Genome Editing in Cowpea Vigna unguiculata Using CRISPR-Cas9
Abstract
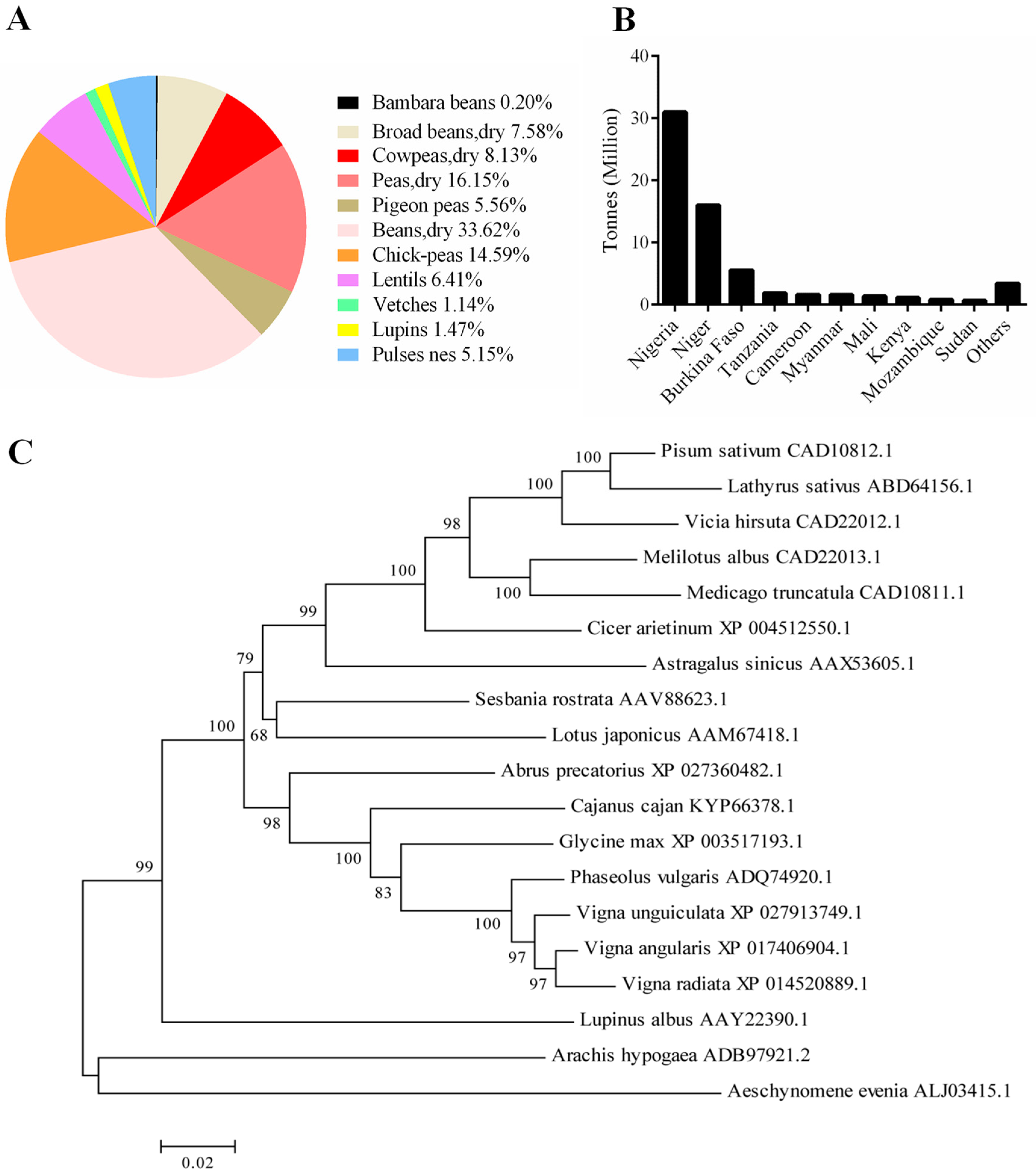
1. Introduction
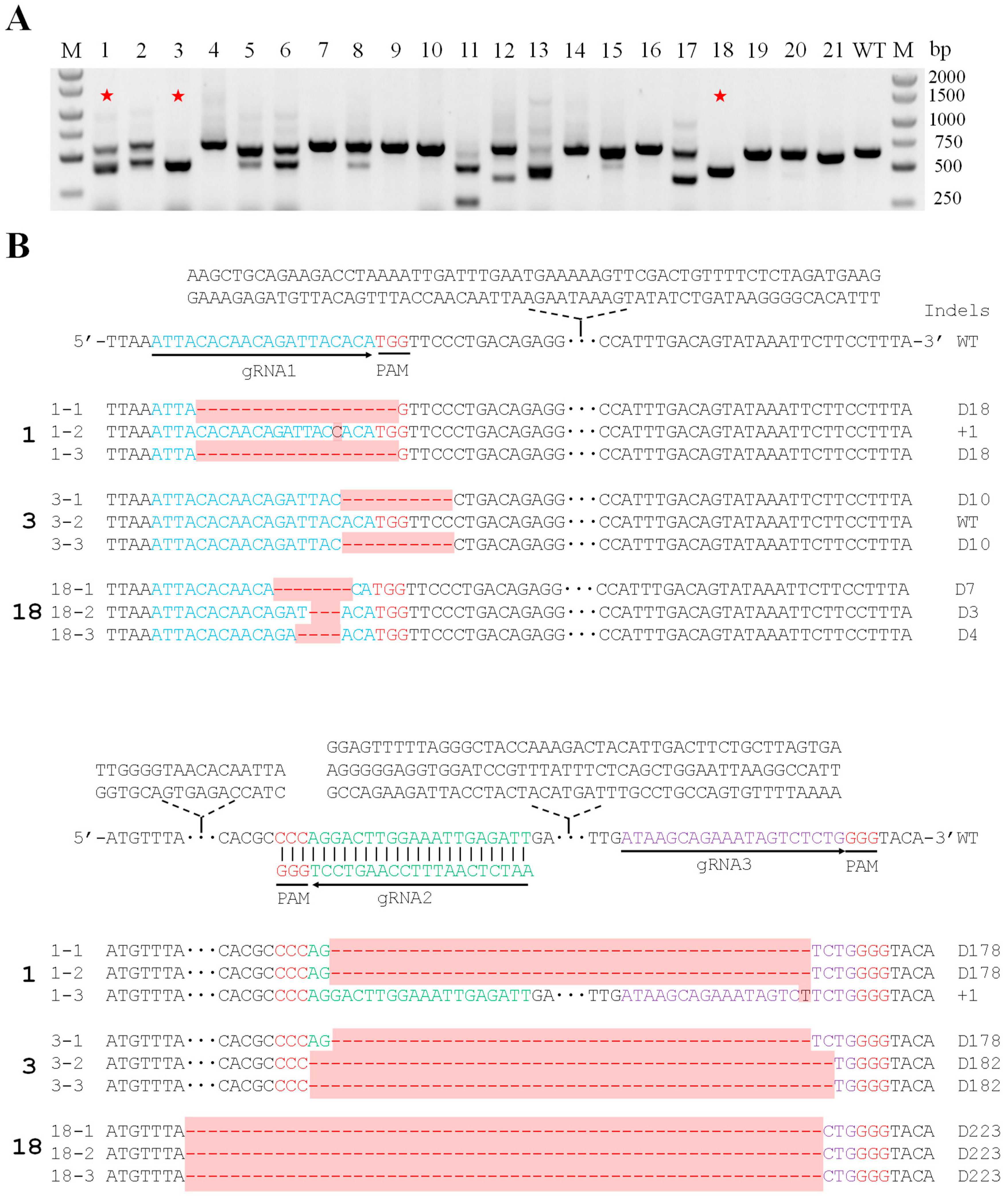
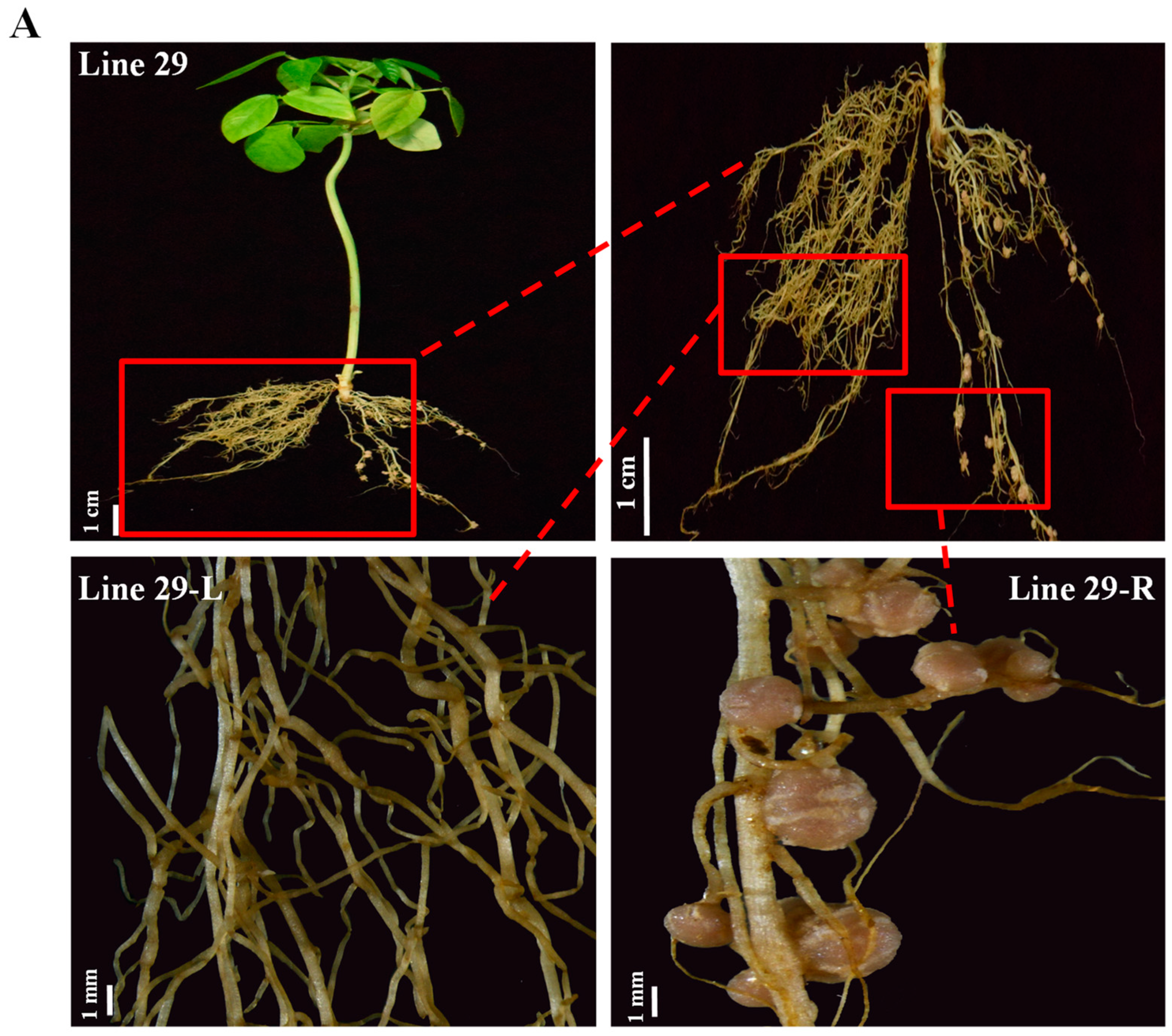
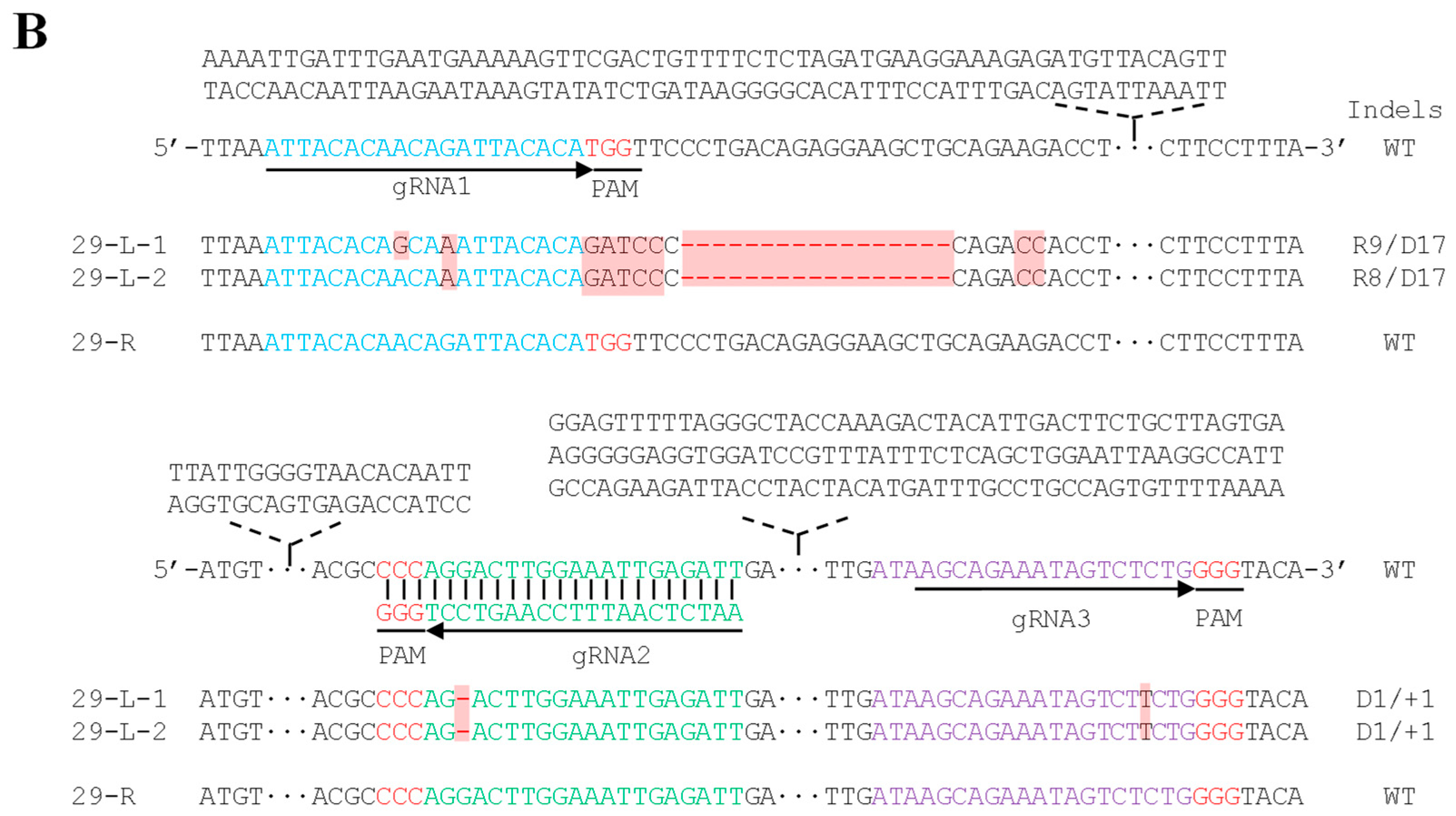
2. Results
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Vector Construction
4.3. Cowpea Hairy Root Transformation
4.4. Rhizobia Inoculation and Phenotypic Analysis
4.5. DNA Isolation, PCR, Plasmid Extraction and Sequencing
