Study of the Effect of Metal Complexes on Morphology and Viability of Embryonated Toxocara canis Eggs. Vector Borne Zoonotic Dis. 2018, 18, 548–553. [Google Scholar] [CrossRef]
- Hurtado, J.; Ibarra, L.; Yepes, D.; García-huertas, P.; Macías, M.A. Synthesis, crystal structure, catalytic and anti-Trypanosoma cruzi activity of a new chromium (III) complex containing bis (3,5-dimethylpyrazol-1-yl) methane. J. Mol. Struct. 2017, 1146, 365–372. [Google Scholar] [CrossRef]
- Le Bail, A.; Duroy, H.F. Ab-initio structure determination. Mat. Res. Bul. 1988, 23, 447–452. [Google Scholar] [CrossRef]
- Grimme, S.; Chemie, T.O.; Münster, O.I.D.U. Semiempirical GGA-Type Density Functional Constructed with a Long-Range Dispersion Correction. Wiley Intersci. 2006, 16, 1787–1799. [Google Scholar] [CrossRef] [PubMed]
- Velde, G.T.E.; Bickelhaupt, F.M.; Baerends, E.J.; Guerra, C.F.; Gisbergen, S.J.A.V.A.N. Chemistry with ADF. J. Comput. Chem. 2001, 22, 931–967. [Google Scholar] [CrossRef]
- Lienx, E.J.; Guo, Z.; Li, R.; Su, C. Dipole Moment as a Parameter. J. Pharm. Sci. 1982, 71, 641–655. [Google Scholar]
- Cheeseright, T.; Mackey, M.; Rose, S.; Vinter, A. Molecular Field Extrema as Descriptors of Biological Activity: Definition and Validation. J. Chem. Inf. Model. 2006, 46, 665–676. [Google Scholar] [CrossRef] [PubMed]
- Bulat, F.A.; Toro-labbé, A.; Brinck, T.; Murray, J.S.; Politzer, P. Quantitative analysis of molecular surfaces: Areas, volumes, electrostatic potentials and average local ionization energies. J. Mol. Model. 2010, 16, 1679–1691. [Google Scholar] [CrossRef] [PubMed]
- Olender, D.; Zwawiak, J.; Zaprutko, L. Multidirectional Efficacy of Biologically Active Nitro Compounds Included in Medicines. Pharmaceuticals 2018, 11, 54. [Google Scholar] [CrossRef] [PubMed]
- Stone, V.; Johnston, H.; Schins, R.P.F. Development of in vitro systems for nanotoxicology: Methodological considerations. Crit. Rev. Toxicol. 2009, 39, 613–626. [Google Scholar] [CrossRef] [PubMed]
- Chohan, Z.H.; Hanif, M. Design, synthesis, and biological properties of triazole derived compounds and their transition metal complexes. J. Enzym. Inhib. Med. Chem. 2010, 25, 737–749. [Google Scholar] [CrossRef] [PubMed]
- Chohan, Z.H.; Hanif, M. Antibacterial and antifungal metal based triazole Schiff bases. J. Enzym. Inhib. Med. Chem. 2013, 28, 944–953. [Google Scholar] [CrossRef]
- Allen, D.; Wilson, D.; Drew, R.; Perfect, J. Azole antifungals: 35 years of invasive fungal infection management. Expert Rev. Anti Infect. Ther. 2015, 13, 787–798. [Google Scholar] [CrossRef]
- Heerding, D.A.; Chan, G.; DeWolf, W.E.; Fosberry, A.P.; Janson, C.A.; Jaworski, D.D.; McManus, E.; Miller, W.H.; Moore, T.D.; Payne, D.J.; et al. 1,4-Disubstituted imidazoles are potential antibacterial agents functioning as inhibitors of enoyl acyl carrier protein reductase (FabI). Bioorganic Med. Chem. Lett. 2001, 11, 2061–2065. [Google Scholar] [CrossRef]
