Breeding for Anthracnose Disease Resistance in Chili: Progress and Prospects
Abstract
1. Introduction
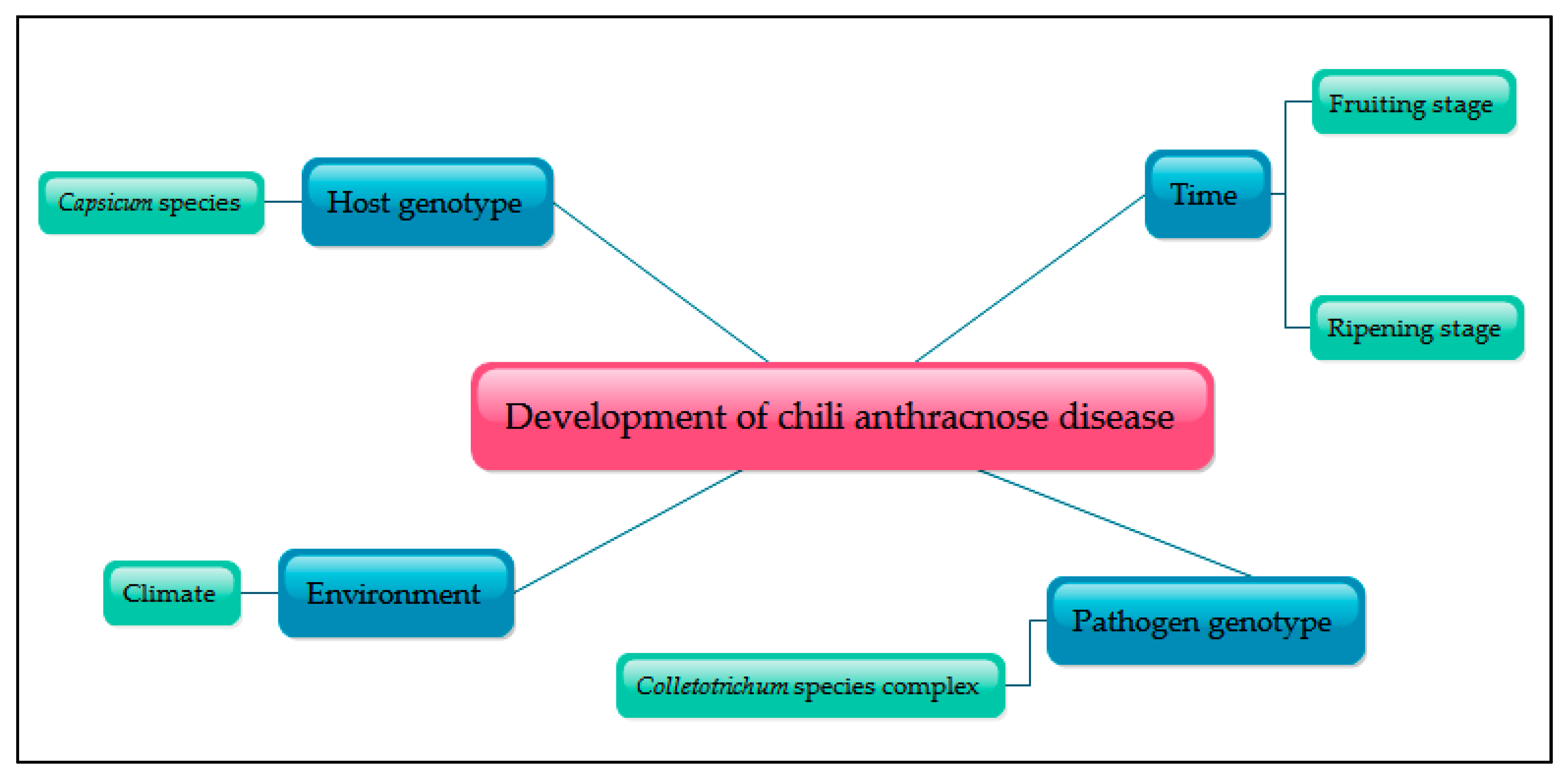
2. Chili Anthracnose
3. Colletotrichum spp. and Occurrence
4. Chemical Components in Capsicum and Their Relation to Anthracnose Disease
5. Screening Method for Chili Anthracnose
6. Markers Available for Disease Resistance
- All markers are not breeder-friendly. This difficulty can be minimized by altering non-breeder-friendly markers to new types of breeder-friendly markers e.g., RAPD to sequence characterized amplified region (SCAR) and RFLP to sequence-tagged sites (STS).
- All markers are not appropriate for whole populations, due to the absence of marker polymorphisms or trustworthy marker-trait association. Multiple mapping populations are used to better identify genetic background effects and marker allelic diversity.
- Incorrect selection may occur due to recombination between the markers and the genes/QTLs of interest. In that case, using flanking markers or many markers for the target gene/QTL can help achieve consistent selection [128].
- Incorrect estimations of QTL locations and effects may result in delayed advancement. High marker density with fine mapping and well-designed phenotyping across various environments and large populations may deliver more precise estimates of QTL locations and effects. The efficiency of QTL detection depends on algorithms, the number of polymorphic markers, mapping methods, and the population type and size [129].
- The marker-assisted backcrossing (MAB) methods and schemes are comprehensible, suitable, and implementable for plant breeders, unless designed for large-scale use in practical breeding programs.
- A huge quantity of breeding programs have not been furnished with satisfactory facilities and situations for a large-scale implementation of MAB in practice.
- In many cases, laboratory expenditures and labor charges are still considerable [130].
