Abstract
Recently, anoctamin1 (ANO1), a calcium-activated chloride channel, has been considered an important drug target, due to its involvement in various physiological functions, as well as its possibility for treatment of cancer, pain, diarrhea, hypertension, and asthma. Although several ANO1 inhibitors have been discovered by high-throughput screening, a discovery of new ANO1 inhibitors is still in the early phase, in terms of their potency and specificity. Moreover, there is no computational model to be able to identify a novel lead candidate of ANO1 inhibitor. Therefore, three-dimensional quantitative structure-activity relationship (3D-QSAR) pharmacophore modeling approach was employed for identifying the essential chemical features to be required in the inhibition of ANO1. The pharmacophore hypothesis 2 (Hypo2) was selected as the best model based on the highest correlation coefficient of prediction on the test set (0.909). Hypo2 comprised a hydrogen bond acceptor, a hydrogen bond donor, a hydrophobic, and a ring aromatic feature with good statistics of the total cost (73.604), the correlation coefficient of the training set (0.969), and the root-mean-square deviation (RMSD) value (0.946). Hypo2 was well assessed by the test set, Fischer randomization, and leave-one-out methods. Virtual screening of the ZINC database with Hypo2 retrieved the 580 drug-like candidates with good potency and ADMET properties. Finally, two compounds were selected as novel lead candidates of ANO1 inhibitor, based on the molecular docking score and the interaction analysis. In this study, the best pharmacophore model, Hypo2, with notable predictive ability was successfully generated, and two potential leads of ANO1 inhibitors were identified. We believe that these compounds and the 3D-QSAR pharmacophore model could contribute to discovering novel and potent ANO1 inhibitors in the future.
1. Introduction
Anoctamin1 (ANO1/TMEM16A) is a calcium-activated chloride channel (CaCC) that responds to an increase of intracellular Ca2+ concentration [1,2,3]. Since the time when the molecular identity of ANO1 was deciphered by the three independent groups in 2008 [1,2,3], various aspects of physiological and pathological relevance of ANO1 have been discovered up to now. ANO1 is ubiquitously expressed in many tissues [4] and it is known to play important roles in fluid secretion, smooth muscle contraction, nociception, insulin secretion, cell proliferation, and migration [5,6]. In addition, ANO1 has emerged as a new drug target for the treatment of cancer, pain, diarrhea, hypertension, and asthma [5,6,7].
Despite the important biological role of ANO1, a discovery of new ANO1 inhibitors is still in the early phase. Due to the absence of structural information of ANO1 until 2017, most of the ANO1 inhibitors have been discovered through high-throughput chemical library screening using a yellow fluorescent protein (YFP)-iodide based sensor [7,8]. To date, several ANO1 inhibitors such as CaCCinh-A01 [9], T16inh-A01 [10], MONNA [11], benzbromarone [12], Ani9 [13], tannic acid [14], eugenol [15], luteolin [16], and crofelemer [17] have been discovered from both the chemical and natural product space. Of these, CaCCinh-A01, T16inh-A01, MONNA, and Ani9 are the most potent chemical inhibitors of ANO1, whose half maximal inhibitory concentration (IC50) values range from 100 nM to 3 μM [9,10,11,13]. Natural product inhibitors have IC50 values up to 10 μM or more, which are higher than those of chemical inhibitors [14,15,16,17]. Among the natural product inhibitors of ANO1, the crofelemer (previously known as Fulyzaq and now as Mytesi) from Napo Pharmaceutical in 2012 is the first FDA-approved drug to be used for anti-human immunodeficiency virus (HIV) associated anti-diarrhea, through targeting the ANO1 [17,18].
Although many ANO1 inhibitors have been experimentally discovered, most of the ANO1 inhibitors still have revealed a low potency and selectivity (μM level) [5]. In addition, many ANO1 inhibitors have revealed the inhibition of the structurally similar ANO2 (62% of amino acid homology) [3,19], and also the other ion channels such as CFTR, ENaC, and BEST1 [5,20]. Therefore, there is a need to find more potent and selective inhibitors as novel lead candidates targeting ANO1. Although high-throughput chemical library screening has been successfully explored several ANO1 inhibitors so far, it is very labor intensive and has a low hit-rate compared to the effort required. In addition, there is still no available structural information regarding the discovery of novel leads of ANO1.
We aimed to generate a chemical feature-based pharmacophore model for identifying novel lead candidates with the potential to be ANO1 inhibitors. The pharmacophore model contains abstract features that define interaction types that are necessary for chemicals’ biological activities [21]. Thus, the virtual screening of a chemical library using the pharmacophore model could usually guide the design of novel lead candidates. A ligand-based pharmacophore modeling approach with subsequent molecular docking study has identified several novel lead candidates of renin, tubulin, PDE4, BACE1, AKR1B10, and so on [22,23,24,25,26]. Moreover, the structures of mouse ANO1 have been elucidated recently by cryo-electron microscopy (cryo-EM) techniques [27,28,29]. Thus, both ligand-based and structure-based drug discovery approaches are able to be applied for identifying the novel ANO1 inhibitor candidates.
In this study, a three-dimensional quantitative structure–activity relationship (3D-QSAR) pharmacophore model was generated based on the chemical features present in known ANO1 inhibitors. Then, virtual screening of large chemical database (ZINC) was carried out to obtain novel lead candidates using the pharmacophore model. The retrieved hit compounds were subjected to molecular docking for further characterization. Finally, final hit compounds with a favorable binding mode to ANO1 were identified as novel lead candidates.
2. Results and Discussion
2.1. Pharmacophore Model Generation
The HypoGen algorithm in Discovery Studio 3.1 (DS) from Accelrys (San Diego, CA, USA) was implemented to generate the 3D-QSAR pharmacophore model that can quantitatively predict the biological activity (IC50) of given compounds against ANO1. The 20 compounds with activities ranging from 0.107 to 29.2 μM in the training set were used to train the pharmacophore models (Figure 1). For identifying the meaningful pharmacophore features from the training set, the Feature mapping of DS was carried out, and it resulted in hydrogen bond donor (HBD), hydrogen bond acceptor (HBA), hydrophobic (HY), and ring aromatic (RA) features. As a result of the HypoGen algorithm with the training set and their pharmacophore features, 10 top-scored hypothetical pharmacophore models were generated with their statistical parameters (Table 1).