- Sanguinetti, M.; Posteraro, B. Antifungal drug resistance among Candida species: Mechanisms and clinical impact. Mycoses 2015, 58, 2–13. [Google Scholar] [CrossRef] [PubMed]
- Taylor, P.; Mayer, F.L.; Wilson, D.; Hube, B.; Mayer, F.L.; Wilson, D.; Hube, B. Candida albicans pathogenicity mechanisms. Virulence 2013, 4, 119–128. [Google Scholar]
- Berman, J.; Sudbery, P.E. Candida albicans: A molecular revolution built on lessons from budding yeast. Nat. Rev. Genet. 2002, 3, 918–930. [Google Scholar] [CrossRef] [PubMed]
- Pukkila-Worley, R.; Peleg, A.Y.; Tampakakis, E.; Mylonakis, E. Candida albicans Hyphal Formation and Virulence Assessed Using a Caenorhabditis elegans Infection Model. Eukaryote Cell 2009, 8, 1750–1758. [Google Scholar] [CrossRef]
- Lo, H.; Ko, J.R.; Didomenico, B.; Loebenberg, D.; Cacciapuoti, A.; Fink, G.R. Nonfilamentous C. albicans Mutants Are Avirulent. Cell 2015, 90, 939–949. [Google Scholar] [CrossRef]
- Taylor, P. Regulation of phenotypic transitions in the fungal Regulation of phenotypic transitions in the fungal pathogen Candida albicans. Virulence 2012, 3, 37–41. [Google Scholar]
- Rocha, C.R.C.; Schro, K.; Harcus, D.; Marcil, A.; Dignard, D.; Taylor, B.N.; Thomas, D.Y.; Leberer, E. Signaling through Adenylyl Cyclase Is Essential for Hyphal Growth and Virulence in the Pathogenic Fungus Candida albicans. Mol. Biol. Cell 2001, 12, 3631–3643. [Google Scholar] [CrossRef]
- Harcus, D.; Marcil, A.; Rigby, T.; Whiteway, M. Transcription Profiling of Cyclic AMP Signaling in. Mol. Biol. Cell 2004, 15, 4490–4499. [Google Scholar] [CrossRef]
- Bockmu, D.P.; Ernst, J.F. A Potential Phosphorylation Site for an A-Type Kinase in the Efg1 Regulator Protein Contributes to Hyphal Morphogenesis of Candida albicans. Genetics 2001, 157, 1523–1530. [Google Scholar]
- Cao, F.; Lane, S.; Raniga, P.P.; Lu, Y.; Zhou, Z.; Ramon, K.; Chen, J.; Liu, H. The Flo8 Transcription Factor Is Essential for Hyphal Development and Virulence in Candida albicans. Mol. Biol. Cell 2006, 17, 295–307. [Google Scholar] [CrossRef] [PubMed]
- Leberer, E.; Harcus, D.; Dignard, D.; Johnson, L.; Ushinsky, S.; Thomas, D.Y.; Schro, K. Ras links cellular morphogenesis to virulence by regulation of the MAP kinase and cAMP signalling pathways in the pathogenic fungus Candida albicans. Mol. Microbiol. 2001, 42, 673–687. [Google Scholar] [CrossRef]
- Lane, S.; Zhou, S.; Pan, T.; Dai, Q.; Liu, H. The Basic Helix-Loop-Helix Transcription Factor Cph2 Regulates Hyphal Development in Candida albicans Partly via Tecmolecular. Cell. Biol. 2001, 21, 6418–6428. [Google Scholar]