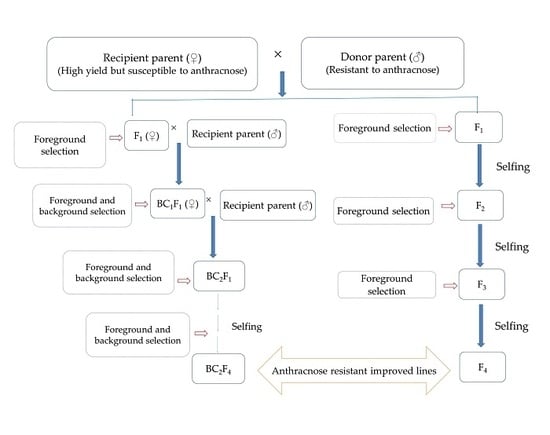
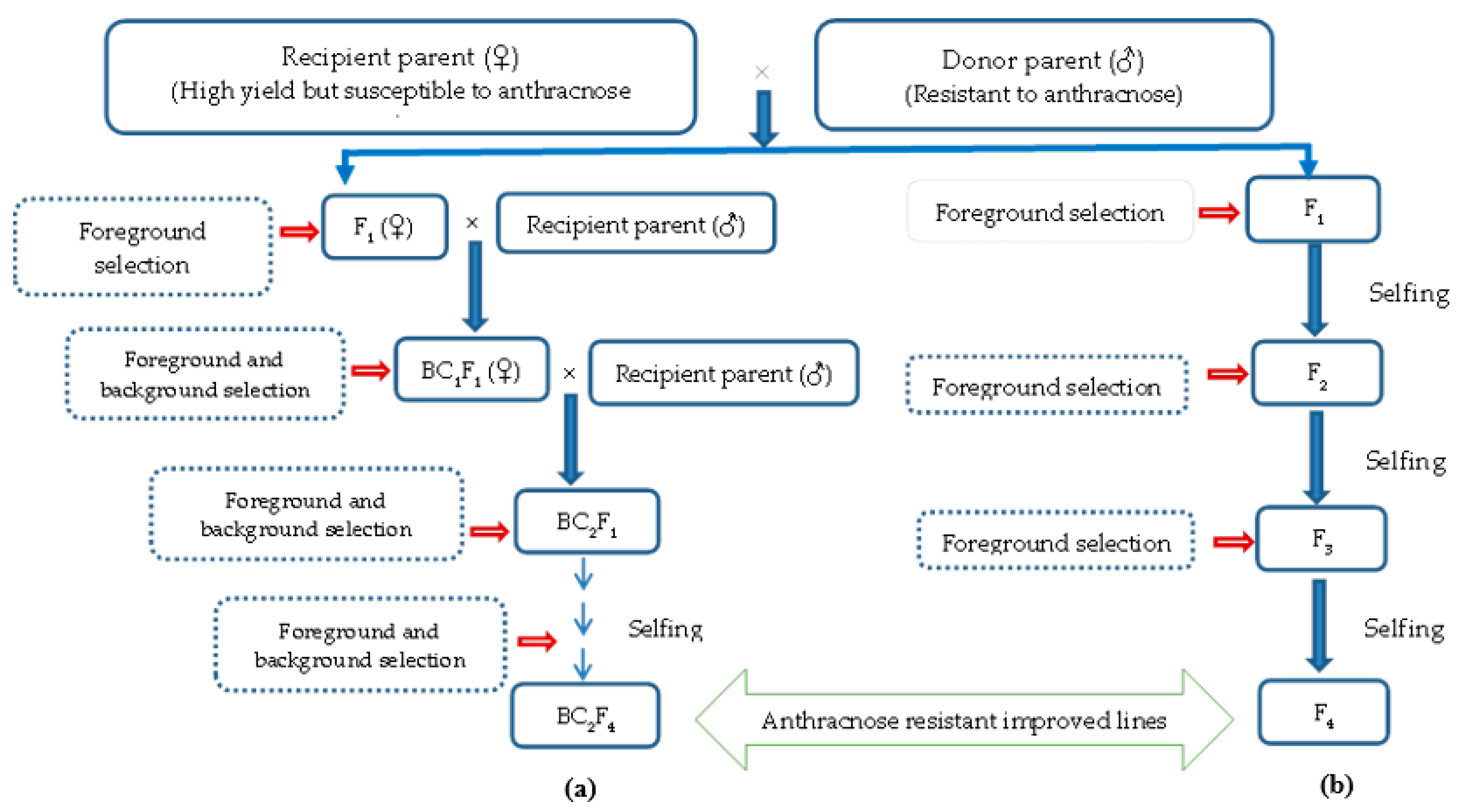
7. Conventional Breeding to Marker-Assisted Breeding for Anthracnose Disease Resistance
8. Present Progress and Outlook
Acknowledgments
Conflicts of Interest
References and Note
- FAOSTAT. Agricultural Production Data. 2016. Available online: http://www.fao.org/ (accessed on 7 March 2018).
- Moscone, E.A.; Scaldaferro, M.A.; Grabiele, M.; Cecchini, N.M.; Sanchez, G.Y.; Jarret, R.; Davina, J.R.; Ducasse, D.A.; Barboza, G.E.; Ehrendorfer, F. The evolution of chili peppers (Capsicum—Solanaceae): A cytogenetic perspective. Acta Hortic. 2007, 745, 137–170. [Google Scholar] [CrossRef]
- Barboza, G.E.; Agra, M.F.; Romero, M.V.; Scaldaferro, M.A.; Moscone, E.A. New endemic species of Capsicum (Solanaceae) from the Brazilian Caatinga: Comparison with the re-circumscribed C. parvifolium. Syst. Bot. 2011, 36, 768–781. [Google Scholar] [CrossRef]
- Mongkolporn, O.; Taylor, P.W.J. Capsicum. In Wild Crop Relatives: Genomic and Breeding Resources; Springer: Berlin/Heidelberg, Germany, 2011; pp. 43–57. [Google Scholar]
- Dias, G.B.; Gomes, V.M.; Moraes, T.M.; Zottich, U.P.; Rabelo, G.R.; Carvalho, A.; OMoulin, M.; Goncalves, L.S.A.; Rodrigues, R.; Da Cunha, M. Characterization of Capsicum species using anatomical and molecular data. Genet. Mol. Res. 2013, 12, 6488–6501. [Google Scholar] [CrossRef] [PubMed]
- Akbar, N.; Ahmad, H.; Ghafoor, S.; Begum, K.; Afridi, S.G.; Muhammad, I.; Khan, I.A. Estimation of genetic diversity in Capsicum germplasm using randomly amplified polymorphic DNA. Asian J. Agric. Sci. 2010, 2, 53–56. [Google Scholar]
- Tong, N.; Bosland, P.W. Capsicum tovarii, a new member of the Capsicum complex. Euphytica 1999, 109, 71–72. [Google Scholar] [CrossRef]
- Bosland, P.W.; Votava, E.J. Peppers: Vegetable and Spice Capsicums; CAB International: Oxfordshire, UK, 2003; p. 333. [Google Scholar]
- Sariah, M. Incidence of Colletotrichum spp. on chili in Malaysia and pathogenicity of C. gloeosporioides. Biotrop Spec. Publ. 1994, 54, 103–120. [Google Scholar]
- Kim, C.H.; Park, K.S. A predictive model of disease progression of red pepper anthracnose. Korean J. Pathol. 1988, 4, 325–331. [Google Scholar]
- Sastrosumarjo, S. Formation of Antracnose-Resistant Chili Varieties with a Conventional Method and Biotechnology Approach. In RUT VIII Research Report; Ministry of Research and Technology RI LIPI: Jakarta, Indonesia, 2003. [Google Scholar]
- Poonpolgul, S.; Kumphai, S. Chilli pepper anthracnose in Thailand. In Abstracts of the First International Symposium on Chilli Anthracnose; Oh, D.G., Kim, K.T., Eds.; National Horticultural Research Institute, Rural Development of Administration: Jeollabukdo, South Korea, 2007; p. 23. [Google Scholar]
- Harp, T.L.; Pernezny, K.; Lewis Ivey, M.L.; Miller, S.A.; Kuhn, P.J.; Datnoff, L. The etiology of recent pepper anthracnose outbreaks in Florida. Crop Prot. 2008, 27, 1380–1384. [Google Scholar] [CrossRef]
- Tozze, H.J., Jr.; Massola, N.M., Jr. First report of Colletotrichum boninense causing anthracnose on pepper in Brazil. Plant Dis. 2009, 93, 106. [Google Scholar] [CrossRef]
- Azad, P. Fate and role of chemical constituents of chilli fruits during infection with Colletotrichum capsici. Indian Phytopathol. 1991, 44, 129–131. [Google Scholar]
- Mistry, D.S.; Sharma, L.P.; Patel, S.T. Bio-chemical parameter of chili fruits as influences by Colletotrichum capsici (Sydow) Butler & Bisby infection. Karnataka J. Agric. Sci. 2010, 21, 586–587. [Google Scholar]
- Hyde, K.D.; Cai, L.; McKenzie, E.H.C.; Yang, Y.L.; Zhang, J.Z.; Prihastuti, H. Colletotrichum: A catalogue of confusion. Fungal Divers. 2009, 39, 1–17. [Google Scholar]
- Montri, P. Pathotypes of Colletotrichum capsici, the causal agent of chili anthracnose in Thailand. Plant Dis. 2009, 93, 17–20. [Google Scholar] [CrossRef]
- Farr, D.F.; Rossman, A.Y.; Fungal Databases, Systematic Mycology and Microbiology Laboratory. USDA: ARS. 2016. Available online: https://nt.ars-grin.gov/fungaldatabases/ (accessed on 9 June 2018).
