Isolation and Characterization of a Green-Tissue Promoter from Common Wild Rice (Oryza rufipogon Griff.)
Abstract
:1. Introduction
2. Results
2.1. Expression Pattern of OrGSE in Common Wild Rice
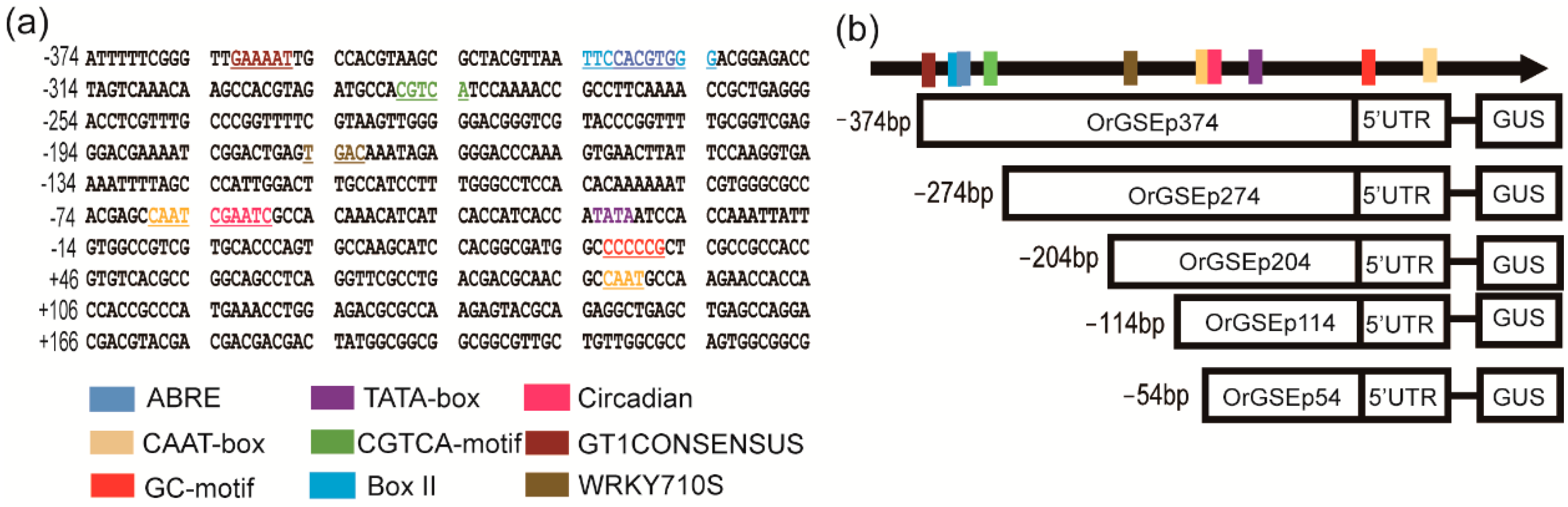
2.2. Cloning and Sequence Analysis of the OrGSE Promoter (OrGSEp-374)
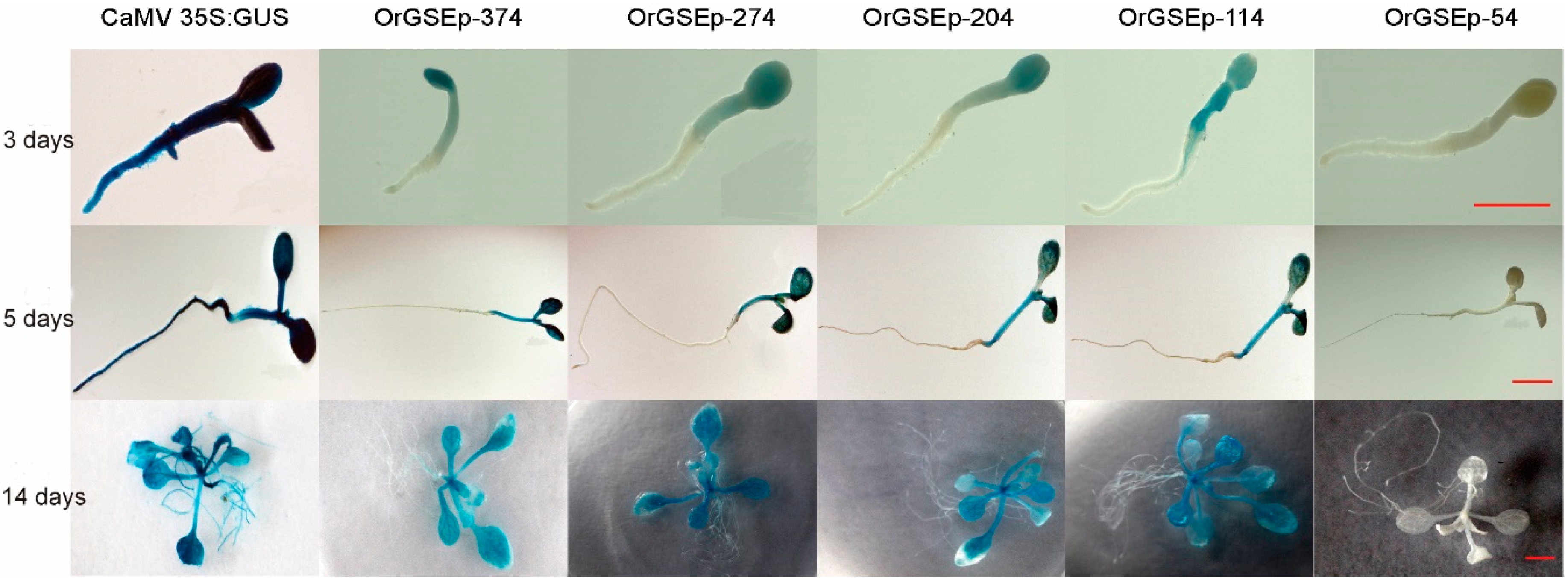
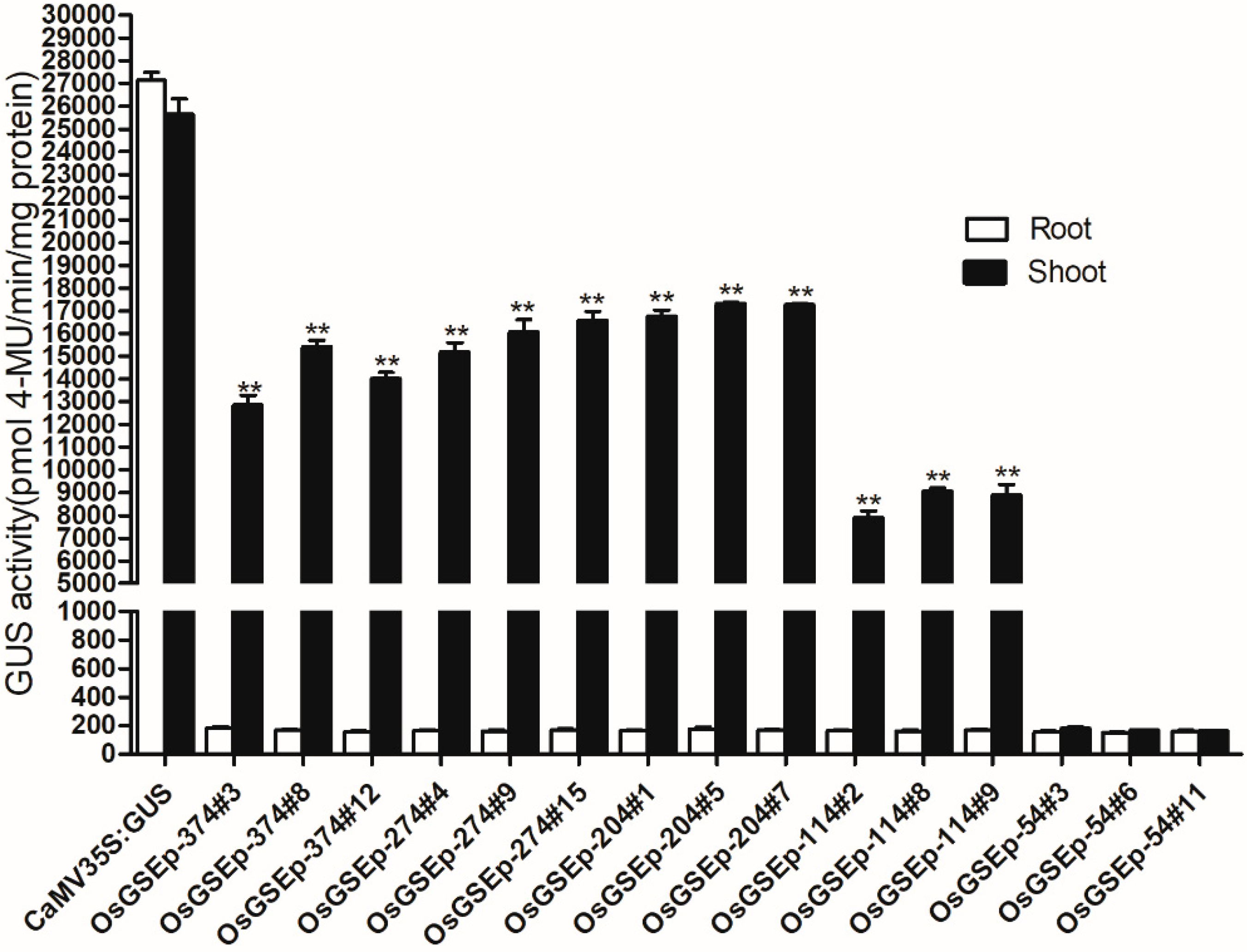
2.3. Spatiotemporal Expression Patterns of OrGSEp-374 and 5’-Deletion Fragments in Arabidopsis
2.4. OrGSEp-374 Confers Light-Responsive Expression
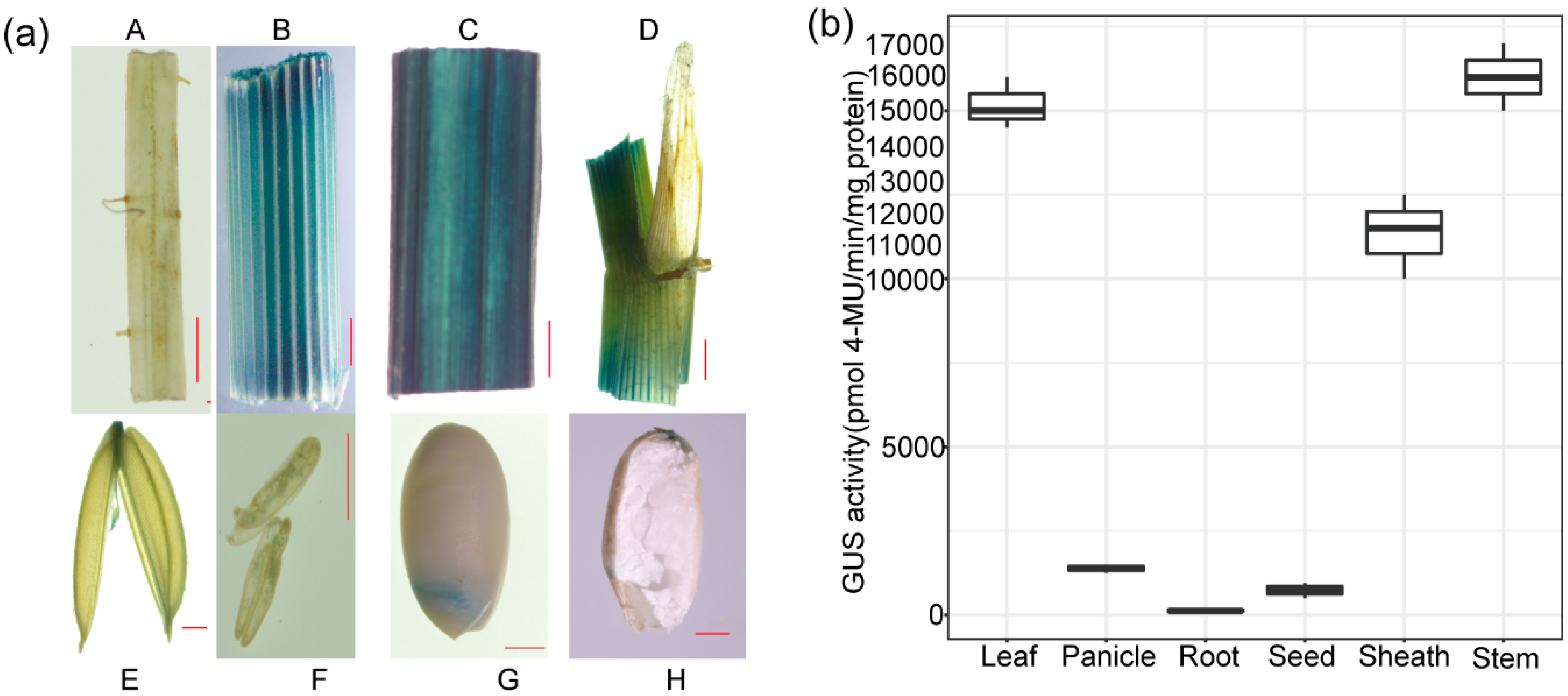
2.5. The Expression Pattern of OrGSEp-374 in Rice
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Screening of Green Tissue-Specific Genes and Expression Analysis
4.3. Cloning of the OrGSE Promoter and Sequence Analysis
4.4. PCR Amplification of 5’-Deletion Fragments of the OrGSE Promoter
4.5. Detection of the Expression Pattern of the OrGSEp Promoter and 5’-Deletion Fragments in Different Organs
4.6. Inducible Activity Analysis of the OrGSEp Promoter and 5’-Deletion Fragments
4.7. GUS Histochemical and Fluorometric Analysis
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Li, Y.; Sun, Y.; Yang, Q.C.; Kang, J.M.; Zhang, T.J.; Gruber, M.Y.; Fang, F. Cloning and function analysis of an alfalfa (Medicago sativa L.) zinc finger protein promoter MsZPP. Mol. Biol. Rep. 2012, 39, 8559–8569. [Google Scholar] [CrossRef] [PubMed]
- Niu, G.L.; Gou, W.; Han, X.L.; Qin, C.; Zhang, L.X.; Abomohra, A.E.; Ashraf, M. Cloning and functional analysis of phosphoethanolamine methyltransferase promoter from maize (Zea mays L.). Int. J. Mol. Sci. 2018, 19, 191. [Google Scholar] [CrossRef] [PubMed]
- Pih, K.T.; Yoo, J.; Fosket, D.E.; Han, I.S. A comparison of the activity of three cauliflower mosaic virus 35s promoters in rice seedlings and tobacco (by-2) protoplasts by analysis of gus reporter gene transient expression. Plant. Sci. 1996, 114, 141–148. [Google Scholar] [CrossRef]
