Disruption of PTPS Gene Causing Pale Body Color and Lethal Phenotype in the Silkworm, Bombyx mori
Abstract
1. Introduction
2. Results
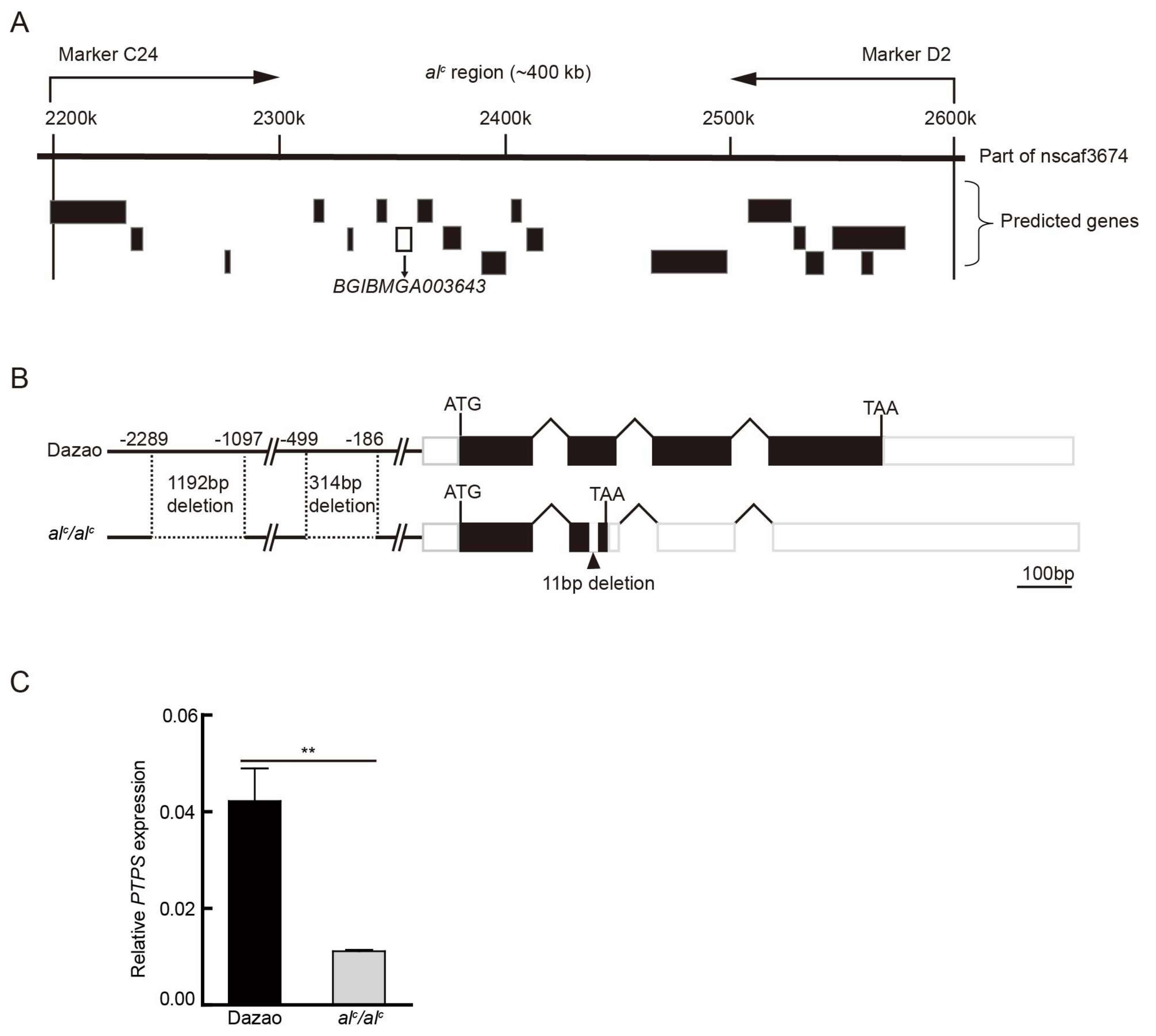
2.1. Fine Mapping and Gene Cloning
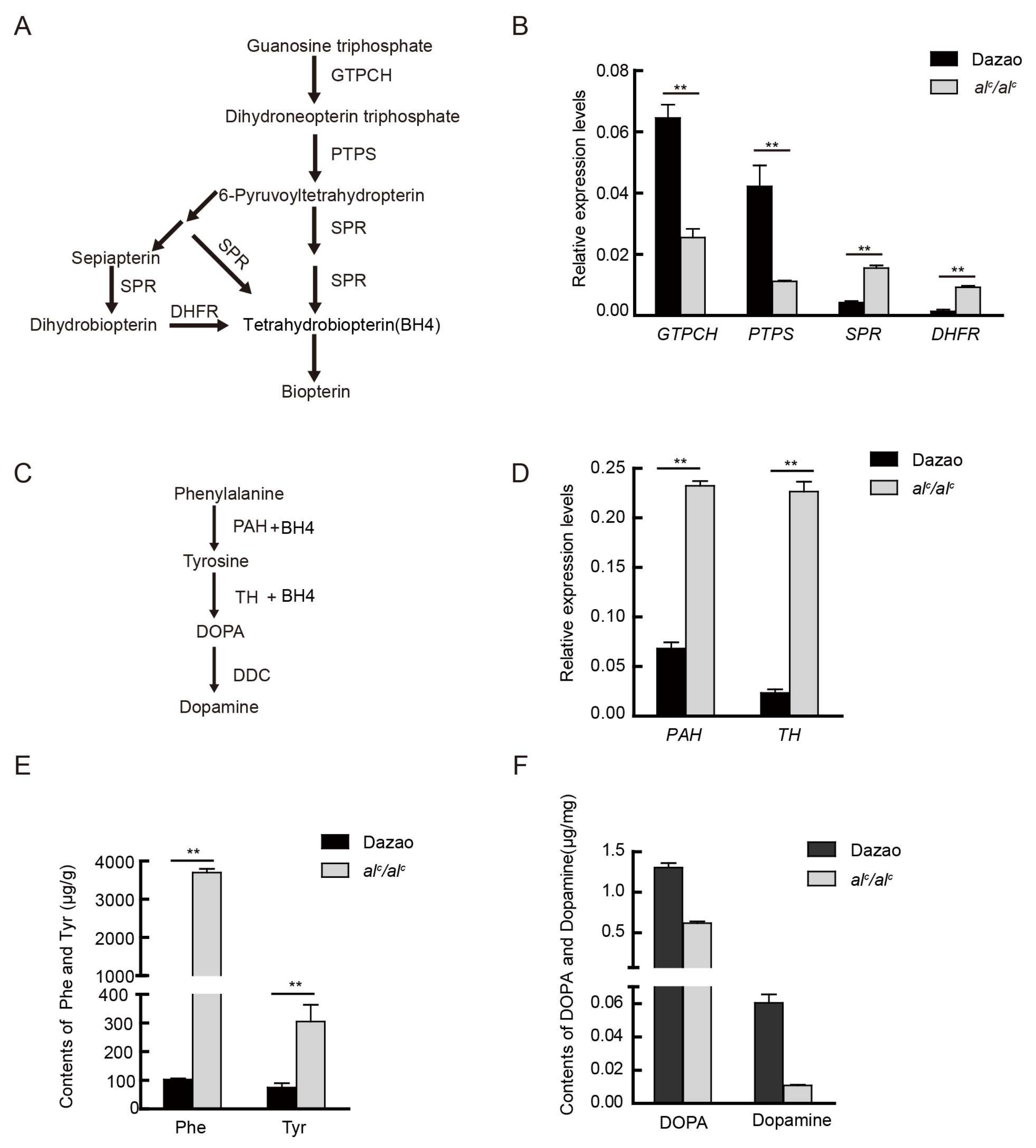
2.2. Expression Levels of Key Genes Involved in the BH4 Pathway
2.3. The Expression Level of Key Genes and Content of Substances in the Melanin Metabolic Pathway
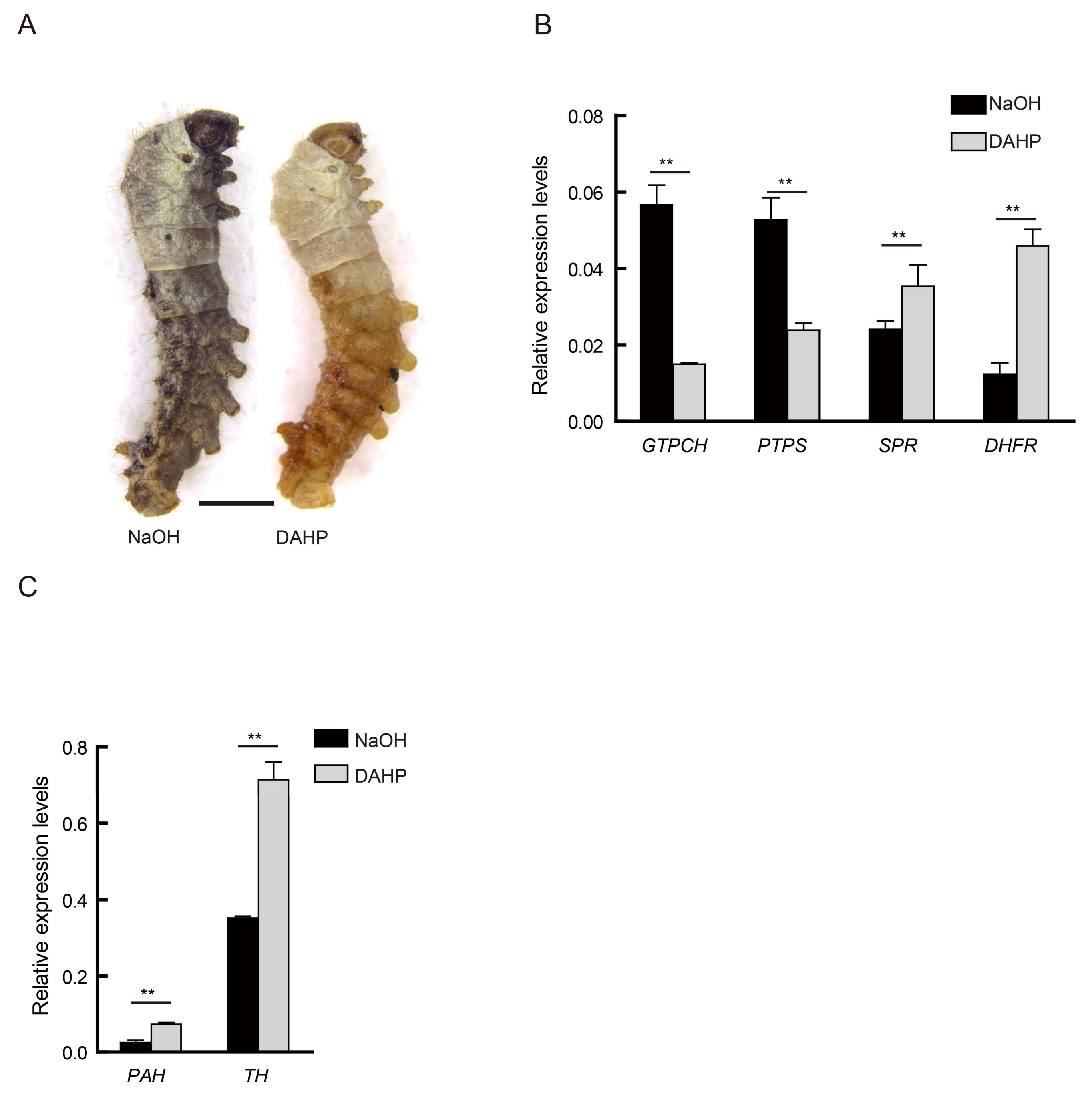
2.4. Treatment Using a BH4 Inhibitor
2.5. Treatment of BH4 Deficiency
3. Discussion
4. Materials and Methods
4.1. Silkworm Strains
4.2. DNA Extraction and Sample Preparation
4.3. Fine Mapping of Albino C Locus
4.4. Cloning of the PTPS Gene
4.5. Quantitative RT-PCR Analysis
4.6. Chemicals
4.7. Quantification of Amino Acids and Catecholamine
4.8. DAHP Feeding Experiments
4.9. BH4 Feeding Experiments
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| PKU | phenylketonuria |
| PAH | phenylalanine hydroxylase |
| BH4 | tetrahydrobiopterin |
| PTPS | 6-pyruvoyl-tetrahydropterin synthase |
| SilkDB | Silkworm Genome Database |
| GTPCH | guanosine triphosphate cyclohydrolase I |
| SPR | sepiapterin reductase |
| DHFR | dihydrofolate reductase |
| NADA | N-acetyl dopamine |
| NBAD | N-β-alanyldopamine |
| DAHP | hydroxypyrimidine |
References
- Blau, N.; van Spronsen, F.J.; Levy, H.L. Phenylketonuria. Lancet 2010, 376, 1417–1427. [Google Scholar] [CrossRef]