- Feng, Q.; Summers, E.; Guo, B.; Fink, G.; Acteriol, J.B. Ras Signaling Is Required for Serum-Induced Hyphal Differentiation in Candida albicans. J. Bacteriol. 1999, 181, 6339–6346. [Google Scholar] [PubMed]
- Sheldrick, G.M. Crystal structure refinement with SHELXL. Acta Crystallogr. Sect. C Struct. Chem. 2015, 71, 3–8. [Google Scholar] [CrossRef]
- Macrae, C.F.; Bruno, I.J.; Chisholm, J.A.; Edgington, P.R.; Mccabe, P.; Pidcock, E.; Rodriguez-monge, L.; Taylor, R.; Streek, J.V.D.; Wood, P.A. Mercury CSD 2.0—New features for the visualization and investigation of crystal structures. J. Appl. Crystallogr. 2008, 41, 466–470. [Google Scholar] [CrossRef]
- Article, R. Crystallographic Computing System JANA2006: General features. Zeitschrift für Kristallographie-Crystalline Materials 2014, 229, 345–352. [Google Scholar]
- Castillo, K.F.; Bello-Vieda, N.J.; Nuñez-Dallos, N.G.; Pastrana, H.F.; Celis, A.M.; Restrepo, S.; Hurtado, J.J.; Ávila, A.G. Metal complex derivatives of azole: A study on their synthesis, characterization, and antibacterial and antifungal activities. J. Braz. Chem. Soc. 2016, 27, 2334–2347. [Google Scholar] [CrossRef]
- Hurtado, J.; Nuñez-Dallos, N.; Movilla, S.; Pietro Miscione, G.; Peoples, B.C.; Rojas, R.; Valderrama, M.; Fröhlich, R. Chromium(III) complexes bearing bis(benzotriazolyl)pyridine ligands: Synthesis, characterization and ethylene polymerization behavior. J. Coord. Chem. 2017, 70, 803–818. [Google Scholar] [CrossRef]
| Compound (Formula) | Molecular Weight/(g mol−1) | Color (Yield)/% | Melting Point/°C | Calculated (Found)/% | ||
|---|---|---|---|---|---|---|
| C | H | N | ||||
| 1 (C14H6N4O12Co) | 481.15 | Purple (98) | 300–302 | 34.95 (34.90) | 1.26 (1.25) | 11.46 (11.51) |
| 2 (C25H22N8O12Co) | 685.43 | Light pink (92) | 324–326 | 43.81 (43.80) | 3.24 (3.22) | 16.35 (16.33) |
| 3 (C25H20N10O16Co) | 775.42 | Dark pink (90) | 347–349 | 38.72 (38.64) | 2.60 (2.60) | 18.06 (17.99) |
| 4 (C33H30N8O12Co) | 789.58 | Purple (87) | 353–355 | 47.95 (47.83) | 3.50 (3.48) | 16.23 (16.18) |
| 5 (C33H28N10O16Co) | 879.57 | Dark blue (84) | 362–364 | 42.97 (42.69) | 2.91 (2.90) | 17.78 (17.73) |
| 6 (C31H27N9O12Co) | 776.54 | Pink (80) | 378–380 | 50.20 (49.96) | 3.83 (3.79) | 14.19 (14.16) |
| 7 (C31H25N11O16Co) | 866.54 | Lilac (82) | 384–386 | 45.06 (45.05) | 3.21 (3.20) | 15.92 (15.90) |
| Complex | FT-IR (cm−1) | Raman (cm−1) | |||||
|---|---|---|---|---|---|---|---|
| υas(NO2) | υs(NO2) | υas(COO−) | υs(COO−) | δring t | υ(Co-O) | ||
| 1 | 1462 | 1346 | 1624 | 1577 | 999 | 346 | 204 |
| 2 | 1458 | 1342 | 1631 | 1589 | 996 | 339 | 201 |
| 3 | 1500 | 1342 | 1631 | 1570 | 998 | 341 | 208 |
| 4 | 1462 | 1346 | 1624 | 1581 | 996 | 345 | 207 |
| 5 | 1465 | 1338 | 1627 | 1604 | 996 | 343 | 206 |
| 6 | 1458 | 1338 | 1643 | 1573 | 993 | 348 | 212 |
| 7 | 1485 | 1342 | 1627 | 1566 | 996 | 348 | 207 |
