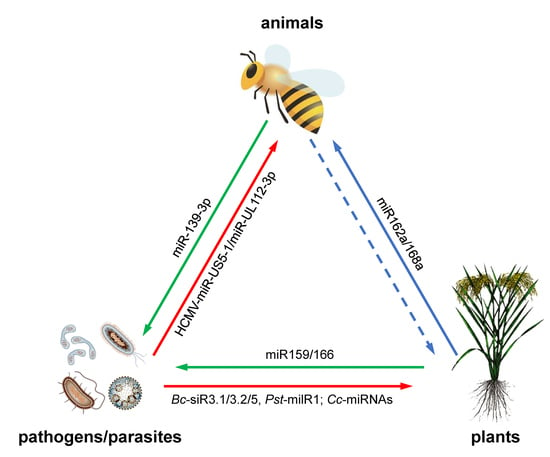
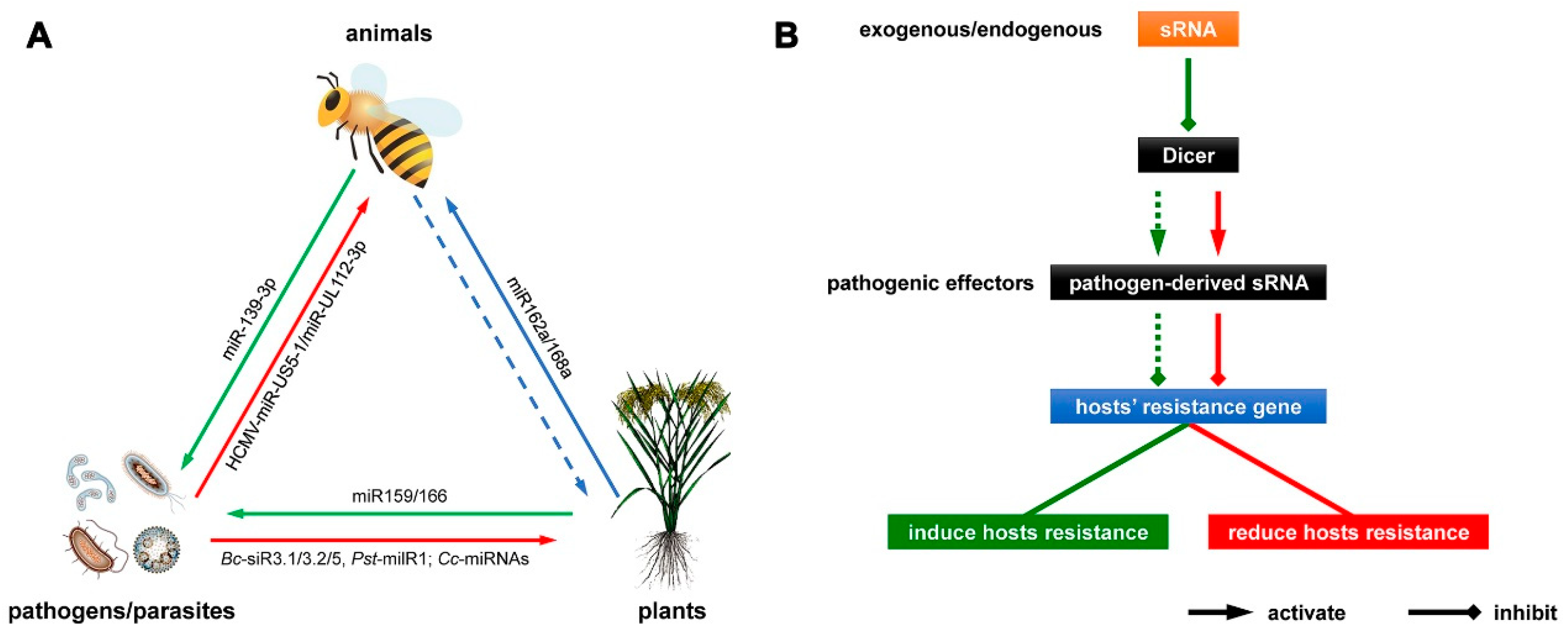
Plant MicroRNAs in Cross-Kingdom Regulation of Gene Expression
Abstract
1. Introduction
2. Plant miRNAs Regulate Gene Expression in Animals
3. Pathogen miRNAs Regulate Gene Expression in Hosts
4. Host miRNAs Regulate Gene Expression in Pathogens
5. Conclusions and Future Prospects
Author Contributions
Funding
Conflicts of Interest
References
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- He, L.; Hannon, G.J. MicroRNAs: Small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004, 5, 522–531. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Chu, C. MicroRNAs in crop improvement: Fine-tuners for complex traits. Nat. Plants 2017, 3, 17077. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.Y.; Hong, P.; Wu, J.Y.; Chen, X.B.; Ye, X.G.; Pan, Y.Y.; Wang, J.; Zhang, X.S. The tae-miR408-mediated control of TaTOC1 gene transcription is required for the regulation of heading time in wheat (Triticum aestivum L.). Plant Physiol. 2016, 170, 1578–1594. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhang, J.; Yan, J.; Gou, F.; Mao, Y.; Tang, G.; Botella, J.R.; Zhu, J.-K. Short tandem target mimic rice lines uncover functions of miRNAs in regulating important agronomic traits. Proc. Natl. Acad. Sci. USA 2017, 114, 5277–5282. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Tu, L.; Wang, L.; Hu, H.; Xu, J.; Zhang, X. MicroRNA 157-targeted SPL genes regulate floral organ size and ovule production in cotton. BMC Plant Biol. 2017, 17, 7. [Google Scholar] [CrossRef] [PubMed]
- Ivashuta, S.I.; Petrick, J.S.; Heisel, S.E.; Zhang, Y.; Guo, L.; Reynolds, T.L.; Rice, J.F.; Allen, E.; Roberts, J.K. Endogenous small RNAs in grain: Semi-quantification and sequence homology to human and animal genes. Food Chem. Toxicol. 2009, 47, 353–360. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Hou, D.; Chen, X.; Li, D.; Zhu, L.; Zhang, Y.; Li, J.; Bian, Z.; Liang, X.; Cai, X.; et al. Exogenous plant MIR168a specifically targets mammalian LDLRAP1: Evidence of cross-kingdom regulation by microRNA. Cell Res. 2012, 22, 107–126. [Google Scholar] [CrossRef] [PubMed]
- Pfeffer, S.; Zavolan, M.; Grässer, F.A.; Chien, M.; Russo, J.J.; Ju, J.; John, B.; Enright, A.J.; Marks, D.; Sander, C.; et al. Identification of virus-encoded microRNAs. Science 2004, 304, 734–736. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.-R.; Mitra, J.; Lee, S.; Gao, S.-J.; Oh, T.-K.; Kim, M.H.; Ha, T.; Jung, J.U. Kaposi’s sarcoma-associated herpesvirus viral interferon regulatory factor 4 (virf4) perturbs the g1-s cell cycle progression via deregulation of the cyclin d1 gene. J. Virol. 2016, 90, 1139–1143. [Google Scholar] [CrossRef] [PubMed]
- Dickinson, B.; Zhang, Y.; Petrick, J.S.; Heck, G.; Ivashuta, S.; Marshall, W.S. Lack of detectable oral bioavailability of plant micrornas after feeding in mice. Nat. Biotechnol. 2013, 31, 965–967. [Google Scholar] [CrossRef] [PubMed]