- Blau, N.; Shen, N.; Carducci, C. Molecular genetics and diagnosis of phenylketonuria: State of the art. Exp. Rev. Mol. Diagn. 2014, 14, 655–671. [Google Scholar] [CrossRef] [PubMed]
- Berry, S.A.; Brown, C.; Grant, M.; Greene, C.L.; Jurecki, E.; Koch, J.; Moseley, K.; Suter, R.; van Calcar, S.C.; Wiles, J.; et al. Newborn screening 50 years later: Access issues faced by adults with PKU. Genet. Med. Off. J. Am. Coll. Med. Genet. 2013, 15, 591–599. [Google Scholar] [CrossRef] [PubMed]
- Guthrie, R.; Susi, A. A Simple Phenylalanine Method for Detecting Phenylketonuria in Large Populations of Newborn Infants. Pediatrics 1963, 32, 338–343. [Google Scholar] [PubMed]
- Christ, S.E.; Moffitt, A.J.; Peck, D.; White, D.A. The effects of tetrahydrobiopterin (BH4) treatment on brain function in individuals with phenylketonuria. Neuroimage Clin. 2013, 3, 539–547. [Google Scholar] [CrossRef] [PubMed]
- Heine, C.L.; Kolesnik, B.; Schmidt, R.; Werner, E.R.; Mayer, B.; Gorren, A.C. Interaction between neuronal nitric-oxide synthase and tetrahydrobiopterin revisited: Studies on the nature and mechanism of tight pterin binding. Biochemistry 2014, 53, 1284–1295. [Google Scholar] [CrossRef] [PubMed]
- Martinez, A.; Knappskog, P.M.; Haavik, J. A structural approach into human tryptophan hydroxylase and its implications for the regulation of serotonin biosynthesis. Curr. Med. Chem. 2001, 8, 1077–1091. [Google Scholar] [CrossRef] [PubMed]
- Thony, B.; Calvo, A.C.; Scherer, T.; Svebak, R.M.; Haavik, J.; Blau, N.; Martinez, A. Tetrahydrobiopterin shows chaperone activity for tyrosine hydroxylase. J. Neurochem. 2008, 106, 672–681. [Google Scholar] [CrossRef] [PubMed]