| Complex | L5 | 1m (Methanol) | 1e (Acetone: Ethanol) |
|---|---|---|---|
| Crystal Data | |||
| Chemical formula | C19H24N4 | C18H22CoN4O16 | C22H30CoN4O16 |
| Mr | 308.42 | 609.33 | 665.43 |
| Crystal system, space group | Monoclinic, C2/c | Triclinic, P | Monoclinic, P21/n |
| Temperature (K) | 298 | 273 | 298 |
| a, b, c (Å) | 15.0005 (8), 8.4989 (6), 13.7852 (8) | 6.4119 (13), 8.7752 (19), 12.154 (2) | 6.7234 (4), 9.0108 (6), 24.6830 (13) |
| α, β, γ (°) | 90.0, 96.778 (3), 90.0 | 90.444 (7), 100.387 (7), 102.212 (8) | 90.0, 93.3622 (19), 90.0 |
| V (Å3) | 1745.16 (18) | 656.7 (2) | 1492.80 (15) |
| Z | 4 | 1 | 2 |
| Radiation type | Mo Kα | Mo Kα | Mo Kα |
| µ (mm−1) | 0.07 | 0.74 | 0.65 |
| Crystal size (mm) | 0.48 × 0.43 × 0.34 | 0.27 × 0.17 × 0.11 | 0.29 × 0.14 × 0.10 |
| Data Collection | |||
| Diffractometer | Bruker D8 Venture/Photon 100 CMOS diffractometer | ||
| Absorption correction | Multi-scan SADABS; Bruker, 2016 | ||
| Tmin, Tmax | 0.715, 0.747 | 0.700, 0.746 | 0.674, 0.746 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 39,902, 3342, 2036 | 24,349, 3987, 3301 | 32,804, 3050, 2439 |
| Rint | 0.052 | 0.037 | 0.046 |
| (sin θ/λ)max (Å−1) | 0.771 | 0.714 | 0.625 |
| Refinement | |||
| R[F2 > 2σ(F2)], wR(F2), S | 0.057, 0.186, 1.03 | 0.042, 0.113, 1.03 | 0.045, 0.127, 1.09 |
| No. of reflections | 3342 | 3987 | 3050 |
| No. of parameters | 109 | 186 | 244 |
| No. of restraints | 0 | 24 | 80 |
| H-atom treatment | H-atom parameters constrained | H atoms treated with a mixture of independent and constrained refinement | H atoms treated with a mixture of independent and constrained refinement |
| Δρmax, Δρmin (e Å−3) | 0.23 (0.91 Å from H2B), −0.24 (1.01 Å from N1) | 0.53 (0.98 Å from O5), −0.39 (0.35 Å from O5) | 0.33 (0.10 Å from C10A), −0.37 (0.19 Å from C11A) |
| Compound | C. albicans | C. tropicalis | C. krusei | Vero Cells | ||||
|---|---|---|---|---|---|---|---|---|
| MIC | MFC | MIC | MFC | MIC | MFC | CC50 ± SD | CC90 ± SD | |
| L1 | 1000 | >2000 | 1000 | >2000 | 1000 | >2000 | 157.38 ± 3.1 | >300 |
| L2 | >2000 | >2000 | >2000 | >2000 | >2000 | >2000 | >300 | >300 |
| L3 | 2000 | >2000 | 2000 | >2000 | >2000 | >2000 | 121.94 ± 2.1 | >300 |
| L4 | >2000 | >2000 | >2000 | >2000 | >2000 | >2000 | 111.43 ± 1.4 | >300 |
| L5 | 2000 | >2000 | 2000 | >2000 | 2000 | >2000 | 44.95 ± 1.8 | >300 |
| L6 | >2000 | >2000 | >2000 | >2000 | >2000 | >2000 | >300 | >300 |
| 1 | 125 | 250 | 125 | >500 | 62.5 | >2000 | 197.05 ± 1.4 | >300 |
| 2 | 125 | 250 | 125 | >500 | 62.5 | >1000 | 162.22 ± 2.5 | >300 |
| 3 | 125 | 500 | 250 | >2000 | 31.25 | 31.25 | 298.73 ± 3.6 | >300 |
| 4 | 250 | 500 | 250 | >2000 | 62.5 | >2000 | >300 | >300 |
| 5 | 250 | 250 | 250 | 1000 | 62.5 | >2000 | >300 | >300 |