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| BAC | bacterial artificial chromosome |
| Cas9 | CRISPR-associated protein 9 |
| Cpf1 | CRISPR-associated protein, subtype Prevotella and Francisella |
| CRISPR | clustered regularly interspaced short palindromic repeats |
| ESTs | expressed sequence tags |
| gRNA | guide RNA |
| LRR | leucine-rich repeats |
| MEGA | molecular evolutionary and genetics analysis |
| PAM | protospacer adjacent motif |
| Ri | root-inducing |
| SNF | symbiotic nitrogen fixation |
| SNPs | single nucleotide polymorphisms |
| SSNs | sequence-specific nucleases |
| SYMRK | symbiosis receptor-like kinase |
| TALENs | transcription activator-like effector nucleases |
| Ti | tumor-inducing |
| YEP | yeast extract peptone |
| ZFNs | zinc finger nucleases |
References
- Carvalho, M.; Lino-Neto, T.; Rosa, E.; Carnide, V. Cowpea: A legume crop for a challenging environment. J. Sci. Food. Agric. 2017, 97, 4273–4284. [Google Scholar] [CrossRef] [PubMed]
- Coulibaly, S.; Pasquet, R.S.; Papa, R.; Gepts, P. AFLP analysis of the phenetic organization and genetic diversity of Vigna unguiculata L. Walp. reveals extensive gene flow between wild and domesticated types. Theor. Appl. Genet. 2002, 104, 358–366. [Google Scholar] [CrossRef] [PubMed]
- Ng, N.Q.; Maréchal, R. Cowpea taxonomy, origin and germplasm. In Cowpea Research, Production, and Utilization; Singh, S., Rachie, K., Eds.; Wiley: Chichester, UK, 1985; pp. 11–21. [Google Scholar]
- Food and Agriculture Organization of the United Nations. FAO. Available online: http://www.fao.org/faostat/en/#data/QC (accessed on 10 March 2019).