- Chin, A.R.; Fong, M.Y.; Somlo, G.; Wu, J.; Swiderski, P.; Wu, X.; Wang, S.E. Cross-kingdom inhibition of breast cancer growth by plant MIR159. Cell Res. 2016, 26, 217–228. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.-C.; Chen, W.L.; Kung, W.-H.; Huang, H.-D. Plant miRNAs found in human circulating system provide evidences of cross kingdom RNAi. BMC Genom. 2017, 18, 112. [Google Scholar] [CrossRef] [PubMed]
- Witwer, K. Alternative miRNAs? Human sequences misidentified as plant miRNAs in plant studies and in human plasma [version 1; referees: 2 approved]. F1000Research 2018, 7. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Roh, J.; Davis, C.D.; Wang, T.T.Y. An improved method to quantitate mature plant microRNA in biological matrices using modified periodate treatment and inclusion of internal controls. PLoS ONE 2017, 12, e0175429. [Google Scholar] [CrossRef] [PubMed]
- Micó, V.; Martín, R.; Lasunción, M.A.; Ordovás, J.M.; Daimiel, L. Unsuccessful detection of plant microRNAs in beer, extra virgin olive oil and human plasma after an acute ingestion of extra virgin olive oil. Plant Foods Hum. Nutr. 2016, 71, 102–108. [Google Scholar] [CrossRef] [PubMed]
- Zhu, K.; Liu, M.; Fu, Z.; Zhou, Z.; Kong, Y.; Liang, H.; Lin, Z.; Luo, J.; Zheng, H.; Wan, P.; et al. Plant microRNAs in larval food regulate honeybee caste development. PLoS Genet. 2017, 13, e1006946. [Google Scholar] [CrossRef] [PubMed]
- Evans, J.D.; Wheeler, D.E. Differential gene expression between developing queens and workers in the honey bee, Apis mellifera. Proc. Natl. Acad. Sci. USA 1999, 96, 5575–5580. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Su, S.; Geir, S.; Li, W.; Li, Z.; Zhang, S.; Chen, S.; Chen, R. Differential expression of miRNAs related to caste differentiation in the honey bee, Apis mellifera. Apidologie 2016, 47, 495–508. [Google Scholar] [CrossRef]
- Li, X.; Zhang, M.; Zhang, H. RNA interference of four genes in adult bactrocera dorsalis by feeding their dsRNAs. PLoS ONE 2011, 6, e17788. [Google Scholar] [CrossRef] [PubMed]
- Ashby, R.; Foret, S.; Searle, I.; Maleszka, R. MicroRNAs in honey bee caste determination. Sci. Rep. 2016, 6, 18794. [Google Scholar] [CrossRef] [PubMed]
- Haydak, M.H. Larval food and development of castes in the honeybee. J. Econ. Entomol. 1943, 36, 778–792. [Google Scholar] [CrossRef]
- Flanders, S.E. Caste in the honey bee. Insectes Soc. 1960, 7, 9–16. [Google Scholar] [CrossRef]
- Hou, D.; He, F.; Ma, L.; Cao, M.; Zhou, Z.; Wei, Z.; Xue, Y.; Sang, X.; Chong, H.; Tian, C.; et al. The potential atheroprotective role of plant MIR156a as a repressor of monocyte recruitment on inflamed human endothelial cells. J. Nutr. Biochem. 2018, 57, 197–205. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Yu, S.; Li, M.; Gui, X.; Li, P. Isolation of exosome-like nanoparticles and analysis of microRNAs derived from coconut water based on small RNA high-throughput sequencing. J. Agric. Food Chem. 2018, 66, 2749–2757. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Hang, C.; Zhang, Y.; Chen, M.; Meng, X.; Cao, Q.; Song, N.; Itkow, J.; Shen, F.; Yu, D. Dietary MIR-451 protects erythroid cells from oxidative stress via increasing the activity of Foxo3 pathway. Oncotarget 2017, 8, 107109–107124. [Google Scholar] [CrossRef] [PubMed]