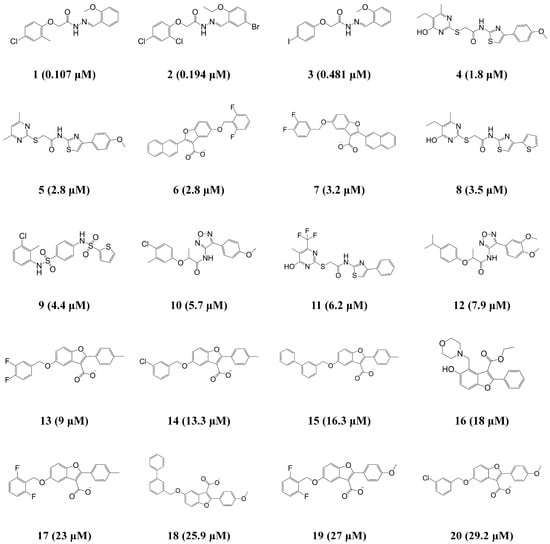
Figure 1.
Two-dimensional (2D) chemical structures of the 20 training set compounds with their experimental half maximal inhibitory concentration (IC50) values.

Table 1.
Statistical results of the top 10 pharmacophore hypotheses generated by HypoGen algorithm.
Then, a cost analysis was conducted to assess the quality of the generated pharmacophore hypotheses. We used the following values for assessing the hypotheses: (i) the cost difference between null cost and fixed cost; (ii) the cost difference between null cost and total cost; (iii) the configuration cost; (iv) the root-mean-square deviation (RMSD); (v) the correlation coefficient of the training set (r2training). First, the 10 hypotheses were developed with the fixed cost of 63.804 and the null cost of 194.608. The larger cost difference (130.804) between the null cost and the fixed cost implied that the first and second models (Hypo1 and Hypo2) are meaningful models that have more statistical significance than 90%. Second, the total cost of the 10 pharmacophore hypotheses ranged from 70.969 to 87.091. Among them, the total cost of Hypo1 and Hypo2 were calculated to be closer to the fixed cost than other models. The cost differences between the null cost and the total cost of the Hypo1 and Hypo2 were 123.64 and 121.00, respectively. Third, the configuration cost in this study was 15.343, which would be less than 17 [23]. This indicates that the correlation from the generated pharmacophores cannot be attributed to chance. Fourth, the RMSD values of the top two pharmacophore hypotheses were less than 1, which illustrates the good regression ability of these hypotheses. Finally, the r2training values of the 10 pharmacophore models were greater than 0.9, and the top two models have the higher r2training values above 0.95 indicating the strong capability to predict the biological activity of the training set compounds. Among the 10 hypotheses, eight of 10 hypotheses including top two hypotheses consist of HBA, HBD, HY, and RA features.
2.2. Pharmacophore Model Evaluation and Selection
Based on the results of the cost analysis, the Hypo1 model should be selected as the best model for further analysis. However, the important factor in order to be the best pharmacophore model is that whether the generated pharmacophore model has the best ability to estimate the activity of new compounds which are retrieved from a database consisting of activity-unknown compounds. Therefore, the correlation coefficient of the test set (r2test) value was used as the primary criterion to select the most reliable pharmacophore model among the 10 generated hypotheses. The 32 compounds with activity ranging from 0.301 to 28.7 μM were selected as the test set (Figure 2). Diverse conformations of the test set compounds were prepared in the same manner as the training set preparation. To assess the regression ability of the generated pharmacophore models about the test set, each generated pharmacophore hypothesis was applied to predict the biological activity of the 32 test set compounds. As a result, the r2test values of each hypothesis were calculated (Table 1), and only one hypothesis (Hypo2) revealed the high correlation coefficient of 0.909. Despite the best results of Hypo1 in the cost analysis, the Hypo1 model showed an r2test value of 0.75. Compared to the cost analysis result of Hypo1, Hypo2 has similar statistical values in terms of the total cost (73.604), the cost difference (121.00), the RMSD (0.946), and the r2training (0.969). Hence, Hypo2, which consists of each of HBA, HBD, HY, and RA was determined as the best hypothesis model for further analysis (Figure 3A). The inter-spatial constraints are shown in Figure 3B.
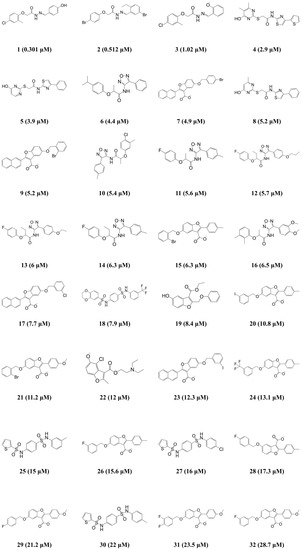
Figure 2.
2D chemical structures of the 32 test set compounds with their experimental IC50 values.
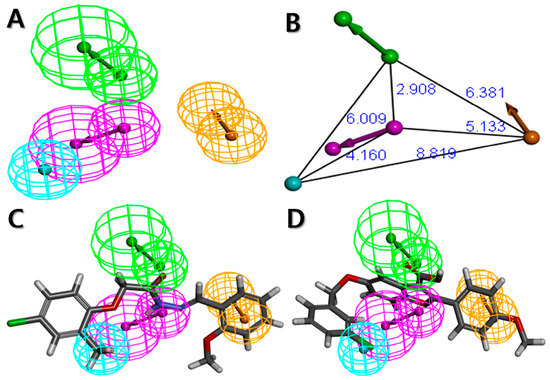
Figure 3.
The best HypoGen pharmacophore model, Hypo2. (A) Pharmacophore features present in Hypo2; (B) 3D spatial relationship and geometric parameters of Hypo2; (C) Mapping of the most active compound (compound 1) on Hypo2; (D) Mapping of the least active compound (compound 20) on Hypo2. Pharmacophore features are color-coded: magenta, hydrogen bond donor (HBD); green, hydrogen bond acceptor (HBA); cyan, hydrophobic (HY); orange, ring aromatic (RA).