- Chai, A.L.; Zhao, Y.J.; Shi, Y.X.; Xie, X.W.; Li, B.J. Identification of Colletotrichum capsici (Syd.) Butler causing anthracnose on pumpkin in China. Can. J. Plant Pathol. 2014, 36, 121–124. [Google Scholar] [CrossRef]
- Tarnowski, T.L.B.; Ploetz, R.C. First report of Colletotrichum capsici causing postharvest anthracnose on papaya in South Florida. Plant Dis. 2010, 94, 1065. [Google Scholar] [CrossRef]
- Mclean, K.S.; Roy, K.W. Weeds as a source of Colletotrichum capsici causing anthracnose on tomato fruit and cotton seedlings. Can. J. Plant Pathol. 1991, 13, 131–134. [Google Scholar] [CrossRef]
- Rashid, M.M.; Kabir, M.H.; Hossain, M.M.; Bhuiyan, M.R.; Khan, M.A.I. Eco-friendly management of chilli anthracnose (Colletotrichum capsici). Int. J. Plant Pathol. 2015, 6, 1–11. [Google Scholar] [CrossRef]
- Dale, S.M.; Narkprasert, U.; Diewvanich, D. Efficacy of Amistar 25 SC (azoxystrobin) against anthracnose disease (Colletotrichum capsici) in chilli. In Proceedings of the National Plant Protection Conference, Chonburi, Thailand, 27–29 October 1999; pp. 27–29. [Google Scholar]
- Ahiladevi, P.; Prakasam, V. Bioefficacy of Azoxystrobin 25 SC along with bioagents against chilli anthracnose diseases under field conditions. Pest Manag. Hortic. Ecosyst. 2013, 19, 57–62. [Google Scholar]
- Harp, T.; Kuhn, P.; Roberts, P.D.; Pernezny, K.L. Management and cross-infectivity potential of Colletotrichum acutatum causing anthracnose on bell pepper in Florida. Phytoparasitica 2014, 42, 31–39. [Google Scholar] [CrossRef]
- Gopinath, K.; Radhakrishnan, N.V.; Jayaraj, J. Effect of propiconazole and difenoconazole on the control of anthracnose of chilli fruits caused by Colletotrichum capsici. Crop Prot. 2006, 25, 1024–1031. [Google Scholar] [CrossRef]
- Yadav, O.P.; Gaur, L.B.; Gaur, S.C. Chemical management of anthracnose of chilli (Capsicum annuum L.). Int. J. Plant Prot. 2014, 7, 96–98. [Google Scholar]
- Linu, M.S.; Jisha, M.S. In vitro control of Colletotrichum capsici induced chilli anthracnose by fungicides and biocontrol agent. Int. J. Appl. Pure Sci. Agric. 2006, 3, 27–33. [Google Scholar]
- Gao, Y.Y.; He, L.F.; Li, B.X.; Mu, W.; Lin, J.; Liu, F. The potential of fludioxonil for anthracnose control on China chili fruit. Phytoparasitica 2017, 45, 281–292. [Google Scholar] [CrossRef]
- Suwan, N.; Boonying, W.; Nalumpang, S. Antifungal activity of soil actinomycetes to control chilli anthracnose caused by Colletotrichum gloeosporioides. J. Agric. Technol. 2012, 8, 725–737. [Google Scholar]
- Setiawati, W.; Sutarya, R.; Sumiarta, K.; Kamandalu, A.; Suryawan, I.B.; Latifah, E.; Luther, G. Incidence and severity of pest and diseases on vegetables in relation to climate change (with emphasis on East Java and Bali). In Proceedings of the Prosiding Seminar Nasional Perhimpunan Hortikultura Indonesia, Balitsa, Lembang, Indonesia, 23–24 November 2011; pp. 88–99. [Google Scholar]
- Sharma, P.N.; Kaur, M.; Sharma, O.P.; Sharma, P.; Pathania, A. Morphological, pathological and molecular variability in Colletotrichum capici, the cause of fruit rot of peppers in the subtropical region of north western India. J. Phytopathol. 2005, 153, 232–237. [Google Scholar] [CrossRef]
- Than, P.P.; Prihastuti, H.; Phoulivong, S.; Taylor, P.W.J.; Hyde, K.D. Chili anthracnose disease caused by Colletotrichum species. J. Zhejiang Univ. Sci. B 2008, 9, 764–778. [Google Scholar] [CrossRef] [PubMed]
- Saxena, A.; Raghuvanshi, R.; Singh, H.B. Molecular, phenotypic and pathogenic variability in Colletotrichum isolates of subtropical region in North Eastern India, causing fruit rot of chillies. J. Appl. Microbiol. 2014, 117, 1422–1434. [Google Scholar] [CrossRef] [PubMed]
- Agrios, G.N. Plant Pathology, 5th ed.; Academic Press: San Diego, CA, USA, 2005; p. 922. [Google Scholar]
- Tenaya, I.M.N.; Setiamihardja, R.; Natasasmita, S. Correlation of capsaicin content, fructose, and peroxidase activity with anthracnose disease in chili pepper × red pepper (Abstract). Zuriat 2001, 12, 73–83. [Google Scholar]
- Isaac, S. Fungal Plant Interaction; Chapman and Hall Press: London, UK, 1992; p. 115. [Google Scholar]
- Shin, H.J.; Xu, T.; Zhang, C.L.; Chen, Z.J. The comparative study of Capsicum anthracnose pathogens from Korea with that of China. J. Zhejiang Univ. (Agric. Life Sci.) 2000, 26, 629–634. [Google Scholar]
- Oanh, L.T.K.; Korpraditskul, V.; Rattanakreetakul, C.A. pathogenicity of anthracnose fungus, Colletotrichum capsici on various Thai chilli varieties. Kasetsart J. (Nat. Sci.) 2004, 38, 103–108. [Google Scholar]
- Taylor, P.W.J.; Mongkolporn, O.; Than, P.P.; Montri, P.; Ranathunge, N.; Kanchana-Udonkan, C.; Ford, R.; Pongsupasamit, S.; Hyde, K.D. Pathotypes of Colletotrichum spp. infecting chili peppers and mechanisms of resistance. In First International Symposium on Chilli Anthracnose; Oh, D.G., Kim, K.T., Eds.; National Horticultural Research Institute: Seoul, South Korea, 2007; p. 29. [Google Scholar]
- Hussain, F.; Abid, M. Pests and diseases of chilli crop in Pakistan: A review. Int. J. Biol. Biotechnol. 2011, 8, 325–332. [Google Scholar]
- Halsted, B.D. A new anthracnose of pepper. Bull. Torrey Bot. Club 1890, 18, 14–15. [Google Scholar] [CrossRef]
- Von Arx, J.A. Die Arten der Gattung Colletotrichum Cda. Phytopathologicshe Zeitschrift 1957, 29, 414–468. [Google Scholar]