- Lukasik, A.; Brzozowska, I.; Zielenkiewicz, U.; Zielenkiewicz, P. Detection of plant miRNAs abundance in human breast milk. Int. J. Mol. Sci. 2018, 19, 37. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Davis, C.D.; Wang, T.T.Y. Extensive degradation and low bioavailability of orally consumed corn miRNAs in mice. Nutrients 2018, 10, 215. [Google Scholar] [CrossRef] [PubMed]
- Masood, M.; Everett, C.P.; Chan, S.Y.; Snow, J.W. Negligible uptake and transfer of diet-derived pollen microRNAs in adult honey bees. RNA Biol. 2016, 13, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wiggins, B.E.; Lawrence, C.; Petrick, J.; Ivashuta, S.; Heck, G. Analysis of plant-derived miRNAs in animal small RNA datasets. BMC Genom. 2012, 13, 381. [Google Scholar] [CrossRef] [PubMed]
- Perge, P.; Nagy, Z.; Decmann, Á.; Igaz, I.; Igaz, P. Potential relevance of microRNAs in inter-species epigenetic communication, and implications for disease pathogenesis. RNA Biol. 2017, 14, 391–401. [Google Scholar] [CrossRef] [PubMed]
- Lukasik, A.; Zielenkiewicz, P. Plant microRNAs—Novel players in natural medicine? Int. J. Mol. Sci. 2017, 18, 9. [Google Scholar] [CrossRef] [PubMed]
- Rafiqi, M.; Ellis, J.G.; Ludowici, V.A.; Hardham, A.R.; Dodds, P.N. Challenges and progress towards understanding the role of effectors in plant-fungal interactions. Curr. Opin. Plant Biol. 2012, 15, 477–482. [Google Scholar] [CrossRef] [PubMed]
- Hilbi, H.; Haas, A. Secretive bacterial pathogens and the secretory pathway. Traffic 2012, 13, 1187–1197. [Google Scholar] [CrossRef] [PubMed]
- Weiberg, A.; Wang, M.; Lin, F.-M.; Zhao, H.; Zhang, Z.; Kaloshian, I.; Huang, H.-D.; Jin, H. Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science 2013, 342, 118–123. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Weiberg, A.; Lin, F.-M.; Thomma, B.P.H.J.; Huang, H.-D.; Jin, H. Bidirectional cross-kingdom RNAi and fungal uptake of external RNAs confer plant protection. Nat. Plants 2016, 2, 16151. [Google Scholar] [CrossRef] [PubMed]
- Schwessinger, B. Fundamental wheat stripe rust research in the 21st century. New Phytol. 2017, 213, 1625–1631. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Sun, Y.; Song, N.; Zhao, M.; Liu, R.; Feng, H.; Wang, X.; Kang, Z. Puccinia striiformis f. sp. Tritici microRNA-like RNA 1 (pst-milR1), an important pathogenicity factor of pst, impairs wheat resistance to pst by suppressing the wheat pathogenesis-related 2 gene. New Phytol. 2017, 215, 338–350. [Google Scholar] [CrossRef] [PubMed]
- Hayden, M.S.; Ghosh, S. Signaling to NF-κB. Genes Dev. 2004, 18, 2195–2224. [Google Scholar] [CrossRef] [PubMed]
- Hancock, M.H.; Hook, L.M.; Mitchell, J.; Nelson, J.A. Human cytomegalovirus microRNAs MIR-us5-1 and MIR-ul112-3p block proinflammatory cytokine production in response to NF-κB-activating factors through direct downregulation of IKKα and IKKβ. mBio 2017, 8, e00109-17. [Google Scholar] [CrossRef] [PubMed]