To further explore the model performance of Hypo2 on the training set, the training set compounds were primarily categorized into following four groups based on experimental activities: most active (IC50 < 1 μM, ++++), active (1 ≤ IC50 < 8 μM, +++), moderately active (8 ≤ IC50 < 18 μM, ++), and inactive (IC50 ≥ 18 μM, +). The activities and its scales of the training set compounds were then estimated based on Hypo2. As shown in Table 2, only one compound (compound 17) was estimated wrongly as being moderately active (++), which is overestimated than the activity scale of experimental value. The rest of the compounds were estimated perfectly as their corresponding activity scales of experimental values, indicating the good predictability of the Hypo2. In addition, none of the training set compounds had an error value above 2.5, signifying that the activity scale difference is below one order of magnitude. Error value indicates the ratio between the estimated and experimental activities. Then, pharmacophore features of the Hypo2 were mapped to the training set compounds. The mapping results of the most and the least active compounds on the Hypo2 are shown in Figure 3C and D, respectively. The most active compound (compound 1) were well fitted to all the pharmacophore features of the Hypo2, consisting of each one of HBA, HBD, HY, and RA, and the fit value was 5.9. On the other hand, the least active compound (compound 20) was mapped to the three features of the Hypo2, except for HBD, and the fit value was 3.58. These results signified that Hypo2 is a significant model to accurately predict the experimental activity of the training set compounds.

Table 2.
Experimental and estimated IC50 values of the training set compounds based on the best pharmacophore hypothesis, Hypo2.
2.3. Pharmacophore Model Validation
To evaluate the best pharmacophore model (Hypo2) further, we used the test set, Fischer randomization, and leave-one-out methods.
2.3.1. Test Set Method
The prediction ability of Hypo2 had already evaluated at the best model selection step. The r2test value of the regression using Hypo2 revealed value of 0.909 (Figure 4). The test set compounds were also categorized into the four groups, which were at the same activity range as the training set compounds. The activities and its scales of the test set compounds were then estimated based on Hypo2 as well. As shown in Table 3, 29 of the 32 test set compounds revealed error values of less than 2. Five compounds (compound 3, 15, 22, 23, and 30) out of 32 test set compounds were estimated in a different activity scale resulting in a prediction rate of 84.4%. Among these five compounds, compound 15 was only underestimated, while the rest of the four compounds were overestimated compared to the scale of experimental activity. This overestimation tendency of the Hypo2 model implied that the Hypo2 model might act to minimize the false-negative compounds, in spite of increment of false-positive candidates. According to the above results, these results suggested that Hypo2 has a good prediction performance of ANO1 inhibitors from newly queried compounds.
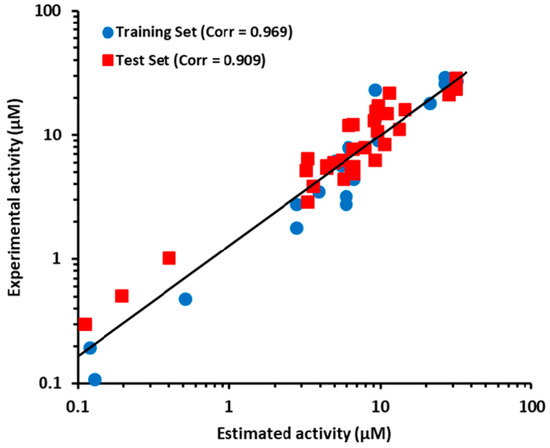
Figure 4.
The correlation graph between experimental and estimated activity values for the training set and test set compounds based on Hypo2.

Table 3.
Experimental and estimated IC50 values of the test set compounds based on the best pharmacophore hypothesis, Hypo2.
2.3.2. Fischer Randomization Method
To validate the statistical significance of the Hypo2 model, the Fischer randomization method was carried out based on the randomized activity data of the training set compounds. This method could test that the pharmacophore model was not generated by the random correlation between biological activities and structures of training set compounds. To reveal that the Hypo2 model was not generated by chance with a 95% confidence level, 19 hypotheses were generated from spreadsheets consisting of random relationships between chemical structures and biological activities (Figure 5). None of the 19 randomly generated hypotheses scored a lower total cost than that of Hypo2. This Fischer randomization result clearly indicates that Hypo2 is statistically robust and not generated by chance because Hypo2 has represented a true correlation in the training set.
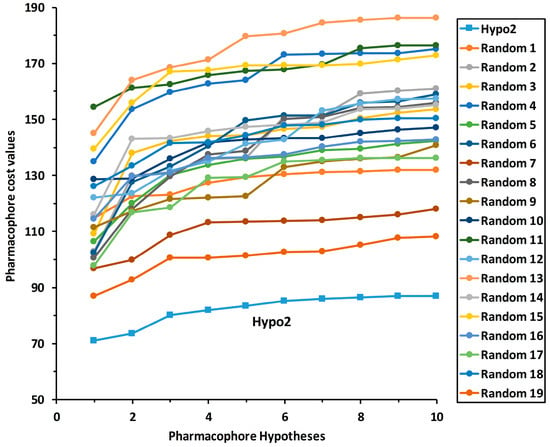
Figure 5.
Fischer randomization test results of Hypo2 for 95% confidence level.
2.3.3. Leave-One-Out Method
The leave-one-out (LOO) method was performed for validating Hypo2. The purpose of this validation method is to verify that the r2training value of Hypo2 is not only dependent on one particular compound in the training set. To this end, the 20 pharmacophore hypotheses were regenerated under the same parameter setting of the Hypo2 by omitting one compound at a time from the training set. If there is no meaningful difference of r2training value between Hypo2 and the newly generated hypothesis in the LOO step, the LOO test is passed. To assess the statistical significance of the Hypo2, a RMSD value between the r2training of the Hypo2 (0.969) and the mean value of r2training of each hypothesis from LOO was calculated. The RMSD of r2training from the LOO step was 0.0112, which was very small value, signifying that there is no significant difference between Hypo2 and each hypothesis from LOO. This result suggested that the Hypo2 is a robust model that is not affected by only one particular compound in the training set. Based on the aforementioned results, Hypo2 was determined for use in identifying potent candidates to be ANO1 inhibitors from a large chemical database.
2.4. Virtual Screening
The Hypo2 model was used to retrieve novel hit compounds from the ZINC database [32]. First, the Lipinski’s rule and the absorption, distribution, metabolism, excretion, and toxicity (ADMET) properties of 309,149 compounds were calculated. As a result of Lipinski’s rule and ADMET descriptor filtering, a total 19,701 compounds remained as drug-like candidates with good ADMET properties. Then, these filtered compounds were subsequently applied to the Ligand Pharmacophore Mapping of DS with the Hypo2 pharmacophore model. A total of 17,678 compounds were retrieved by pharmacophore mapping of the Hypo2. Among these 17,678 compounds, we finally selected 580 compounds as hit candidates by restricting the estimated activity to less than 100 nM. These 580 compounds that met all the above criteria were applied to molecular docking studies for further characterization. The flowchart of virtual screening is described in Figure 6.