- Li, Y.; Tian, C.F.; Chen, W.F.; Wang, L.; Sui, X.H.; Chen, W.X. High-resolution transcriptomic analyses of Sinorhizobium sp. NGR234 bacteroids in determinate nodules of Vigna unguiculata and indeterminate nodules of Leucaena leucocephala. PLoS ONE 2013, 8, e70531. [Google Scholar] [CrossRef] [PubMed]
- Pueppke, S.G.; Broughton, W.J. Rhizobium Sp. Strain NGR234 and R. fredii USDA257 share exceptionally broad, nested host ranges. Mol. Plant Microbe Interact. 1999, 12, 293–318. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.T.; Yang, J.K.; Yuan, T.Y.; Zhou, J.C. Genetic diversity phylogeny of indigenous rhizobia from cowpea (Vigna unguiculata (L.) Walp.). Biol. Fertil. Soils 2007, 44, 201–210. [Google Scholar] [CrossRef]
- Hernandez-Lucas, I.; Segovia, L.; Martinez-Romero, E.; Pueppke, S.G. Phylogenetic relationships and host range of Rhizobium spp. that nodulate Phaseolus vulgaris L. Appl. Environ. Microbiol. 1995, 61, 2775–2779. [Google Scholar]
- Wang, L.; Wang, L.; Zhou, Y.; Duanmu, D. Use of CRISPR/Cas9 for Symbiotic Nitrogen Fixation Research in Legumes. Prog. Mol. Biol. Transl. 2017, 149, 187. [Google Scholar]
- Muchero, W.; Diop, N.N.; Bhat, P.R.; Fenton, R.D.; Wanamaker, S.; Pottorff, M.; Hearne, S.; Cisse, N.; Fatokun, C.; Ehlers, J.D.; et al. A consensus genetic map of cowpea Vigna unguiculata (L.) Walp. and synteny based on EST derived SNPs. Proc. Natl. Acad. Sci. USA 2009, 106, 18159–18164. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-Amatriaín, M.; Mirebrahim, H.; Xu, P.; Wanamaker, S.I.; Luo, M.; Alhakami, H.; Alpert, M.; Atokple, I.; Batieno, B.J.; Boukar, O.; et al. Genome resources for climate resilient cowpea, an essential crop for food security. Plant J. 2017, 89, 1042–1054. [Google Scholar] [CrossRef]
- Urnov, F.D.; Miller, J.C.; Lee, Y.L.; Beausejour, C.M.; Rock, J.M.; Augustus, S.; Jamieson, A.C.; Porteus, M.H.; Gregory, P.D.; Holmes, M.C. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature 2005, 435, 646–651. [Google Scholar] [CrossRef] [PubMed]
- Osakabe, K.; Osakabe, Y.; Toki, S. Site-directed mutagenesis in Arabidopsis using custom-designed zinc finger nucleases. Proc. Natl. Acad. Sci. USA 2010, 107, 12034–12039. [Google Scholar] [CrossRef]
- Li, T.; Liu, B.; Spalding, M.H.; Weeks, D.P.; Yang, B. High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat. Biotechnol. 2012, 30, 390–392. [Google Scholar] [CrossRef]
- Xie, K.; Yang, Y. RNA-guided genome editing in plants using a CRISPR–Cas system. Mol. Plant 2013, 6, 1975–1983. [Google Scholar] [CrossRef] [PubMed]
- Demirci1, Y.; Zhang, B.; Unver, T. CRISPR/Cas9: An RNA-guided highly precise synthetic tool for plant genome editing. Cell Physiol. 2017, 233, 1844–1859. [Google Scholar] [CrossRef] [PubMed]
- Shah, T.; Andleeb, T.; Lateef, S.; Noor, M.A. Genome editing in plants: Advancing crop transformation and overview of tools. Plant Physiol. Biochem. 2018, 131, 12–21. [Google Scholar] [CrossRef]