- McElroy, D.; Blowers, A.D.; Jenes, B.; Wu, R. Construction of expression vectors based on the rice actin 1 (act1) 5' region for use in monocot transformation. Mol. Gen. Genet. 1991, 231, 150–160. [Google Scholar] [CrossRef] [PubMed]
- Christensen, A.H.; Quail, P.H. Ubiquitin promoter-based vectors for high-level expression of selectable and/or screenable marker genes in monocotyledonous plants. Transgenic Res. 1996, 5, 213–218. [Google Scholar] [CrossRef] [PubMed]
- Eva, C.; Teglas, F.; Zelenyanszki, H.; Tamas, C.; Juhasz, A.; Meszaros, K.; Tamas, L. Cold inducible promoter driven cre-lox system proved to be highly efficient for marker gene excision in transgenic barley. J. Biotechnol. 2018, 265, 15–24. [Google Scholar] [CrossRef] [PubMed]
- Qu, D.; Song, Y.; Li, W.M.; Pei, X.W.; Wang, Z.X.; Jia, S.R.; Zhang, Y.Q. Isolation and characterization of the organ-specific and light-inducible promoter of the gene encoding rubisco activase in potato (Solanum tuberosum). Genet. Mol. Res. 2011, 10, 621–631. [Google Scholar] [CrossRef] [PubMed]
- Thomson, B.; Graciet, E.; Wellmer, F. Inducible promoter systems for gene perturbation experiments in Arabidopsis. Methods Mol. Biol. 2017, 1629, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Ding, X.; Chen, J.; Shen, Y.; Kong, L.; Li, N.; Chu, Z. Functional analysis of the GRMZM2g174449 promoter to identify rhizoctonia solani-inducible cis-elements in maize. BMC Plant. Biol. 2017, 17, 233. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.I.; Lee, B.H. Generation of a stress-inducible luminescent Arabidopsis and its use in genetic screening for stress-responsive gene deregulation mutants. Methods Mol. Biol. 2017, 1631, 109–119. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Qin, R.; Xu, R.; Li, H.; Yang, Y.; Li, L.; Wei, P.; Yang, J. Isolation and identification of five cold-inducible promoters from Oryza sativa. Planta 2017, 247, 99–111. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Jing, R.; Mao, X. Functional characterization of Tasnrk2.8 promoter in response to abiotic stresses by deletion analysis in transgenic Arabidopsis. Front. Plant. Sci. 2017, 8, 1198. [Google Scholar] [CrossRef] [PubMed]
- Kakrana, A.; Kumar, A.; Satheesh, V.; Abdin, M.Z.; Subramaniam, K.; Bhattacharya, R.C.; Srinivasan, R.; Sirohi, A.; Jain, P.K. Identification, validation and utilization of novel nematode-responsive root-specific promoters in Arabidopsis for inducing host-delivered RNAi mediated root-knot nematode resistance. Front. Plant. Sci. 2017, 8, 2049. [Google Scholar] [CrossRef] [PubMed]
- Nanjareddy, K.; Arthikala, M.K.; Aguirre, A.L.; Gomez, B.M.; Lara, M. Plant promoter analysis: Identification and characterization of root nodule specific promoter in the common bean. J. Vis. Exp. 2017, 23, 130. [Google Scholar] [CrossRef] [PubMed]
- Chavez-Barcenas, A.T.; Valdez-Alarcon, J.J.; Martinez-Trujillo, M.; Chen, L.; Xoconostle-Cazares, B.; Lucas, W.J.; Herrera-Estrella, L. Tissue-specific and developmental pattern of expression of the rice SPS1 gene. Plant. Physiol. 2000, 124, 641–654. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Lee, H.J.; Jung, Y.J.; Kang, K.K.; Tyagi, W.; Kovach, M.; Sweeney, M.; McCouch, S.; Cho, Y.G. Functional properties of an alternative, tissue-specific promoter for rice nadph-dependent dihydroflavonol reductase. PLoS ONE 2017, 12, e0183722. [Google Scholar] [CrossRef] [PubMed]
- Panguluri, S.K.; Sridhar, J.; Jagadish, B.; Sharma, P.C.; Kumar, P.A. Isolation and characterization of a green tissue-specific promoter from pigeonpea [Cajanus cajan (L.) millsp.]. Indian J. Exp. Biol. 2005, 43, 369–372. [Google Scholar] [PubMed]