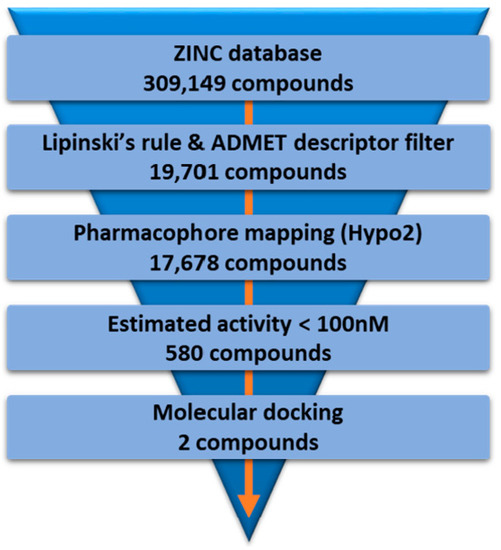
Figure 6.
The flowchart of virtual screening using Hypo2.
2.5. Molecular Docking
To reduce the false-positive candidates from the results of ligand-based virtual screening and to refine the novel lead candidates, the retrieved hit compounds, along with the most active (++++) and active (+++) groups compounds in the dataset were docked into ANO1, using the Libdock algorithm in DS. Although the protein structure of ANO1 is recently deciphered [27,28,29], the binding mode of ANO1 inhibitors on ANO1 is still unclear. Since the ANO1 is a CaCC class channel, many researchers have focused on the putative residues of calcium-binding site through the mutagenesis studies for deciphering association between each residue and calcium-dependent activity changes of ANO1 [29,33,34,35,36]. The influences of the following mutants to the activity of ANO1 have been reported so far: K588, K645, E654, E702, E705, E734, and D738 residues [29,33,34,35,36]. Then, these residues finally have been identified as the core residues of calcium-binding site and intracellular vestibule site [27,28]. Therefore, in this study, the active site was defined, based on the vicinity of the calcium-binding site in the structure of ANO1.
The Libdock scores (LS) were calculated for all 610 compounds, which consist of the 580 retrieved hit compounds along with the 30 compounds in the most active (++++) and active (+++) groups of the dataset. Interestingly, the most active (++++) group compounds including compound 1 of the training set, which were derived from the same scaffold structure, showed a much lower average LS value (102.35) compared to the average LS value of the other dataset compounds (114.49). This implied that the most active (++++) group compounds may interact with different binding sites for the strong inhibition of ANO1. The highest LS value of the dataset compounds was 135.61. To find a promising novel lead candidate of ANO1 inhibitors, hit compounds were selected based on the strict threshold greater than LS value of 155. As a result, only five compounds (ZINC8643627, ZINC225516955, ZINC8624466, ZINC225516884, and ZINC225519862) showed the LS values above 155. Among the five compounds, ZINC8643627 and ZINC8624466 had the same scaffold structure, and ZINC225516955, ZINC225516884, and ZINC225519862 had the same scaffold structure as well. Thus, we decided to analyze further only one compound per each scaffold structure, which had the highest LS value among the compounds of the same scaffold. Finally, two compounds (ZINC8643627 and ZINC225516955) with different scaffold structures were selected as lead candidate compounds of ANO1. ZINC8643627 and ZINC225516955 revealed LS values of 162.29 and 160.92, respectively. The binding modes of the two compounds are depicted in Figure 7. The calcium-binding site of ANO1 contains Q646, K650, N651, E654, E702, E705, N730, E734, and D738 [28]. The intracellular vestibule site of ANO1 contains E555, K588, K645, E654, E702, and K741 [28]. ZINC8643627 has formed hydrogen bond interactions with N650 and E702, which are the critical residues for calcium-ion binding according to the mutagenesis studies [33,34]. It has also formed Pi–cation interactions with K588 and K741, which are located in the intracellular vestibule site of ANO1 (Figure 7A). The ZINC225516955 formed hydrogen bond interactions with A697 and K741 as well as Pi–cation interaction with K741 (Figure 7B). The mapping results of the two final hit compounds on Hypo2 are shown in Figure 8. ZINC8643627 revealed a fit value of 6.29 and an estimated activity value of 0.052 μM. ZINC225516955 revealed a fit value of 6.04 and an estimated activity value of 0.092 μM. The 2D representations of the two final hit compounds with corresponding estimated scores are shown in Table 4. Interestingly, these two final hit compounds were both natural products, and they did not have any reports on the inhibitory activity of ANO1 based on the PubChem [37] search, signifying the novelty of the two final hit compounds. All of these results suggested that ZINC8643627 and ZINC225516955 may highly affect the calcium-binding and the intracellular vestibule site with low IC50 values, resulting in the current disturbance of ANO1.
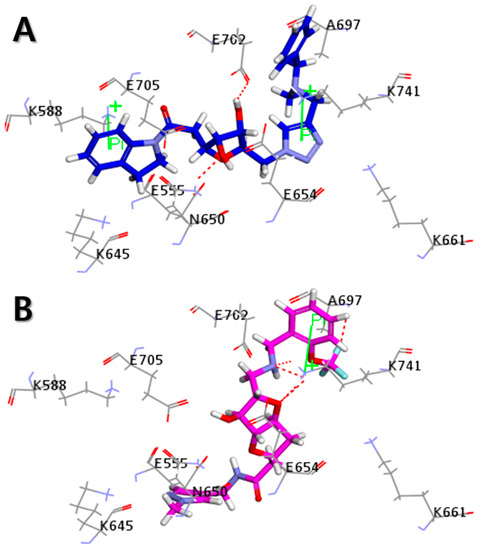
Figure 7.
Molecular docking results. 3D interaction diagram for the calcium-binding site of ANO1 with (A) ZINC8643627 (blue) and (B) ZINC225516955 (magenta). Hydrogen bond interactions and Pi–cation interactions are described in red dotted line and green line, respectively.