- Wang, L.; Wang, L.; Tan, Q.; Fan, Q.; Zhu, H.; Hong, Z.; Zhang, Z.; Duanmu, D. Efficient inactivation of symbiotic nitrogen fixation related genes in Lotus japonicas using CRISPR-Cas9. Front Plant Sci. 2016, 7, 1333. [Google Scholar] [PubMed]
- Li, Z.; Liu, Z.; Xing, A.; Moon, B.P.; Koellhoffer, J.P.; Huang, L.; Ward, R.T.; Clifton, E.; Falco, S.C.; Cigan, A.M. Cas9-Guide RNA Directed Genome Editing in Soybean. Plant Physiol. 2015, 169, 960–970. [Google Scholar] [CrossRef]
- Curtin, S.J. Editing the Medicago truncatula Genome: Targeted Mutagenesis Using the CRISPR-Cas9 Reagent. In Methods in Molecular Biology; Cañas, L., Beltrán, J., Eds.; Humana Press: New York, NY, USA, 2018; Volume 1822, pp. 161–174. [Google Scholar]
- Endre, G.; Kereszt, A.; Kevei, Z.; Mihacea, S.; Kalo, P.; Kiss, G.B. A receptor kinase gene regulating symbiotic nodule development. Nature 2002, 417, 962–966. [Google Scholar] [CrossRef]
- Indrasumunar, A.; Wilde, J.; Hayashi, S.; Li, D.; Gresshoff, P.M. Functional analysis of duplicated Symbiosis Receptor Kinase (SymRK) genes during nodulation and mycorrhizal infection in soybean (Glycine max). Plant Physiol. 2015, 176, 157–168. [Google Scholar] [CrossRef]
- Stracke, S.; Kistner, C.; Yoshida, S.; Mulder, L.; Sato, S.; Kaneko, T.; Tabata, S.; Sandal, N.; Stougaard, J.; Szczyglowski, K.; et al. A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature 2002, 417, 959–962. [Google Scholar] [CrossRef]
- Zetsche, B.; Gootenberg, J.S.; Abudayyeh, O.O.; Slaymaker, I.M.; Makerova, K.S.; Essletzbichler, P.; Volz, S.E.; Joung, J.; van der Oost, J.; Regev, A.; et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 2015, 163, 759–777. [Google Scholar] [CrossRef] [PubMed]
- Begemann, M.B.; Gray, B.N.; January, E.; Gorgon, G.C.; He, Y.; Liu, H.; Wu, X.; Brutnell, T.P.; Mockler, T.C.; Oufattole, M. Precise insertion and guided editing of higher plant genomes using Cpf1 CRISPR nucleases. Scientific Reports 2017, 7, 11606. [Google Scholar] [CrossRef] [PubMed]
- Jia, H.; Orbovic, V.; Wang, N. CRISPR-LbCas12a-mediated modification of citrus. Plant Biotechnolol. J. 2019. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Kim, J.; Hur, J.K.; Been, K.W.; Yoon, S.H. Genome-wide analysis reveals specificities of Cpf1 endonucleases in human cells. Nat. Biotechnol. 2016, 34, 863–868. [Google Scholar] [CrossRef] [PubMed]
- Chilton, M.D.; Tepfer, D.A.; Petit, A.; David, C.; Casse-Delbart, F.; Tempe, J. Agrobacterium rhizogenes inserts T-DNA into the genomes of the host plant root cells. Nature 1982, 295, 432–434. [Google Scholar] [CrossRef]
- White, F.F.; Nester, E.W. Hairy root: Plasmids encode virulence traits in Agrobacterium rhizogenes. J. Bacteriol. 1980, 141, 1134. [Google Scholar] [PubMed]
- Makhzoum, A.B.; Sharma, P.; Bernards, M.A.; Trémouillaux-Guiller, J. Hairy Roots: An Ideal Platform for Transgenic Plant Production and Other Promising Applications. In Recent Advances in Phytochemistry; Gang, D., Ed.; Springer: New York, NY, USA, 2013; Volume 42, pp. 95–142. [Google Scholar]