- Roberts, P.D.; Pernezny, K.; Kucharek, T.A. Anthracnose Caused by Colletotrichum sp. on Pepper; University of Florida Press: Gainesville, FL, USA, 2001; p. 178. [Google Scholar]
- Hasyim, A.; Setiawati, W.; Sutarva, R. Screening for resistance to anthracnose caused by Colletotrichum acutatum in chili pepper (Capsicum annuum L.) in Kediri, East Java. Adv. Agric. Bot. 2014, 6, 104–118. [Google Scholar]
- Manandhar, J.B.; Hartman, G.L.; Wang, T.C. Anthracnose development on pepper fruits inoculated with Colletotrichum gloeosporioides. Plant Dis. 1995, 79, 380–383. [Google Scholar] [CrossRef]
- Rajapakse, R.G.A.S.; Ranasinghe, J.A.D.A.R. Development of variety screening method for anthracnose disease of chili (Capsicum annuum L.) under field conditions. Trop. Agric. Res. Ext. 2002, 5, 7–11. [Google Scholar]
- Tode, H.J. Fungi Mecklenburgenses Selecti. Fasc. 1. Nova Fungorum Genera Complectens. 1790. [Google Scholar]
- Dean, R.; Van Kan, J.A.; Pretorius, Z.A.; Hammond-Kosack, K.E.; Di Pietro, A.; Spanu, P.D.; Rudd, J.J.; Dickman, M.; Kahmann, R.; Ellis, J.; et al. The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 414–430. [Google Scholar] [CrossRef] [PubMed]
- Garg, R.; Loganathan, M.; Saha, S.; Roy, B.K. Chilli Anthracnose: A review of causal organism, resistance source and mapping of gene. In Microbial Diversity and Biotechnology in Food Security; Springer: New Delhi, India, 2014; pp. 589–610. [Google Scholar]
- Syukur, M.; Sujiprihati, S.; Koswara, J.; Widodo, W. Inheritance of resistance to anthracnose caused by Colletotrichum acutatum in Chili pepper (Capsicum annuum L.). Bull. Agron. 2007, 35, 112–117. [Google Scholar]
- Than, P.P.; Rajesh, J.; Hyde, K.D.; Pongsupasamit, S.; Mongkolporn, O.; Taylor, P.W.J. Characterization and pathogenicity of Colletotrichum species associated with anthracnose infection on chili (Capsicum spp.). Plant Pathol. 2008b, 57, 562–572. [Google Scholar] [CrossRef]
- Mahmodi, F.; Kadir, J.; Puteh, A. Genetic diversity and pathogenic variability of Colletotrichum truncatum causing anthracnose of pepper in Malaysia. J. Phytopathol. 2014, 162, 456–465. [Google Scholar] [CrossRef]
- Voorrips, R.E.; Finkers, R.; Sanjaya, L.; Groenwold, R. QTL mapping of anthracnose (Colletotrichum spp.) resistance in a cross between Capsicum annum and C. chinense. Theor. Appl. Genet. 2004, 109, 1275–1282. [Google Scholar] [CrossRef] [PubMed]
- Thind, T.S.; Jhooty, J.S. Studies on variability in two Colletotrichum spp. causing anthracnose nose and fruit rot of chilli in Punjab. Indian Phytopathol. 1990, 43, 53–58. [Google Scholar]
- Hegde, G.M.; Anahosur, K.H.; Srikant, K. Biological control of Colletotrichum capsici causing fruit rot of chilli. Plant Pathol. Newsl. 2002, 20, 4–5. [Google Scholar]
- Paul, Y.S.; Behl, M.K. Some studies on bell pepper anthracnose caused by Colletotrichum capsici and its control. Seed Res. 1990, 1, 656–659. [Google Scholar]
- Susheela, K. Evaluation of screening method for anthracnose disease in chili. Pest Manag. Hortic. Ecosyst. 2012, 18, 188–193. [Google Scholar]
- Selvakumar, R. Variability among Colletotrichum capsici causing chilli anthracnose in north east India. In Proceedings of the First International Symposium on Chilli Anthracnose, Hoam Faculty House, Seoul National University, Seoul, South Korea, 17–19 September 2007; p. 35. [Google Scholar]
- Kaur, S.; Singh, J. C. acutatum, a threat to chilli crop in Punjab. Indian Phytopathol. 1990, 43, 108–110. [Google Scholar]
- Ramachandran, N.; Rathnamma, K. Colletotrichum acutatum—A new addition to the species of chilli anthracnose pathogen in India. In Proceedings of the Annual Meeting & Symposium of Indian Phytopathological Society, Central Planation Crops Research Institute, Kasaragod, India, 27–28 November 2006. [Google Scholar]
- Sharma, G.; Shenoy, B.D. Colletotrichum fructicola and C. siamense are involved in chilli anthracnose in India. Arch. Phytopathol. Plant Protect. 2014, 47, 1179–1194. [Google Scholar] [CrossRef]
- Park, K.S.; Kim, C.H. Identification, distribution, and etiological characteristics of anthracnose fungi of red pepper in Korea. Korean J. Plant Pathol. 1992, 8, 61–69. [Google Scholar]
- Pearson, M.N.; Bull, P.B.; Speke, H. Anthracnose of Capsicum in Papua, New Guinea, varietal reaction and associated fungi. Trop. Pest Manag. 1984, 30, 230–233. [Google Scholar] [CrossRef]
- Johnston, P.R.; Jones, D. Relationship among Colletotrichum isolates from fruit-rots assessed using rDNA sequences. Mycologia 1997, 89, 420–430. [Google Scholar] [CrossRef]
- Simmonds, J.H. A study of the species of Colletotrichum causing ripe fruit rots in Queenslands. Qld. J. Agric. Anim. Sci. 1965, 22, 437–459. [Google Scholar]
- Adikaram, N.K.B.; Brown, A.; Swinburne, T.R. Observations on Infection of Capsicum annuum fruit by Glomerella cingulata and Colletotrichum capsici. Trans. Br. Mycol. Soc. 1983, 80, 395–401. [Google Scholar] [CrossRef]
- Don, L.D.; Van, T.T.; Phuong, V.T.T.; Kieu, P.T.M. Colletotichum spp. attacking on chilli pepper growing in Vietnam. Country Report. In Abstracts of First Internationaal Symposium on Chilli Anthracnose; Oh, D.G., Kim, K.T., Eds.; National Horticultural Research Institute, Rural Development of Administration: Jeollabukdo, Korea, 2007; p. 24. [Google Scholar]