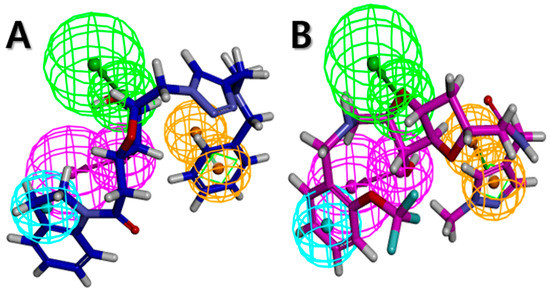
Figure 8.
Pharmacophore mapping of two final hit compounds on Hypo2. (A) ZINC8643627 (blue) (B) ZINC225516955 (magenta). Pharmacophore features are color-coded: magenta, hydrogen bond donor (HBD); green, hydrogen bond acceptor (HBA); cyan, hydrophobic (HY); orange, ring aromatic (RA).

Table 4.
2D chemical structures of the final hit compounds with corresponding estimated scores.
3. Materials and Methods
3.1. Pharmacophore Model Generation
3.1.1. Selection of Dataset Compounds
To represent the structural diversity and broad activity range, the 52 compounds were retrieved from the four literature resources [10,13,30,31]. To achieve a significant pharmacophore modeling, among the 52 dataset compounds with the experimental activity values (IC50), 20 compounds were selected as a training set and the remaining 32 compounds were used as a test set during pharmacophore model validation. The above dataset was selected based on the following criteria [25,38]: (1) All 52 compounds have an inhibitory activity against the ANO1 channel. The apical membrane current of all 52 compounds was determined by an ANO1 functional assay using Fischer rat thyroid (FRT) cells, which can stably express human ANO1 and the halide sensor YFP–H148Q/I152L/F46L. (2) The dataset compounds should be widely populated and less redundant. The IC50 values of both training set and test set compounds consist of a wide activity range from 0.107 to 29.2 μM. (3) To avoid using the different standard of IC50 values measured by different assays and research groups, all IC50 values of 52 compounds were collected from the same biological assays (YFP fluorescence plate reader assay) and the research groups (the A.S. Verkman group and their colleagues, UCSF, San Francisco, CA, USA).
3.1.2. Dataset Compound Preparation
The 2D structures of all the compounds in the dataset were drawn by using ChemDraw Ultra 12.0 (Cambridge Soft Corp., Cambridge, MA, USA), and subsequently converted to 3D structures in DS. Then, hydrogen atoms were added to these 3D structures of compounds as their proper ionized forms under the pH environment of 7.4. Energy minimization of all compounds was carried out using the smart minimizer that carried out 1000 steps of the steepest descent algorithm, followed by a conjugate gradient algorithm with a convergence gradient of 0.001 kcal/mol. After the energy minimization step, the Poling algorithm in the Generate Conformations of DS with the Best conformer generation option under the Chemistry at Harvard Macromolecular Mechanics (CHARMM) force filed was used to generate multiple conformers. A maximum number of 255 conformations with an energy threshold of 10 kcal/mol above the global energy minimum were generated for each compound in the dataset [22,25,38].
3.1.3. Generation of Pharmacophore Models
The HypoGen algorithm [38] in DS was used to perform the 3D-QSAR pharmacophore modeling. This algorithm can generate hypothetical pharmacophore models that correlate between a 3D spatial arrangement of features in a submitted training set compounds and their respective biological activities based on following three steps: the constructive phase, the subtractive phase, and the optimization phase [38]. For establishing the hypothetical pharmacophore model, the Feature mapping of DS was first used to identify the inherent pharmacophore features of the 20 training set compounds. The resulting features were used to generate 10 hypothetical pharmacophore models using the 3D-QSAR pharmacophore generation of DS. All other control parameters of the HypoGen algorithm were kept to their default settings, except for the uncertainty value and the minimum inter-feature distance, which were set to 1.5 and 2 Å, respectively [22,23].
3.2. Pharmacophore Model Evaluation
3.2.1. Cost Analysis
The generated pharmacophore models were briefly assessed for their quality based on the following three cost parameters, which were calculated in the unit of bits during the pharmacophore model generation: the null cost, the fixed cost, and the total cost [38]. The null cost is equal to the maximum occurring error cost, which represents the highest cost value of a pharmacophore model with no features. The fixed cost is a cost of the simplest ideal model that can predict all the data points perfectly. The total cost represents the cost value of each generated pharmacophore hypotheses calculating by the summation of the weight cost, the configuration cost, and error cost. The details of the above three cost parameters are described in [22,38]. For a significant pharmacophore hypothesis, the total cost should be close to the fixed cost, and far from the null cost. In addition, the higher null cost and the lower fixed cost represent the meaningful pharmacophore hypotheses generation. Along with three cost values, other statistical parameters such as a cost difference, RMSD, and r2training were calculated. The cost difference is defined as a difference between the null cost and the total cost. A cost difference of 40~60 bits implies a 75~90% chance of representing a true correlation in the data [23,38]. The RMSD is a measure for the error between the experimental and estimated activity of the training set compounds. The significant pharmacophore model should have the higher cost difference, the higher r2training, and the lower RMSD.
3.2.2. Test Set Method
The 32 compounds were used as the test set to validate the pharmacophore models. This method is used to investigate whether the generated pharmacophore model is sufficient to predict the biological activities of new compounds other than the training set compounds, and to classify them correctly in their activity scale. The biological activities of 32 test set compounds were predicted using the Ligand Pharmacophore Mapping with the Best/Flexible search option in DS. The r2test was used as a primary criterion to determine the best pharmacophore model.
3.2.3. Fischer Randomization Method
The Fischer randomization test was carried out to verify whether a high correlation exists between the chemical structures of the training set compounds and their biological activities. This method generates pharmacophore models by randomizing the biological activity data of the training set compounds, whereas using the same parameters used to develop the original pharmacophore model. During the pharmacophore hypotheses generation, if the hypothesis generated by a randomized dataset has similar or better various statistic values, including cost values, correlation, and RMSD, it means the original hypothesis has been generated by chance [22,23]. In this study, a 95% confidence level was selected for pharmacophore model validation, and 19 random hypotheses were constructed.
3.2.4. Leave-One-Out Method
Finally, the pharmacophore model was validated by the LOO method. In this method, one compound in the training set was omitted from the building step of a new pharmacophore model, and its biological activity was predicted by that new model. This method can verify whether the generated pharmacophore model is strongly dependent on single particular training set compounds [22,23]. In this study, under the same parameter settings used for the generation of the original pharmacophore model, 20 pharmacophore models were generated by omitting one compound at a time from the 20 training set compounds.