- Estrada-Navarrete, G.; Alvarado-Affantranger, X.; Olivares, J.E.; Díaz-Camino, C.; Santana, O.; Murillo, E.; Guillén, G.; Sánchez-Guevara, N.; Acosta, J.; Quinto, C.; et al. Agrobacterium rhizogenes transformation of the Phaseolus spp.: A tool for functional genomics. Mol Plant Microbe Interact. 2006, 19, 1385–1393. [Google Scholar] [CrossRef]
- Kereszt, A.; Li, D.; Indrasumunar, A.; Nguyen, C.D.T.; Nontachaiyapoom, S.; Kinkema, M.; Gresshoff, P.M. Agrobacterium rhizogenes-mediated transformation of soybean to study root biology. Nat. Protocols 2007, 2, 948–952. [Google Scholar] [CrossRef]
- Savka, M.A.; Ravillion, B.; Noel, G.R.; Farrand, S.K. Induction of hairy roots on cultivated soybean genotypes and their use to propagate the soybean cyst nematode. Phytopathology 1990, 80, 503–508. [Google Scholar] [CrossRef]
- Bett, B.; Gollasch, S.; Moore, A.; Harding, R.; Higgins, T.J.V. An Improved Transformation System for Cowpea (Vigna unguiculata L. Walp) via Sonication and a Kanamycin-Geneticin Selection Regime. Front. Plant Sci. 2019, 10, 219. [Google Scholar] [CrossRef]
- Zhou, G.Y.; Weng, J.; Zeng, Y.; Huang, J.; Qian, S.; Liu, G. Introduction of exogenous DNA into cotton embryos. Methods Enzymol. 1983, 101, 433–481. [Google Scholar] [PubMed]
- Chaudhury, D.; Madanpotra, S.; Jaiwal, R.; Saini, R.; Kumar, P.A.; Jaiwal, P.K. Agrobacterium tumefaciens-mediated high frequency genetic transformation of an Indian cowpea (Vigna unguiculata L. Walp.) cultivar and transmission of transgenes into progeny. Plant Sci. 2007, 172, 692–700. [Google Scholar] [CrossRef]
- Ivo, N.L.; Nascimento, C.P.; Vieira, L.S.; Campos, F.A.P.; Aragão, F.J.L. Biolistic-mediated genetic transformation of cowpea (Vigna unguiculata) and stable Mendelian inheritance of transgenes. Plant Cell Rep. 2008, 27, 1475–1483. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Ding, Y.; Zhou, Y.; Jin, W.; Xie, K.; Chen, L.-L. CRISPR-P 2.0: An improved CRISPR/Cas9 tool for genome editing in plants. Mol. Plant 2017, 10, 530–532. [Google Scholar] [CrossRef] [PubMed]
- Broughton, W.J.; Dilworth, M.J. Control of leghaemoglobin synthesis in snake beans. Biochem. J. 1971, 125, 1075–1080. [Google Scholar] [CrossRef] [PubMed]


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ji, J.; Zhang, C.; Sun, Z.; Wang, L.; Duanmu, D.; Fan, Q. Genome Editing in Cowpea Vigna unguiculata Using CRISPR-Cas9. Int. J. Mol. Sci. 2019, 20, 2471. https://doi.org/10.3390/ijms20102471
Ji J, Zhang C, Sun Z, Wang L, Duanmu D, Fan Q. Genome Editing in Cowpea Vigna unguiculata Using CRISPR-Cas9. International Journal of Molecular Sciences. 2019; 20(10):2471. https://doi.org/10.3390/ijms20102471
Chicago/Turabian StyleJi, Jie, Chunyang Zhang, Zhongfeng Sun, Longlong Wang, Deqiang Duanmu, and Qiuling Fan. 2019. "Genome Editing in Cowpea Vigna unguiculata Using CRISPR-Cas9" International Journal of Molecular Sciences 20, no. 10: 2471. https://doi.org/10.3390/ijms20102471
APA StyleJi, J., Zhang, C., Sun, Z., Wang, L., Duanmu, D., & Fan, Q. (2019). Genome Editing in Cowpea Vigna unguiculata Using CRISPR-Cas9. International Journal of Molecular Sciences, 20(10), 2471. https://doi.org/10.3390/ijms20102471