- Perry, L.; Dickau, R.; Zarrillo, S.; Holst, I.; Pearsall, D.M.; Piperno, D.R.; Berman, M.J.; Cooke, R.G.; Rademaker, K.; Ranere, A.J.; et al. Starch fossils and the domestication and dispersal of chili peppers (Capsicum spp. L.) in the Americas. Science 2007, 315, 986–988. [Google Scholar] [CrossRef] [PubMed]
- Nadeem, M.; Anjum, F.M.; Khan, M.R.; Saeed, M.; Riaz, A. Antioxidant potential of bell pepper (Capsicum annum L.) a review. Pak. J. Food Sci. 2011, 21, 45–51. [Google Scholar]
- Raghavan, S. Handbook of Spices, Seasonings, and Flavorings; CRC Press: Boca Raton, FL, USA, 2007; pp. 87–91. [Google Scholar]
- Wesolowska, A.; Jadczak, D.; Grzeszczuk, M. Chemical composition of the pepper fruit extracts of hot cultivars Capsicum annuum L. Acta Sci. Pol. Hortorum Cultus 2011, 10, 171–184. [Google Scholar]
- Diaz, J.; Pomar, F.; Bernal, A.; Merino, F. Peroxidases and the metabolism of capsaicin in Capsicum annuum L. Phytochem. Rev. 2004, 3, 141–157. [Google Scholar] [CrossRef]
- Pruthi, J.S. Spices and Condiments; National Book Trust: New Delhi, India, 1976; p. 269. [Google Scholar]
- Tapia, J.C.; Garcia, R.; Eleazar, M.; Calva, G.; Rocha, J.A. Capsaicin recovery from a cell culture broth. Ind. Eng. Chem. Res. 1993, 32, 2242–2246. [Google Scholar] [CrossRef]
- Prasad, B.C.N.; Gururaj, H.B.; Kumar, V.; Giridhar, P.; Ravishankar, G.A. Valine pathway is more crucial than phenyl propanoid pathway in regulating capsaicin biosynthesis in Capsicum frutescens mill. J. Agric. Food Chem. 2006, 54, 6660–6665. [Google Scholar] [CrossRef] [PubMed]
- Higashiguchi, H.; Nakamura, H.; Hayashi, H.; Kometani, T. Purification and structure determination of glucosides of capsaicin and dihydrocapsaicin from various Capsicum fruits. J. Agric. Food Chem. 2006, 54, 5948–5953. [Google Scholar] [CrossRef] [PubMed]
- Davis, C.B.; Carolyn, E.M.; Marianna, A.B.; Kenneth, W.B. Determination of capsaicinoids in Habanero peppers by Chemometric analysis of UV spectral data. J. Agric. Food Chem. 2007, 55, 5925–5933. [Google Scholar] [CrossRef] [PubMed]
- Madhumathy, A.P.; Aivazi, A.A.; Vijayan, V.A. Larvicidal efficacy of Capsicum annum against Anopheles stephensi and Culex quinquefasciatus. J. Vector Borne Dis. 2007, 44, 223–226. [Google Scholar] [PubMed]
- Gahungu, A.; Ruganintwali, E.; Karangwa, E.; Zhang, X.; Mukunzi, D. Volatile compounds and capsaicinoid content of fresh hot peppers (Capsicum chinense) Scotch Bonnet variety at red stage. Adv. J. Food Sci. Technol. 2011, 3, 211–218. [Google Scholar]
- Barbero, G.F.; Palma, M.; Barroso, C.G. Pressurized liquid extraction of capsaicinoids from peppers. J. Agric. Food Chem. 2006, 54, 3231–3236. [Google Scholar] [CrossRef] [PubMed]
- Materska, M.; Perucka, I. Antioxidant activity of the main phenolic compounds isolated from hot pepper fruit (Capsicum annuum L.). J. Agric. Food Chem. 2005, 53, 1750–1756. [Google Scholar] [CrossRef] [PubMed]
- Walter, W.R. Wax and Capsaicin Based Pesticide; Wilder Agricultural Product Co Inc.: New Wilmington, PA, USA, 1995. [Google Scholar]
- Macho, A.; Lucena, C.; Sancho, R.; Daddario, N.; Minassi, A.; Munoz, E.; Appendino, G. Nonpungent capsaicinoids from sweet pepper synthesis and evaluation of the chemopreventive and anticancer potential. Eur. J. Nutr. 2003, 42, 2–9. [Google Scholar] [CrossRef] [PubMed]
- Ohnuki, K.; Niwa, S.; Maeda, S.; Inoue, N.; Yazawa, S.; Fushiki, T. CH-19 sweet, a non-pungent cultivar of red pepper, increased body temperature and oxygen consumption in humans. Biosci. Biotechnol. Biochem. 2001, 65, 2033–2036. [Google Scholar] [CrossRef] [PubMed]
- Sancho, R.; Lucena, C.; Macho, A.; Calzado, M.A.; Blanco-Molina, M.; Minassi, A.; Appendino, G.; Munoz, E. Immunosuppressive activity of capsaicinoids: Capsiate derived from sweet peppers inhibits NF-κB activation and is a potent anti-inflammatory compound in vivo. Eur. J. Immunol. 2002, 32, 1753–1763. [Google Scholar] [CrossRef]
- Krajewska, A.M.; Powers, J.J. Sensory properties of naturally occurring capsaicinoids. J. Food Sci. 1988, 53, 902–905. [Google Scholar] [CrossRef]
- Usman, M.G.; Rafii, M.Y.; Ismail, M.R.; Malek, M.A.; Latif, M.A. Capsaicin and dihydrocapsaicin determination in chili pepper genotypes using ultra-fast liquid chromatography. Molecules 2014, 19, 6474–6488. [Google Scholar] [CrossRef] [PubMed]
- Saxena, A.; Raghuwanshi, R.; Gupta, V.K.; Singh, H.B. Chilli anthracnose: The epidemiology and management. Front. Microbiol. 2016, 7, 1527. [Google Scholar] [CrossRef] [PubMed]
- Gudeva, L.K.; Mitrev, S.; Maksimova, V.; Spasov, D. Content of capsaicin extracted from hot pepper (Capsicum annuum ssp. microcarpum L.) and its use as an ecopesticide. Hem. Ind. 2013, 67, 671–675. [Google Scholar] [CrossRef]
- Ko, M.K.; Jeon, W.B.; Kim, K.S.; Lee, H.H.; Seo, H.H.; Kim, Y.S.; Oh, B.J. A Colletotrichum gloeosporioides induced esterase gene of non-climacteric pepper (Capsicum annuum) fruit during ripening plays a role is resistance against fungal infection. Plant Mol. Biol. 2005, 58, 529–541. [Google Scholar] [CrossRef] [PubMed]
- Oh, B.J.; Kim, K.D.; Kim, Y.S. A microscopic characterization of the infection of green and red pepper fruits by an isolate of Colletotrichum gloeosporioides. J. Phytopathol. 1998, 146, 301–303. [Google Scholar] [CrossRef]
- Kim, K.D.; Oh, B.J.; Yang, J. Differential interactions of a Colletotrichum gloeosporioides isolate with green and red pepper fruits. Phytoparasitica 1999, 27, 1–10. [Google Scholar] [CrossRef]