- Xu, W.; Liu, W.; Ye, R.; Mazarei, M.; Huang, D.; Zhang, X.; Stewart, C.N., Jr. A profilin gene promoter from switchgrass (Panicum virgatum L.) directs strong and specific transgene expression to vascular bundles in rice. Plant. Cell. Rep. 2018, 37, 587–597. [Google Scholar] [CrossRef] [PubMed]
- Zaidi, M.A.; O’Leary, S.J.; Wu, S.; Chabot, D.; Gleddie, S.; Laroche, A.; Eudes, F.; Robert, L.S. Investigating triticeae anther gene promoter activity in transgenic brachypodium distachyon. Planta 2017, 245, 385–396. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Li, J.; Xu, R.; Qin, R.; Song, F.; Li, L.; Wei, P.; Yang, J. Isolation of five rice non-endosperm tissue-expressed promoters and evaluation of their activities in transgenic rice. Plant. Biotechnol. J. 2017, 16, 1138–1147. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.; Fowler, T.J.; Tierney, M.L. Deletion analysis and localization of SbPRP1, a soybean cell wall protein gene, in roots of transgenic tobacco and cowpea. Plant. Mol. Biol. 1993, 21, 109–119. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Jiang, B.; Wu, C.; Sun, S.; Hou, W.; Han, T. GmPRP2 promoter drives root-preferential expression in transgenic Arabidopsis and soybean hairy roots. BMC Plant. Biol. 2014, 14, 245. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Fan, M.; Wang, G.; Zhang, C.; Shi, L.; Wei, Z.; Ma, W.; Chang, J.; Huang, S.; Lin, F. Isolation and characterization of a novel pollen-specific promoter in maize (Zea mays L.). Genome 2017, 60, 485–495. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; El-Mezawy, A.; Shah, S. A seed coat outer integument-specific promoter for Brassica napus. Plant. Cell. Rep. 2011, 30, 75–80. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, A.; Khan, A.; Mishra, D.R.; Bhuyan, K.; Sahoo, B.; Maiti, I.B.; Dey, N. WRKY71 and TGA1A physically interact and synergistically regulate the activity of a novel promoter isolated from petunia vein-clearing virus. Biochim. Biophys. Acta 2018, 1861, 133–146. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Zhu, Y.; Sun, L.; Li, L.; Jin, S.; Zhang, X. Transgenic Bt cotton driven by the green tissue-specific promoter shows strong toxicity to lepidopteran pests and lower Bt toxin accumulation in seeds. Sci. China Life Sci. 2016, 59, 172–182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manikandan, R.; Balakrishnan, N.; Sudhakar, D.; Udayasuriyan, V. Development of leaffolder resistant transgenic rice expressing cry2ax1 gene driven by green tissue-specific rbcS promoter. World J. Microb. Biot. 2016, 32, 37. [Google Scholar] [CrossRef] [PubMed]
- Menguer, P.K.; Sperotto, R.A.; Ricachenevsky, F.K. A walk on the wild side: Oryza species as source for rice abiotic stress tolerance. Genet. Mol. Biol. 2017, 40, 238–252. [Google Scholar] [CrossRef] [PubMed]
- Mao, D.; Yu, L.; Chen, D.; Li, L.; Zhu, Y.; Xiao, Y.; Zhang, D.; Chen, C. Multiple cold resistance loci confer the high cold tolerance adaptation of dongxiang wild rice (Oryza rufipogon) to its high-latitude habitat. Theor. Appl. Genet. 2015, 128, 1359–1371. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Long, Y.; Wang, J.; Zhang, J.; Wang, Y.; Li, W.; Peng, Y.; Yuan, Q.; Pei, X. De novo transcriptome assembly of common wild rice (Oryza rufipogon griff.) and discovery of drought-response genes in root tissue based on transcriptomic data. PLoS ONE 2015, 10, e0131455. [Google Scholar] [CrossRef] [PubMed]
- Civan, P.; Svec, M. Genome-wide analysis of rice (Oryza sativa L. Subsp. Japonica) tata box and y patch promoter elements. Genome 2009, 52, 294–297. [Google Scholar] [CrossRef] [PubMed]
- Conley, T.R.; Park, S.C.; Kwon, H.B.; Peng, H.P.; Shih, M.C. Characterization of cis-acting elements in light regulation of the nuclear gene encoding a subunit of chloroplast isozymes of glyceraldehyde-3-phosphate dehydrogenase from Arabidopsis thaliana. Mol. Cell. Biol. 1994, 14, 2525–2533. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.Q.; Lu, X.; Zhao, F.Y.; Li, Q.T.; Niu, S.L.; Wei, W.; Zhang, W.K.; Ma, B.; Chen, S.Y.; Zhang, J.S. Soybean GmDREBl increases lipid content in seeds of transgenic Arabidopsis. Sci. Rep. 2016, 6, 34307. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Wang, R.X.; Qian, Q.; Yan, M.X.; Meng, X.B.; Fu, Z.M.; Yan, C.Y.; Jiang, B.; Su, Z.; Li, J.Y.; et al. DWARF27, an Iron-Containing Protein Required for the Biosynthesis of Strigolactones, Regulates Rice Tiller Bud Outgrowth. Plant Cell 2009, 21, 1512–1525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waters, M.T.; Brewer, P.B.; Bussell, J.D.; SmitSh, S.M.; Beveridge, C.A. The Arabidopsis ortholog of rice dwarf27 acts upstream of max1 in the control of plant development by strigolactones. Plant. Physiol. 2012, 159, 1073–1085. [Google Scholar] [CrossRef] [PubMed]
- Ye, R.; Zhou, F.; Lin, Y. Two novel positive cis-regulatory elements involved in green tissue-specific promoter activity in rice (Oryza sativa L. ssp.). Plant. Cell. Rep. 2012, 31, 1159–1172. [Google Scholar] [CrossRef] [PubMed]
- Eberhard, S.; Finazzi, G.; Wollman, F.A. The dynamics of photosynthesis. Annu. Rev. Genet. 2008, 42, 463–515. [Google Scholar] [CrossRef] [PubMed]
- Tanabe, N.; Tamoi, M.; Shigeoka, S. The sweet potato RbcS gene (IbRbcS1) promoter confers high-level and green tissue-specific expression of the gus reporter gene in transgenic Arabidopsis. Gene 2015, 567, 244–250. [Google Scholar] [CrossRef] [PubMed]
- Karmakar, S.; Molla, K.A.; Chanda, P.K.; Sarkar, S.N.; Datta, S.K.; Datta, K. Green tissue-specific co-expression of chitinase and oxalate oxidase 4 genes in rice for enhanced resistance against sheath blight. Planta 2016, 243, 115–130. [Google Scholar] [CrossRef] [PubMed]