3.3. Virtual Screening
The SDF files of 309,149 compounds were downloaded from the ZINC database, including both chemicals and natural products [32]. All the compounds in the ZINC database were primarily filtered based on the Lipinski’s rule of five [39], and the ADMET properties that were predicted by the ADMET Descriptors of DS. The selection criteria of Lipinski’s rule used in this study are as follows [39]: (i) less than five hydrogen bond donors; (ii) less than 10 hydrogen bond acceptors; (iii) a molecular weight of less than 500 Da; (iv) a logarithm of an octanol/water partition coefficient (Log P) value of less than 5. The selection criteria of ADMET used in this study are as follows [40,41,42,43,44,45]: (i) aqueous solubility level = 3 (yes, good) or 4 (yes, optimal); (ii) blood brain barrier (BBB) penetration level = 3 (low); (iii) intestinal absorption level = 0 (good); (iv) CYP2D6 = false (non-inhibitor); (v) hepatotoxicity = false (non-toxic). Conformers were generated for each molecule in the ZINC database using the Best conformer generation method that allows a maximum energy of 10 kcal/mol above that of the most stable conformation. Then, the best pharmacophore model validated using several different methods was used as a 3D query in database screening. The Ligand Pharmacophore Mapping with the Best/Flexible search option was applied to the database screening, in order to identify novel hit compounds that fit all the pharmacophore features. Finally, the retrieved compounds were further filtered by the criterion that compounds have estimated activity values of less than 100 nM.
3.4. Molecular Docking
The Libdock algorithm [46] in DS was used to perform molecular docking. Docking experiments on the final candidate sets of ligand-based virtual screening were carried out against the ANO1 protein. Cryo-EM structure complex of ANO1 with Ca2+ obtained at 3.75 Å was downloaded from the protein data bank (PDB ID: 5OYB) [28]. Protein preparation and minimization were carried out using the Prepare protein of DS. Hydrogen atoms were added to the protein–ligand complex under the CHARMM force field. All water molecules were removed, and the pH environment was adjusted to neutral. The active site was defined at a vicinity of the calcium-binding site with a 10 Å radius. The Libdock scores were obtained by the Libdock algorithm with the default setting except for calculating ligand conformations within an energy range of 10 kcal/mol above the global energy minimum. The docking pose and interaction between the ANO1 and the final hit compounds were analyzed using the Analyze Complexes panel of DS.
4. Conclusions
In this study, a ligand-based 3D-QSAR pharmacophore modeling with a subsequent molecular docking approach was employed to discover the novel lead candidates of ANO1 inhibitor. The best quantitative pharmacophore model, Hypo2, was generated based on the chemical features present in known ANO1 inhibitors with characterizing by the total cost (73.604), the cost difference (121.00), the RMSD (0.946), and the r2training (0.969). Hypo2 has one HBA, one HBD, one HY, and one RA feature. The Hypo2 model was further assessed by the test set prediction, Fischer randomization, and leave-one-out method. The activity values of test set compounds were predicted well by Hypo2, resulting in a r2test of 0.909. The other two methods also have revealed the solid validation results of Hypo2. Then, virtual screening of large chemical database (ZINC) was carried out using Hypo2. The retrieved hit compounds were filtered based on the estimated activity threshold of 100 nM, and these filtered 580 compounds were subjected to molecular docking for further characterization. Finally, two lead candidate compounds from the natural products space were identified with strong interactions with the calcium-binding site of ANO1. On the other hand, a docking study on the other putative active sites and a molecular dynamics simulation are needed to identify more reliable lead candidates. Moreover, further biological validation of the two lead candidates would be necessary to absolutely determine the inhibitory activity, as well as their binding mechanism on ANO1. However, to the best of our knowledge, this is the first study where both the ligand and structure-based approach were applied to identify a novel lead candidate of ANO1 inhibitors, although one pharmacophore model of CaCCinh-A01 and its derivatives was reported [47]. Therefore, the identified novel lead candidates and pharmacophore model in this study could be further employed in designing novel and potent ANO1 inhibitors in the future.
Author Contributions
Y.H.L. has designed the study, performed the experiments, analyzed the data, and written the manuscript. G.-S.Y. supervised the work and edited the manuscript. All authors have read and approved the final manuscript.
Funding
This work was supported by the Bio-Synergy Research Project (NRF-2012M3A9C4048759) of the Ministry of Science, ICT and Future Planning through the National Research Foundation.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| ZINC | ZINC Is Not Commercial database |
| ADMET | Absorption, Distribution, Metabolism, Excretion, and Toxicity |
| MONNA | N-((4-methoxy)-2-naphthyl)-5-nitroanthranilic acid |
| FDA | The Food and Drug Administration |
| ANO2 | Anoctamin 2 |
| CFTR | Cystic fibrosis transmembrane conductance regulator |
| ENaC | Epithelial sodium channel |