- Wilson, L.; Madden, L.; Ellis, M. Influence of temperature and wetness duration on infection of immature and mature strawberry fruit by Colletotrichum acutatum. Phytopathology 1990, 80, 111–116. [Google Scholar] [CrossRef]
- Prusky, D.; Koblier, I.; Ardi, R.; Beno-Moalem, D.; Yakoby, N.; Keen, N. Resistance mechanisms of subtropical fruits to Colletotrichum gloeosporioides. In Colletotrichum: Host Specificity, Pathology, and Host-Pathogen Interaction; Prusky, D., Freeman, S., Dickman, M.B., Eds.; American Phytopathological Society: St. Paul, MN, USA, 2000; pp. 232–244. [Google Scholar]
- Chillet, M.; Hubert, O.; de Bellaire, L.D.L. Relationship between physiological age, ripening and susceptibility of banana to wound anthracnose. Crop Prot. 2007, 26, 1078–1082. [Google Scholar] [CrossRef]
- Moral, J.; Bouhmidi, K.; Trapero, A. Influence of fruit maturity, cultivar susceptibility, and inoculation method on infection of olive fruit by Colletotrichum acutatum. Plant Dis. 2008, 92, 1421–1426. [Google Scholar] [CrossRef]
- Prasath, D.; Ponnuswami, V. Heterosis and combining ability for morphological, yield and quality characters in paprika type chilli hybrids. Indian J. Hortic. 2008, 65, 441–445. [Google Scholar]
- Park, S.; Jeong, W.Y.; Lee, J.H.; Kim, Y.H.; Jeong, S.W.; Kim, G.S.; Bae, D.W.; Lim, C.S.; Jin, J.S.; Lee, S.J.; et al. Determination of polyphenol levels variation in Capsicum annuum L. cv. Chelsea (yellow bell pepper) infected by anthracnose (Colletotrichum gloeosporioides) using liquid chromatography–tandem mass spectrometry. Food Chem. 2012, 130, 981–985. [Google Scholar] [CrossRef]
- Pedras, M.S.C.; Zheng, Q.A.; Gadagi, R.S.; Rimmer, S.R. Phytoalexins and polar metabolites from the oilseeds canola and rapeseed: Differential metabolic responses to the biotroph Albugo candida and to abiotic stress. Phytochemistry 2008, 69, 894–910. [Google Scholar] [CrossRef] [PubMed]
- Mongkolporn, O.; Montri, P.; Supakaew, T.; Taylor, P.W.J. Differential reactions on mature green and ripe chili fruit infected by three Colletotrichum spp. Plant Dis. 2010, 94, 306–310. [Google Scholar] [CrossRef]
- Silva, S.A.M.; Rodrigues, R.; Goncalves, L.S.A.; Sudre, C.P.; Bento, C.S.; Carmo, M.G.F.; Medeiros, A.M. Resistance in Capsicum spp. to anthracnose affected by different stages of fruit development during pre- and postharvest. Trop. Plant Pathol. 2014, 39, 335–341. [Google Scholar] [CrossRef]
- Mahasuk, P.; Chinthaisong, J.; Mongkolporn, O. Differential resistances to anthracnose in Capsicum baccatum as responding to two Colletotrichum pathotypes and inoculation methods. Breed. Sci. 2013, 63, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.K.; Hwang, B.K. Influence of inoculum density, wetness duration, plant age, inoculation method, and cultivar resistance on infection of pepper plants by Colletotrichum cocodes. Plant Dis. 1998, 82, 1079–1083. [Google Scholar] [CrossRef]
- Ansari, A.M. Molecular markers in vegetable improvement. Hortic. Biotechnol. Res. 2015, 1, 5–10. [Google Scholar]
- Messeguer, R.; Ganal, M.; de Vicente, M.C.; Young, N.D.; Bolkan, H.; Tanksley, S.D. High resolution RFLP map around the root knot nematode resistance gene (Mi) in tomato. Theor. Appl. Genet. 1991, 82, 529–536. [Google Scholar] [CrossRef] [PubMed]
- Mather, K.; Jink, J.L. Biometrical Genetics; Chapman and Hall: London, UK, 1971; p. 382. [Google Scholar]
- Geldermann, H. Investigations on inheritance of quantitative characters in animals by gene markers I. Methods. Theor. Appl. Genet. 1975, 46, 319–330. [Google Scholar] [CrossRef] [PubMed]
- East, E.M. Studies on Size Inheritance in Nicotiana. Genetics 1916, 1, 164–176. [Google Scholar] [PubMed]
- Yule, G.V. On the theory of inheritance of quantitative compound characters on the basis of Mendel’s laws—A preliminary note. In Proceedings of the 3rd International Conference on Genetics, London, UK; 1906; pp. 140–142. [Google Scholar]
- Kwon, Y.S.; Kim, K.M.; Kim, D.H.; Eun, M.Y.; Sohn, J.K. Marker-assisted introgression of quantitative trait loci associated with plant regeneration ability in anther culture of rice (Oryza sativa L.). Mol. Cell 2002, 14, 24–28. [Google Scholar]
- Kang, B.C.; Nahm, S.H.; Huh, J.H.; Yoo, H.S.; Yu, J.W.; Lee, M.H.; Kim, B.D. An interspecific Capsicum annuum × C. chinense F2 linkage map in pepper using RFLP and AFLP markers. Theor. Appl. Genet. 2001, 102, 531–539. [Google Scholar] [CrossRef]
- Nahm, S.H.; Yu, J.W.; Kang, B.C.; Kim, B.D. Selection of parental line for hot pepper mapping population using RFLP and AFLP analyses. J. Korean Soc. Hortic. Sci. 1997, 38, 693–697. [Google Scholar]
- Kim, K.H.; Yoon, J.B.; Park, H.G.; Park, E.W.; Kim, Y.H. Structural modifications and programmed cell death of chili pepper fruit related to resistance responses to Colletotrichum gloeosporioides infection. Phytopathology 2004, 94, 1295–1304. [Google Scholar] [CrossRef] [PubMed]
- Ratanacherdchai, K.; Wang, H.K.; Lin, F.C.; Soytong, K. RAPD analysis of Colletotrichum species causing chili anthracnose disease in Thailand. J. Agric. Technol. 2007, 3, 211–219. [Google Scholar]
- Miah, G.; Rafii, M.Y.; Ismail, M.R.; Puteh, A.B.; Rahim, H.A.; Islam, K.N.; Latif, M.A. A review of microsatellite markers and their applications in rice breeding programs to improve blast disease resistance. Int. J. Mol. Sci. 2013, 14, 22499–22528. [Google Scholar] [CrossRef] [PubMed]