- Xia, Q.; Zhou, Z.; Lu, C.; Cheng, D.; Dai, F.; Li, B.; Zhao, P.; Zha, X.; Cheng, T.; Chai, C.; et al. A draft sequence for the genome of the domesticated silkworm (Bombyx mori). Science 2004, 306, 1937–1940. [Google Scholar] [PubMed]
- Goldsmith, M.R.; Shimada, T.; Abe, H. The genetics and genomics of the silkworm, Bombyx mori. Ann. Rev. Entomol. 2005, 50, 71–100. [Google Scholar] [CrossRef] [PubMed]
- Duan, J.; Li, R.; Cheng, D.; Fan, W.; Zha, X.; Cheng, T.; Wu, Y.; Wang, J.; Mita, K.; Xiang, Z.; et al. SilkDB v2.0: A platform for silkworm (Bombyx mori) genome biology. Nucleic Acids Res. 2010, 38, D453–D456. [Google Scholar] [CrossRef] [PubMed]
- Meng, Y.; Katsuma, S.; Daimon, T.; Banno, Y.; Uchino, K.; Sezutsu, H.; Tamura, T.; Mita, K.; Shimada, T. The silkworm mutant lemon (Lemon lethal) is a potential insect model for human sepiapterin reductase deficiency. J. Biol. Chem. 2009, 284, 11698–11705. [Google Scholar] [CrossRef] [PubMed]
- Delgado-Esteban, M.; Almeida, A.; Medina, J.M. Tetrahydrobiopterin deficiency increases neuronal vulnerability to hypoxia. J. Neurochem. 2002, 82, 1148–1159. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, B.M.; Dorrance, A.M.; Webb, R.C. GTP cyclohydrolase 1 inhibition attenuates vasodilation and increases blood pressure in rats. Am. J. Physiol. Heart Circ. Physiol. 2003, 285, H2165–H2170. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Opladen, T.; Hoffmann, G.F.; Blau, N. An international survey of patients with tetrahydrobiopterin deficiencies presenting with hyperphenylalaninaemia. J. Inherit. Metab. Dis. 2012, 35, 963–973. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Teng, X.; Chen, M.; Li, F. Orthologs of human disease associated genes and RNAi analysis of silencing insulin receptor gene in Bombyx mori. Int. J. Mol. Sci. 2014, 15, 18102–18116. [Google Scholar] [CrossRef] [PubMed]
- Tabunoki, H.; Ono, H.; Ode, H.; Ishikawa, K.; Kawana, N.; Banno, Y.; Shimada, T.; Nakamura, Y.; Yamamoto, K.; Satoh, J.; et al. Identification of key uric acid synthesis pathway in a unique mutant silkworm Bombyx mori model of Parkinson’s disease. PLoS ONE 2013, 8, e69130. [Google Scholar] [CrossRef] [PubMed]
- Fujii, T.; Daimon, T.; Uchino, K.; Banno, Y.; Katsuma, S.; Sezutsu, H.; Tamura, T.; Shimada, T. Transgenic analysis of the BmBLOS2 gene that governs the translucency of the larval integument of the silkworm, Bombyx mori. Insect Mol. Biol. 2010, 19, 659–667. [Google Scholar] [CrossRef] [PubMed]
- Hanada, Y.; Sekimizu, K.; Kaito, C. Silkworm apolipophorin protein inhibits Staphylococcus aureus virulence. J. Biol. Chem. 2011, 286, 39360–39369. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Song, J.; Chen, M.; Li, Z.; Tong, X.; Hu, H.; Xiang, Z.; Lu, C.; Dai, F. Rhodiola rosea extends lifespan and improves stress tolerance in silkworm, Bombyx mori. Biogerontology 2016, 17, 373–381. [Google Scholar] [CrossRef] [PubMed]