| PDE4 | Type 4 cyclic adenosine monophosphate (cAMP) phosphodiesterase |
| BACE1 | β-Site amyloid precursor protein-cleaving enzyme |
| AKR1B10 | Aldo-keto reductase family 1 B 10 |
| CYP2D6 | Cytochrome P450 2D6 |
References
- Caputo, A.; Caci, E.; Ferrera, L.; Pedemonte, N.; Barsanti, C.; Sondo, E.; Pfeffer, U.; Ravazzolo, R.; Zegarra-Moran, O.; Galietta, L.J. TMEM16A, a membrane protein associated with calcium-dependent chloride channel activity. Science 2008, 322, 590–594. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, B.C.; Cheng, T.; Jan, Y.N.; Jan, L.Y. Expression cloning of TMEM16A as a calcium-activated chloride channel subunit. Cell 2008, 134, 1019–1029. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.D.; Cho, H.; Koo, J.Y.; Tak, M.H.; Cho, Y.; Shim, W.S.; Park, S.P.; Lee, J.; Lee, B.; Kim, B.M.; et al. TMEM16A confers receptor-activated calcium-dependent chloride conductance. Nature 2008, 455, 1210–1215. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.; Rock, J.R.; Harfe, B.D.; Cheng, T.; Huang, X.; Jan, Y.N.; Jan, L.Y. Studies on expression and function of the TMEM16A calcium-activated chloride channel. Proc. Natl. Acad. Sci. USA 2009, 106, 21413–21418. [Google Scholar] [CrossRef] [PubMed]
- Pedemonte, N.; Galietta, L.J. Structure and function of TMEM16 proteins (anoctamins). Physiol. Rev. 2014, 94, 419–459. [Google Scholar] [CrossRef] [PubMed]
- Oh, U.; Jung, J. Cellular functions of TMEM16/anoctamin. Pflugers Arch. 2016, 468, 443–453. [Google Scholar] [CrossRef] [PubMed]
- Verkman, A.S.; Galietta, L.J. Chloride channels as drug targets. Nat. Rev. Drug Discov. 2009, 8, 153–171. [Google Scholar] [CrossRef] [PubMed]
- Galietta, L.J.; Haggie, P.M.; Verkman, A.S. Green fluorescent protein-based halide indicators with improved chloride and iodide affinities. FEBS Lett. 2001, 499, 220–224. [Google Scholar] [CrossRef]
- De La Fuente, R.; Namkung, W.; Mills, A.; Verkman, A.S. Small-molecule screen identifies inhibitors of a human intestinal calcium-activated chloride channel. Mol. Pharmacol. 2008, 73, 758–768. [Google Scholar] [CrossRef] [PubMed]
- Namkung, W.; Phuan, P.W.; Verkman, A.S. TMEM16A inhibitors reveal TMEM16A as a minor component of calcium-activated chloride channel conductance in airway and intestinal epithelial cells. J. Biol. Chem. 2011, 286, 2365–2374. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.J.; Hwang, S.J.; Jung, J.; Yu, K.; Kim, J.; Choi, J.Y.; Hartzell, H.C.; Roh, E.J.; Lee, C.J. MONNA, a potent and selective blocker for transmembrane protein with unknown function 16/anoctamin-1. Mol. Pharmacol. 2013, 84, 726–735. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.; Zhang, H.; Wu, M.; Yang, H.; Kudo, M.; Peters, C.J.; Woodruff, P.G.; Solberg, O.D.; Donne, M.L.; Huang, X.; et al. Calcium-activated chloride channel TMEM16A modulates mucin secretion and airway smooth muscle contraction. Proc. Natl. Acad. Sci. USA 2012, 109, 16354–16359. [Google Scholar] [CrossRef] [PubMed]
- Seo, Y.; Lee, H.K.; Park, J.; Jeon, D.K.; Jo, S.; Jo, M.; Namkung, W. Ani9, A Novel Potent Small-Molecule ANO1 Inhibitor with Negligible Effect on ANO2. PLoS ONE 2016, 11, e0155771. [Google Scholar] [CrossRef] [PubMed]
- Namkung, W.; Thiagarajah, J.R.; Phuan, P.W.; Verkman, A.S. Inhibition of Ca2+-activated Cl− channels by gallotannins as a possible molecular basis for health benefits of red wine and green tea. FASEB J. 2010, 24, 4178–4186. [Google Scholar] [CrossRef] [PubMed]
- Yao, Z.; Namkung, W.; Ko, E.A.; Park, J.; Tradtrantip, L.; Verkman, A.S. Fractionation of a herbal antidiarrheal medicine reveals eugenol as an inhibitor of Ca2+-Activated Cl− channel TMEM16A. PLoS ONE 2012, 7, e38030. [Google Scholar] [CrossRef] [PubMed]
- Seo, Y.; Ryu, K.; Park, J.; Jeon, D.K.; Jo, S.; Lee, H.K.; Namkung, W. Inhibition of ANO1 by luteolin and its cytotoxicity in human prostate cancer PC-3 cells. PLoS ONE 2017, 12, e0174935. [Google Scholar] [CrossRef] [PubMed]
- Tradtrantip, L.; Namkung, W.; Verkman, A.S. Crofelemer, an antisecretory antidiarrheal proanthocyanidin oligomer extracted from Croton lechleri, targets two distinct intestinal chloride channels. Mol. Pharmacol. 2010, 77, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Cottreau, J.; Tucker, A.; Crutchley, R.; Garey, K.W. Crofelemer for the treatment of secretory diarrhea. Expert Rev. Gastroenterol. Hepatol. 2012, 6, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Berg, J.; Yang, H.; Jan, L.Y. Ca2+-activated Cl− channels at a glance. J. Cell Sci. 2012, 125 Pt 6, 1367–1371. [Google Scholar] [CrossRef]
- Hartzell, C.; Putzier, I.; Arreola, J. Calcium-activated chloride channels. Annu. Rev. Physiol. 2005, 67, 719–758. [Google Scholar] [CrossRef] [PubMed]
- Kaserer, T.; Beck, K.R.; Akram, M.; Odermatt, A.; Schuster, D. Pharmacophore Models and Pharmacophore-Based Virtual Screening: Concepts and Applications Exemplified on Hydroxysteroid Dehydrogenases. Molecules 2015, 20, 22799–22832. [Google Scholar] [CrossRef] [PubMed]
- John, S.; Thangapandian, S.; Arooj, M.; Hong, J.C.; Kim, K.D.; Lee, K.W. Development, evaluation and application of 3D QSAR Pharmacophore model in the discovery of potential human renin inhibitors. BMC Bioinform. 2011, 12 (Suppl. 14), S4. [Google Scholar] [CrossRef] [PubMed]
- John, S.; Thangapandian, S.; Sakkiah, S.; Lee, K.W. Potent BACE-1 inhibitor design using pharmacophore modeling, in silico screening and molecular docking studies. BMC Bioinform. 2011, 12 (Suppl. 1), S28. [Google Scholar] [CrossRef] [PubMed]