- Becher, S.A.; Steinmetz, K.; Weising, K.; Boury, S.; Peltier, D.; Renou, J.P.; Kahl, G.; Wolff, K. Microsatellites for variety identification in Pelargonium. Theor. Appl. Genet. 2000, 101, 643–651. [Google Scholar] [CrossRef]
- Litt, M.; Luty, J.M. A hypervariable microsatellite revealed by in vitro amplification of a dinucleotide repeat within the cardiac muscle actin gene. Am. J. Hum. Genet. 1989, 44, 397–401. [Google Scholar] [PubMed]
- Jacob, H.J.; Lindpaintner, K.; Lincoln, S.E.; Kusumi, K.; Bunker, R.K.; Mao, Y.P.; Ganten, D.; Dzau, V.J.; Lander, E.S. Genetic mapping of a gene causing hypertensive rat. Cell 1991, 67, 213–224. [Google Scholar] [CrossRef]
- Edwards, A.; Civitello, A.; Hammond, H.A.; Caskey, C.T. DNA typing and genetic mapping with trimeric and tetrameric tandem repeats. Am. J. Hum. Genet. 1991, 49, 746–756. [Google Scholar] [PubMed]
- McCouch, S.R.; Cho, Y.G.; Yano, M.; Paul, E.; Blinstrub, M.; Morishima, H.; Kinoshita, T. Report on QTL nomenclature. Rice Genet. Newsl. 1997, 14, 11–131. [Google Scholar]
- Mimura, Y.; Inoue, T.; Minamiyama, Y.; Kubo, N. An SSR based genetic map of pepper (Capsicum annuum L.) serves as an anchor for the alignment of major pepper maps. Breed. Sci. 2012, 62, 93–98. [Google Scholar] [CrossRef] [PubMed]
- Dhaliwal, M.S.; Yadav, A.; Jindal, S.K. Molecular characterization and diversity analysis in chilli pepper using simple sequence repeats (SSR) markers. Afr. J. Biotechnol. 2014, 13, 3137–3143. [Google Scholar]
- Lee, Y.H.; Kim, H.S.; Kim, J.Y.; Jung, M.; Park, Y.S.; Lee, J.S.; Choi, S.H.; Her, N.H.; Lee, J.H.; Hyung, N.I.; et al. A new selection method for pepper transformation: Callus-mediated shoot formation. Plant Cell Rep. 2004, 23, 50–58. [Google Scholar] [CrossRef] [PubMed]
- Minamiyama, Y.; Tsuro, M.; Hirai, M. An SSR-based linkage map of Capsicum annuum. Mol. Breed. 2006, 18, 157–169. [Google Scholar] [CrossRef]
- Geleta, L.F.; Labuschagne, M.T.; Viljoen, C.D. Relationship between heterosis and genetic distance based on morphological traits and AFLP markers in pepper. Plant Breed. 2004, 123, 467–473. [Google Scholar] [CrossRef]
- Heffner, E.L.; Sorrells, M.E.; Jannink, J.L. Genomic selection for crop improvement. Crop Sci. 2009, 49, 1–12. [Google Scholar] [CrossRef]
- Wang, X.; Jiang, G.L.; Green, M.; Scott, R.A.; Hyten, D.L.; Cregan, P.B. Quantitative trait locus analysis of saturated fatty acids in a population of recombinant inbred lines of soybean. Mol. Breed. 2012, 30, 1163–1179. [Google Scholar] [CrossRef]
- Jiang, G.L. Plant marker-assisted breeding and conventional breeding: Challenges and perspectives. Adv. Crop Sci. Technol. 2013, 1, e106. [Google Scholar] [CrossRef]
- Stapley, J.; Reger, J.; Feulner, P.G.D.; Smadja, C.; Galindo, J.; Ekblom, R.; Bennison, C.; Ball, A.D.; Beckerman, A.P.; Slate, J. Adaptation genomics: The next generation. Trends Ecol. Evol. 2010, 25, 705–712. [Google Scholar] [CrossRef] [PubMed]
- Henry, R.J. Next-generation sequencing for understanding and accelerating crop domestication. Brief. Funct. Genom. 2012, 11, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Ray, S.; Satya, P. Next generation sequencing technologies for next generation plant breeding. Front. Plant Sci. 2014, 5, 367. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Tao, Y.; Zheng, Z.; Li, C.; Sweetingham, M.W.; Howieson, J.G. Application of next-generation sequencing for rapid marker development in molecular plant breeding: A case study on anthracnose disease resistance in Lupinus angustifolius L. BMC Genom. 2012, 13, 318. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Yin, Y.; Yang, K.; Li, J.; Sang, Y.; Huang, L.; Fan, S. Construction of a high-density genetic map using specific length amplified fragment markers and identification of a quantitative trait locus for anthracnose resistance in walnut (Juglans regia L.). BMC Genom. 2015, 16, 614. [Google Scholar] [CrossRef] [PubMed]
- Julier, B.; Meusnier, I.; Alaux, L.; Flajoulot, S.; Barre, P.; Gouzy, J. Role of the RCT1 gene in anthracnose resistance in alfalfa. In Breeding Strategies for Sustainable Forage and Turf Grass Improvement; Springer: Dordrecht, The Netherlands, 2013; pp. 203–208. [Google Scholar]
- Kim, S.; Park, M.; Yeom, S.I.; Kim, Y.M.; Lee, J.M.; Lee, H.A.; Seo, E.; Choi, J.; Cheong, K.; Kim, K.T.; et al. Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. Nat. Genet. 2014, 46, 270–278. [Google Scholar] [CrossRef] [PubMed]
- Miah, G.; Rafii, M.Y.; Ismail, M.R.; Puteh, A.B.; Rahim, H.A.; Asfaliza, R.; Latif, M.A. Blast resistance in rice: A review of conventional breeding to molecular approaches. Mol. Biol. Rep. 2013, 40, 2369–2388. [Google Scholar] [CrossRef] [PubMed]
- Doshi, K.M.; Shukal, R.T. Genetics and its components in chilli (Capsicum annuum L.). Capsicum Eggplant Newsl. 2000, 19, 78–81. [Google Scholar]
- Collard, B.C.; Mackill, D.J. Marker-assisted selection: An approach for precision plant breeding in the twenty-first century. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2008, 363, 557–572. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.Y.; Mao, S.L.; Zhang, Z.H.; Palloix, A.; Wang, L.H.; Zhang, B.X. Resistances to anthracnose (Colletotrichum acutatum) of Capsicum mature green and ripe fruit are controlled by a major dominant cluster of QTLs on chromosome P5. Sci. Hortic. 2015, 181, 81–88. [Google Scholar] [CrossRef]