- Koch, P.B.; Behnecke, B.; Weigmann-Lenz, M.; Ffrench-Constant, R.H. Insect pigmentation: Activities of β-alanyldopamine synthase in wing color patterns of wild-type and melanic mutant swallowtail butterfly Papilio glaucus. Pigment Cell Melanoma Res. 2000, 13 (Suppl. 8), 54–58. [Google Scholar] [CrossRef]


© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tong, X.; Liang, P.; Wu, S.; Li, Y.; Qiao, L.; Hu, H.; Xiang, Z.; Lu, C.; Dai, F. Disruption of PTPS Gene Causing Pale Body Color and Lethal Phenotype in the Silkworm, Bombyx mori. Int. J. Mol. Sci. 2018, 19, 1024. https://doi.org/10.3390/ijms19041024
Tong X, Liang P, Wu S, Li Y, Qiao L, Hu H, Xiang Z, Lu C, Dai F. Disruption of PTPS Gene Causing Pale Body Color and Lethal Phenotype in the Silkworm, Bombyx mori. International Journal of Molecular Sciences. 2018; 19(4):1024. https://doi.org/10.3390/ijms19041024
Chicago/Turabian StyleTong, Xiaoling, Pingfeng Liang, Songyuan Wu, Yuanhao Li, Liang Qiao, Hai Hu, Zhonghuai Xiang, Cheng Lu, and Fangyin Dai. 2018. "Disruption of PTPS Gene Causing Pale Body Color and Lethal Phenotype in the Silkworm, Bombyx mori" International Journal of Molecular Sciences 19, no. 4: 1024. https://doi.org/10.3390/ijms19041024
APA StyleTong, X., Liang, P., Wu, S., Li, Y., Qiao, L., Hu, H., Xiang, Z., Lu, C., & Dai, F. (2018). Disruption of PTPS Gene Causing Pale Body Color and Lethal Phenotype in the Silkworm, Bombyx mori. International Journal of Molecular Sciences, 19(4), 1024. https://doi.org/10.3390/ijms19041024