- Shahid, S.; Kim, G.; Johnson, N.R.; Wafula, E.; Wang, F.; Coruh, C.; Bernal-Galeano, V.; Phifer, T.; dePamphilis, C.W.; Westwood, J.H.; et al. MicroRNAs from the parasitic plant Cuscuta campestris target host messenger RNAs. Nature 2018, 553, 82–85. [Google Scholar] [CrossRef] [PubMed]
- Jekat, S.; Ernst, A.; von Bohl, A.; Zielonka, S.; Twyman, R.; Noll, G.; Prüfer, D. P-proteins in Arabidopsis are heteromeric structures involved in rapid sieve tube sealing. Front. Plant Sci. 2013, 4, 225. [Google Scholar] [CrossRef] [PubMed]
- Froelich, D.R.; Mullendore, D.L.; Jensen, K.H.; Ross-Elliott, T.J.; Anstead, J.A.; Thompson, G.A.; Pélissier, H.C.; Knoblauch, M. Phloem ultrastructure and pressure flow: Sieve-element-occlusion-related agglomerations do not affect translocation. Plant Cell 2011, 23, 4428–4445. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.; Lu, D.; Gao, X.; Jiang, S.; Ma, X.; Wang, Z.; Mengiste, T.; He, P.; Shan, L. Inverse modulation of plant immune and brassinosteroid signaling pathways by the receptor-like cytoplasmic kinase BIK1. Proc. Natl. Acad. Sci. USA 2013, 110, 12114–12119. [Google Scholar] [CrossRef] [PubMed]
- Veronese, P.; Nakagami, H.; Bluhm, B.; AbuQamar, S.; Chen, X.; Salmeron, J.; Dietrich, R.A.; Hirt, H.; Mengiste, T. The membrane-anchored botrytis-induced kinase1 plays distinct roles in Arabidopsis resistance to necrotrophic and biotrophic pathogens. Plant Cell 2006, 18, 257–273. [Google Scholar] [CrossRef] [PubMed]
- Dharmasiri, N.; Dharmasiri, S.; Weijers, D.; Lechner, E.; Yamada, M.; Hobbie, L.; Ehrismann, J.S.; Jürgens, G.; Estelle, M. Plant development is regulated by a family of auxin receptor F box proteins. Dev. Cell 2005, 9, 109–119. [Google Scholar] [CrossRef] [PubMed]
- Sunkar, R.; Li, Y.F.; Jagadeeswaran, G. Functions of microRNAs in plant stress responses. Trends Plant Sci. 2012, 17, 196–203. [Google Scholar] [CrossRef] [PubMed]
- Wu, J. MiRNAs as a secret weapon in the battlefield of haustoria, the interface between parasites and host plants. Mol. Plant 2018, 11, 354–356. [Google Scholar] [CrossRef] [PubMed]
- Trenkmann, M. Plant genetics: Parasites plant microRNAs in the host. Nat. Rev. Genet. 2018, 19, 127. [Google Scholar] [CrossRef] [PubMed]
- Bhat, R.G.; Subbarao, K.V. Host range specificity in Verticillium dahliae. Phytopathology 1999, 89, 1218–1225. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Zhao, Y.-L.; Zhao, J.-H.; Wang, S.; Jin, Y.; Chen, Z.-Q.; Fang, Y.-Y.; Hua, C.-L.; Ding, S.-W.; Guo, H.-S. Cotton plants export microRNAs to inhibit virulence gene expression in a fungal pathogen. Nat. Plants 2016, 2, 16153. [Google Scholar] [CrossRef] [PubMed]
- Bzhalava, D.; Guan, P.; Franceschi, S.; Dillner, J.; Clifford, G. A systematic review of the prevalence of mucosal and cutaneous human papillomavirus types. Virology 2013, 445, 224–231. [Google Scholar] [CrossRef] [PubMed]