- Niu, M.; Dong, F.; Tang, S.; Fida, G.; Qin, J.; Qiu, J.; Liu, K.; Gao, W.; Gu, Y. Pharmacophore modeling and virtual screening for the discovery of new type 4 cAMP phosphodiesterase (PDE4) inhibitors. PLoS ONE 2013, 8, e82360. [Google Scholar] [CrossRef] [PubMed]
- Niu, M.M.; Qin, J.Y.; Tian, C.P.; Yan, X.F.; Dong, F.G.; Cheng, Z.Q.; Fida, G.; Yang, M.; Chen, H.Y.; Gu, Y.Q. Tubulin inhibitors: Pharmacophore modeling, virtual screening and molecular docking. Acta Pharmacol. Sin. 2014, 35, 967–979. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Son, M.; Bavi, R.; Lee, Y.; Park, C.; Arulalapperumal, V.; Cao, G.P.; Kim, H.H.; Suh, J.K.; Kim, Y.S.; et al. Novel chemical scaffolds of the tumor marker AKR1B10 inhibitors discovered by 3D QSAR pharmacophore modeling. Acta Pharmacol. Sin. 2015, 36, 998–1012. [Google Scholar] [CrossRef] [PubMed]
- Dang, S.; Feng, S.; Tien, J.; Peters, C.J.; Bulkley, D.; Lolicato, M.; Zhao, J.; Zuberbuhler, K.; Ye, W.; Qi, L.; et al. Cryo-EM structures of the TMEM16A calcium-activated chloride channel. Nature 2017, 552, 426–429. [Google Scholar] [CrossRef] [PubMed]
- Paulino, C.; Kalienkova, V.; Lam, A.K.M.; Neldner, Y.; Dutzler, R. Activation mechanism of the calcium-activated chloride channel TMEM16A revealed by cryo-EM. Nature 2017, 552, 421–425. [Google Scholar] [CrossRef] [PubMed]
- Paulino, C.; Neldner, Y.; Lam, A.K.; Kalienkova, V.; Brunner, J.D.; Schenck, S.; Dutzler, R. Structural basis for anion conduction in the calcium-activated chloride channel TMEM16A. eLife 2017, 6, e26232. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Namkung, W.; Verkman, A.S.; Sharma, P.K. Novel 5-substituted benzyloxy-2-arylbenzofuran-3-carboxylic acids as calcium activated chloride channel inhibitors. Bioorg. Med. Chem. 2012, 20, 4237–4244. [Google Scholar] [CrossRef] [PubMed]
- Piechowicz, K.A.; Truong, E.C.; Javed, K.M.; Chaney, R.R.; Wu, J.Y.; Phuan, P.W.; Verkman, A.S.; Anderson, M.O. Synthesis and evaluation of 5,6-disubstituted thiopyrimidine aryl aminothiazoles as inhibitors of the calcium-activated chloride channel TMEM16A/Ano1. J. Enzyme Inhib. Med. Chem. 2016, 31, 1362–1368. [Google Scholar] [CrossRef] [PubMed]
- Sterling, T.; Irwin, J.J. ZINC 15--Ligand Discovery for Everyone. J. Chem. Inf. Model. 2015, 55, 2324–2337. [Google Scholar] [CrossRef] [PubMed]
- Yu, K.; Duran, C.; Qu, Z.; Cui, Y.Y.; Hartzell, H.C. Explaining calcium-dependent gating of anoctamin-1 chloride channels requires a revised topology. Circ. Res. 2012, 110, 990–999. [Google Scholar] [CrossRef] [PubMed]
- Tien, J.; Peters, C.J.; Wong, X.M.; Cheng, T.; Jan, Y.N.; Jan, L.Y.; Yang, H. A comprehensive search for calcium binding sites critical for TMEM16A calcium-activated chloride channel activity. eLife 2014, 3, e02772. [Google Scholar] [CrossRef] [PubMed]
- Jeng, G.; Aggarwal, M.; Yu, W.P.; Chen, T.Y. Independent activation of distinct pores in dimeric TMEM16A channels. J. Gen. Physiol. 2016, 148, 393–404. [Google Scholar] [CrossRef] [PubMed]
- Lim, N.K.; Lam, A.K.; Dutzler, R. Independent activation of ion conduction pores in the double-barreled calcium-activated chloride channel TMEM16A. J. Gen. Physiol. 2016, 148, 375–392. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Thiessen, P.A.; Bolton, E.E.; Chen, J.; Fu, G.; Gindulyte, A.; Han, L.; He, J.; He, S.; Shoemaker, B.A.; et al. PubChem Substance and Compound databases. Nucleic Acids Res. 2016, 44, D1202–D1213. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Sutter, J.; Hoffmann, R. HypoGen: An automated system for generating 3D predictive pharmacophore models. In Pharmacophore Perception Development, and Use in Drug Design; Guner, O.F., Ed.; International University Line: La Jolla, CA, USA, 2000; pp. 172–189. [Google Scholar]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2001, 46, 3–26. [Google Scholar] [CrossRef]
- Egan, W.J.; Merz, K.M., Jr.; Baldwin, J.J. Prediction of drug absorption using multivariate statistics. J. Med. Chem. 2000, 43, 3867–3877. [Google Scholar] [CrossRef] [PubMed]
- Egan, W.J.; Lauri, G. Prediction of intestinal permeability. Adv. Drug Deliv. Rev. 2002, 54, 273–289. [Google Scholar] [CrossRef]
- Cheng, A.; Dixon, S.L. In silico models for the prediction of dose-dependent human hepatotoxicity. J. Comput. Aided Mol. Des. 2003, 17, 811–823. [Google Scholar] [CrossRef] [PubMed]
- Cheng, A.; Merz, K.M., Jr. Prediction of aqueous solubility of a diverse set of compounds using quantitative structure-property relationships. J. Med. Chem. 2003, 46, 3572–3580. [Google Scholar] [CrossRef] [PubMed]
- Susnow, R.G.; Dixon, S.L. Use of robust classification techniques for the prediction of human cytochrome P450 2D6 inhibition. J. Chem. Inf. Comput. Sci. 2003, 43, 1308–1315. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.; Maliski, E.G.; Gallant, P.; Rogers, D. Classification of kinase inhibitors using a Bayesian model. J. Med. Chem. 2004, 47, 4463–4470. [Google Scholar] [CrossRef] [PubMed]
- Rao, S.N.; Head, M.S.; Kulkarni, A.; LaLonde, J.M. Validation studies of the site-directed docking program LibDock. J. Chem. Inf. Model. 2007, 47, 2159–2171. [Google Scholar] [CrossRef] [PubMed]
- Bill, A.; Hall, M.L.; Borawski, J.; Hodgson, C.; Jenkins, J.; Piechon, P.; Popa, O.; Rothwell, C.; Tranter, P.; Tria, S.; et al. Small molecule-facilitated degradation of ANO1 protein: A new targeting approach for anticancer therapeutics. J. Biol. Chem. 2014, 289, 11029–11041. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).