- Ghasimi Hagh, Z.; Rahnama, H.; Panahandeh, J.; Baghban Kohneh Rouz, B.; Arab Jafari, K.M.; Mahna, N. Green-tissue-specific, c(4)-pepc-promoter-driven expression of Cry1ab makes transgenic potato plants resistant to tuber moth (phthorimaea operculella, zeller). Plant. Cell. Rep. 2009, 28, 1869–1879. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Dehais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouze, P.; Rombauts, S. Plantcare, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for agrobacterium-mediated transformation of Arabidopsis thaliana. Plant. J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Zhu, M.; Ye, R.; Liu, Z.; Zhou, F.; Chen, H.; Lin, Y. Novel green tissue-specific synthetic promoters and cis-regulatory elements in rice. Sci. Rep. 2015, 5, 18256. [Google Scholar] [CrossRef] [PubMed] [Green Version]



© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xue, M.; Long, Y.; Zhao, Z.; Huang, G.; Huang, K.; Zhang, T.; Jiang, Y.; Yuan, Q.; Pei, X. Isolation and Characterization of a Green-Tissue Promoter from Common Wild Rice (Oryza rufipogon Griff.). Int. J. Mol. Sci. 2018, 19, 2009. https://doi.org/10.3390/ijms19072009
Xue M, Long Y, Zhao Z, Huang G, Huang K, Zhang T, Jiang Y, Yuan Q, Pei X. Isolation and Characterization of a Green-Tissue Promoter from Common Wild Rice (Oryza rufipogon Griff.). International Journal of Molecular Sciences. 2018; 19(7):2009. https://doi.org/10.3390/ijms19072009
Chicago/Turabian StyleXue, Mande, Yan Long, Zhiqiang Zhao, Gege Huang, Ke Huang, Tianbao Zhang, Ying Jiang, Qianhua Yuan, and Xinwu Pei. 2018. "Isolation and Characterization of a Green-Tissue Promoter from Common Wild Rice (Oryza rufipogon Griff.)" International Journal of Molecular Sciences 19, no. 7: 2009. https://doi.org/10.3390/ijms19072009
APA StyleXue, M., Long, Y., Zhao, Z., Huang, G., Huang, K., Zhang, T., Jiang, Y., Yuan, Q., & Pei, X. (2018). Isolation and Characterization of a Green-Tissue Promoter from Common Wild Rice (Oryza rufipogon Griff.). International Journal of Molecular Sciences, 19(7), 2009. https://doi.org/10.3390/ijms19072009