- Lecellier, C.H.; Dunoyer, P.; Arar, K.; Lehmann-Che, J.; Eyquem, S.; Himber, C.; Saib, A.; Voinnet, O. A cellular microRNA mediates antiviral defense in human cells. Science 2005, 308, 557–560. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Xie, J.; Jia, L.; Liu, N.; Liang, Y.; Wu, F.; Liang, B.; Li, Y.; Wang, J.; Sheng, C.; et al. Analysis of miRNAs involved in mouse brain damage upon enterovirus 71 infection. Front. Cell. Infect. Microbiol. 2017, 7, 133. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Ouyang, J.; Wei, J.; Maarouf, M.; Chen, J.L. Involvement of host non-coding RNAs in the pathogenesis of the influenza virus. Int. J. Mol. Sci. 2016, 18, 39. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, I.M.; Cheng, G.; Wieland, S.; Volinia, S.; Croce, C.M.; Chisari, F.V.; David, M. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature 2007, 449, 919–922. [Google Scholar] [CrossRef] [PubMed]
- Sannigrahi, M.K.; Sharma, R.; Singh, V.; Panda, N.K.; Rattan, V.; Khullar, M. Role of host miRNA hsa-mir-139-3p in hpv-16–induced carcinomas. Clin. Cancer Res. 2017, 23, 3884–3895. [Google Scholar] [CrossRef] [PubMed]
- Cappelle, K.; de Oliveira, C.F.; Van Eynde, B.; Christiaens, O.; Smagghe, G. The involvement of clathrin-mediated endocytosis and two SID-1-like transmembrane proteins in double-stranded RNA uptake in the colorado potato beetle midgut. Insect Mol. Biol. 2016, 25, 315–323. [Google Scholar] [CrossRef] [PubMed]
- Hinas, A.; Wright, A.J.; Hunter, C.P. SID-5 is an endosome-associated protein required for efficient systemic RNAi in C. elegans. Curr. Biol. 2012, 22, 1938–1943. [Google Scholar] [CrossRef] [PubMed]
- Van der Pol, E.; Boing, A.N.; Harrison, P.; Sturk, A.; Nieuwland, R. Classification, functions, and clinical relevance of extracellular vesicles. Pharmacol. Rev. 2012, 64, 676–705. [Google Scholar] [CrossRef] [PubMed]
- Cocucci, E.; Racchetti, G.; Meldolesi, J. Shedding microvesicles: Artefacts no more. Trends Cell Biol. 2009, 19, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Sharples, R.A.; Scicluna, B.J.; Hill, A.F. Exosomes provide a protective and enriched source of miRNA for biomarker profiling compared to intracellular and cell-free blood. J. Extracell. Vesicles 2014, 3, 23743. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Yuan, T.; Tschannen, M.; Sun, Z.; Jacob, H.; Du, M.; Liang, M.; Dittmar, R.L.; Liu, Y.; Liang, M.; et al. Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genom. 2013, 14, 319. [Google Scholar] [CrossRef] [PubMed]
- Mu, J.; Zhuang, X.; Wang, Q.; Jiang, H.; Deng, Z.B.; Wang, B.; Zhang, L.; Kakar, S.; Jun, Y.; Miller, D.; et al. Interspecies communication between plant and mouse gut host cells through edible plant derived exosome-like nanoparticles. Mol. Nutr. Food Res. 2014, 58, 1561–1573. [Google Scholar] [CrossRef] [PubMed]
- Ju, S.; Mu, J.; Dokland, T.; Zhuang, X.; Wang, Q.; Jiang, H.; Xiang, X.; Deng, Z.-B.; Wang, B.; Zhang, L.; et al. Grape exosome-like nanoparticles induce intestinal stem cells and protect mice from DSS-induced colitis. Mol. Ther. 2013, 21, 1345–1357. [Google Scholar] [CrossRef] [PubMed]