| 6 | 250 | >2000 | 125 | >500 | 62.5 | 500 | 299.87 ± 3.2 | >300 |
| 7 | 125 | 500 | 125 | 500 | 62.5 | 250 | 258.45 ± 1.4 | >300 |
| Co(OAc)2∙ 4H2O | 125 | 125 | 125 | 250 | 31.25 | >1000 | 103.97 ± 3.7 | >300 |
| Itraconazole | 1.0 | 4.0 | >16 | ND | 0.5 | 1.0 | 59.84 ± 4.5 | >300 |
| Compound | Concentrations (µg mL−1) ± SD | |||
|---|---|---|---|---|
| 500 | 250 | 125 | 62.5 | |
| L1 | 54.16 *,† ± 6.82 | 16.35 ± 5.16 | 9.87 ± 1.48 | NI |
| L2 | 43.47 * ± 7.18 | 39.55 * ± 2.86 | 7.82 ± 3.30 | NI |
| L3 | 45.24 * ± 5.16 | 21.66 ± 3.83 | NI | NI |
| L4 | 47.32 * ± 5.88 | 44.92 * ± 6.55 | 27.57 ** ± 4.43 | NI |
| L5 | 33.28 * ± 7.03 | 30.65 ** ± 4.59 | 28.09 ** ± 5.99 | 25.21 ± 2.30 |
| 4 | 17.3 ± 1.64 | NI | NI | NI |
| 5 | 30.06 * ± 2.86 | NI | NI | NI |
| 7 | 12.01 ± 2.65 | 5.99 ± 2.21 | 4.56± 3.15 | 4.19 ± 3.18 |
| Co(OAc)2 ∙ 4H2O | 37.73 * ± 4.10 | NI | NI | NI |
| Concentrations (µg mL−1) ± SD | ||||
| Reference Compound | 1 | 0.5 | 0.25 | 0.125 |
| Itraconazole | 44.34 ± 6.15 | 39.91 ± 5.93 | 36.45 ± 2.80 | ND |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fonseca, D.; Leal-Pinto, S.M.; Roa-Cordero, M.V.; Vargas, J.D.; Moreno-Moreno, E.M.; Macías, M.A.; Suescun, L.; Muñoz-Castro, Á.; Hurtado, J.J. Inhibition of C. albicans Dimorphic Switch by Cobalt(II) Complexes with Ligands Derived from Pyrazoles and Dinitrobenzoate: Synthesis, Characterization and Biological Activity. Int. J. Mol. Sci. 2019, 20, 3237. https://doi.org/10.3390/ijms20133237
Fonseca D, Leal-Pinto SM, Roa-Cordero MV, Vargas JD, Moreno-Moreno EM, Macías MA, Suescun L, Muñoz-Castro Á, Hurtado JJ. Inhibition of C. albicans Dimorphic Switch by Cobalt(II) Complexes with Ligands Derived from Pyrazoles and Dinitrobenzoate: Synthesis, Characterization and Biological Activity. International Journal of Molecular Sciences. 2019; 20(13):3237. https://doi.org/10.3390/ijms20133237
Chicago/Turabian StyleFonseca, Daniela, Sandra M. Leal-Pinto, Martha V. Roa-Cordero, José D. Vargas, Erika M. Moreno-Moreno, Mario A. Macías, Leopoldo Suescun, Álvaro Muñoz-Castro, and John J. Hurtado. 2019. "Inhibition of C. albicans Dimorphic Switch by Cobalt(II) Complexes with Ligands Derived from Pyrazoles and Dinitrobenzoate: Synthesis, Characterization and Biological Activity" International Journal of Molecular Sciences 20, no. 13: 3237. https://doi.org/10.3390/ijms20133237
APA StyleFonseca, D., Leal-Pinto, S. M., Roa-Cordero, M. V., Vargas, J. D., Moreno-Moreno, E. M., Macías, M. A., Suescun, L., Muñoz-Castro, Á., & Hurtado, J. J. (2019). Inhibition of C. albicans Dimorphic Switch by Cobalt(II) Complexes with Ligands Derived from Pyrazoles and Dinitrobenzoate: Synthesis, Characterization and Biological Activity. International Journal of Molecular Sciences, 20(13), 3237. https://doi.org/10.3390/ijms20133237