- Regente, M.; Corti-Monzón, G.; Maldonado, A.M.; Pinedo, M.; Jorrín, J.; de la Canal, L. Vesicular fractions of sunflower apoplastic fluids are associated with potential exosome marker proteins. FEBS Lett. 2009, 583, 3363–3366. [Google Scholar] [CrossRef] [PubMed]
- Prado, N.; Alche Jde, D.; Casado-Vela, J.; Mas, S.; Villalba, M.; Rodriguez, R.; Batanero, E. Nanovesicles are secreted during pollen germination and pollen tube growth: A possible role in fertilization. Mol. Plant 2014, 7, 573–577. [Google Scholar] [CrossRef] [PubMed]
- Alder, M.N.; Dames, S.; Gaudet, J.; Mango, S.E. Gene silencing in Caenorhabditis elegans by transitive RNA interference. RNA 2003, 9, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAS: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [PubMed]
- Ashby, B.; Boots, M. Multi-mode fluctuating selection in host-parasite coevolution. Ecol. Lett. 2017, 20, 357–365. [Google Scholar] [CrossRef] [PubMed]
- Allegra, A.; Alonci, A.; Campo, S.; Penna, G.; Petrungaro, A.; Gerace, D.; Musolino, C. Circulating microRNAs: New biomarkers in diagnosis, prognosis and treatment of cancer (review). Int. J. Oncol. 2012, 41, 1897–1912. [Google Scholar] [CrossRef] [PubMed]
- Leonti, M.; Casu, L. Traditional medicines and globalization: Current and future perspectives in ethnopharmacology. Front. Pharmacol. 2013, 4, 92. [Google Scholar] [CrossRef] [PubMed]
- Mochida, K.; Sakurai, T.; Seki, H.; Yoshida, T.; Takahagi, K.; Sawai, S.; Uchiyama, H.; Muranaka, T.; Saito, K. Draft genome assembly and annotation of Glycyrrhiza uralensis, a medicinal legume. Plant J. 2017, 89, 181–194. [Google Scholar] [CrossRef] [PubMed]
- Shao, H.W.; He, M.; Chen, J.S.; Chen, H.; Xiang, J.; Huang, S.L. Extraction of miRNA from Glycyrrhiza uralensis decoction and its effect on immune cells. J. Chin. Med. Mater. 2015, 38, 1449–1453. [Google Scholar]
- Xiang, J.; Huang, J.C.; Xu, C.; He, M.; Xu, P.C.; Zhang, D.; Zhang, W.F.; Shen, H.; Shao, H.W. Effect of miRNA from Glycyrrhiza uralensis decoction on gene expression of human immune cells. China J. Chin. Mater. Med. 2017, 42, 1752–1756. [Google Scholar]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, W.; Liu, D.; Zhang, X.; Chen, D.; Cheng, Y.; Shen, F. Plant MicroRNAs in Cross-Kingdom Regulation of Gene Expression. Int. J. Mol. Sci. 2018, 19, 2007. https://doi.org/10.3390/ijms19072007
Wang W, Liu D, Zhang X, Chen D, Cheng Y, Shen F. Plant MicroRNAs in Cross-Kingdom Regulation of Gene Expression. International Journal of Molecular Sciences. 2018; 19(7):2007. https://doi.org/10.3390/ijms19072007
Chicago/Turabian StyleWang, Wei, Dan Liu, Xiaopei Zhang, Dongdong Chen, Yingying Cheng, and Fafu Shen. 2018. "Plant MicroRNAs in Cross-Kingdom Regulation of Gene Expression" International Journal of Molecular Sciences 19, no. 7: 2007. https://doi.org/10.3390/ijms19072007
APA StyleWang, W., Liu, D., Zhang, X., Chen, D., Cheng, Y., & Shen, F. (2018). Plant MicroRNAs in Cross-Kingdom Regulation of Gene Expression. International Journal of Molecular Sciences, 19(7), 2007. https://doi.org/10.3390/ijms19072007