- Suwor, P.; Thummabenjapone, P.; Sanitchon, J.; Kumar, S.; Techawongstien, S. Phenotypic and genotypic responses of chili (Capsicum annuum L.) progressive lines with different resistant genes against anthracnose pathogen (Colletotrichum spp.). Eur. J. Plant Pathol. 2015, 143, 725–736. [Google Scholar] [CrossRef]
- Lee, J.; Hong, J.; Do, J.; Yoon, J.; Lee, J.D.; Hong, J.H.; Do, J.W.; Yoon, J.B. Identification of QTLs for resistance to anthracnose to two Colletotrichum species in pepper. J. Crop Sci. Biotechnol. 2010, 13, 227–233. [Google Scholar] [CrossRef]
- Mahasuk, P.; Khumpeng, N.; Wasee, S.; Taylor, P.W.J.; Mongkolporn, O. Inheritance of resistance to anthracnose (Colletotrichum capsici) at seedling and fruiting stages in chili pepper (Capsicum spp.). Plant Breed. 2009, 128, 701–706. [Google Scholar] [CrossRef]
- Johnson, L.P.V. Applications of the diallel cross technique to plant breeding. In Statistical Genetics and Plant Breeding; Hanson, W.D., Robinson, H.F., Eds.; National Acad of Sci-National Res Council: Washington, DC, USA, 1963; pp. 561–569. [Google Scholar]
- Fujimaki, H. Recurrent selection by using male sterility for rice improvement. Jpn. Agric. Res. Q. 1979, 13, 153–156. [Google Scholar]
- Bouchez, A.; Hospital, F.; Causse, M.; Gallais, A.; Charcosset, A. Marker-assisted introgression of favorable alleles at quantitative trait loci between maize elite lines. Genetics 2002, 162, 1945–1959. [Google Scholar] [PubMed]
- Oliveira, L.K.; Melo, L.C.; Brondani, C.; Peloso, M.J.D.; Brondani, R.P.V. Backcross assisted by microsatellite markers in common bean. Genet. Mol. Res. 2008, 7, 1000–1010. [Google Scholar] [CrossRef] [PubMed]
- Jiang, G.L. Molecular markers and marker-assisted breeding in plants. In Plant Breeding from Laboratories to Fields; Intech: Rijeka, Croatia, 2013; pp. 45–83. [Google Scholar]
- Ribaut, J.M.; De Vicente, M.C.; Delannay, X. Molecular breeding in developing countries: Challenges and perspectives. Curr. Opin. Plant Biol. 2010, 13, 213–218. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.B.; Kwon, S.W.; Ham, T.H.; Kim, S.; Thomson, M.; Hechanova, S.L.; Park, Y. Marker-Assisted Breeding. In Current Technologies in Plant Molecular Breeding; Springer: Dordrecht, The Netherlands, 2015; pp. 95–144. [Google Scholar]
- Park, H.K.; Kim, B.S.; Lee, W.S. Inheritance of resistance to anthracnose (Colletotrichum spp.) in pepper (Capsicum annuum L.) I. Genetic analysis of anthracnose resistance by diallel crosses. J. Korean Soc. Hortic. Sci. 1990, 31, 91–105. [Google Scholar]
- Park, H.K.; Kim, B.S.; Lee, W.S. Inheritance of resistance to anthracnose (Colletotrichum spp.) in pepper (Capsicum annuum L.) II. Genetic analysis of resistance to Colletotrichum dematium. J. Korean Soc. Hortic. Sci. 1990, 31, 207–212. [Google Scholar]
- Park, S.; Kim, S.H.; Park, H.G.; Yoon, J.B. Capsicum germplasm resistant to pepper anthracnose differentially interact with Colletotrichum isolates. Hortic. Environ. Biotechnol. 2009, 50, 17–23. [Google Scholar]
- Lee, J.; Do, J.W.; Yoon, J.B. Development of STS markers linked to the major QTLs for resistance to the pepper anthracnose caused by Colletotrichum acutatum and C. capsici. Hortic. Environ. Biotechnol. 2011, 52, 596–601. [Google Scholar] [CrossRef]
- Wang, Y.W. Development of Sequence Characterized Amplified Region (SCAR) Markers Associated with Pepper Anthracnose (Colletotrichum acutatum) Resistance. Master’s Thesis, Department of Agronomy, National Chiayi University, Chiayi, Taiwan, 2011. [Google Scholar]
- Temiyakul, P.; Taylor, P.W.J.; Mongkolporn, O. Differential fruit maturity plays an important role in chili anthracnose infection. J. Kmutnb 2012, 22, 494–504. [Google Scholar]
- Yoon, J.B.; Park, H.G. Trispecies bridge crosses (Capsicum annuum × C. chinense) × C. baccatum, as an alternative for introgression of anthracnose reistance from C. baccatum into C. annuum. Hortic. Environ. Biotechnol. 2005, 46, 5–9. [Google Scholar]
- AVRDC. Host resistance to pepper anthracnose. In AVRDC Report; AVRDC—The World Vegetable Centre: Shanhua, Taiwan, 2003; pp. 29–30. [Google Scholar]
- Pakdeevaraporn, P.; Wasee, S.; Taylor, P.W.J.; Mongkolporn, O. Inheritance of resistance to anthracnose caused by Colletotrichum capsici in Capsicum. Plant Breed. 2005, 124, 206–208. [Google Scholar] [CrossRef]
- Setiawati, W.; Udiarto, B.K.; Soetiarso, T.A. The effect of variety and planting system of chili pepper on incidence of whiteflies. J. Hortic. 2008, 18, 55–61. [Google Scholar]
- Reddy, M.K.; Srivastava, A.; Kumar, S.; Kumar, R.; Chawda, N.; Ebert, A.W.; Vishwakarma, M. Chili (Capsicum annuum L.) breeding in India: An overview. J. Breed. Genet. 2014, 46, 160–173. [Google Scholar]
- Garg, R.; Kumar, S.; Kumar, R.; Loganathan, M.; Saha, S.; Kumar, S.; Rai, A.B.; Roy, B.K. Novel source of resistance and differential reactions on chilli fruit infected by Colletotrichum capsici. Australas. Plant Pathol. 2013, 42, 227–233. [Google Scholar] [CrossRef]
- Kaur, N.; Singh, D.J.; Singh, K.D. Physiological and biochemical traits analysis of Capsicum annuum L. germplasm for resistance to Colletotrichum capsici. J. Cell Plant Sci. 2011, 2, 12–21. [Google Scholar]
- Kim, S.H.; Yoon, J.B.; Do, J.W.; Park, H.G. A major recessive gene associated with anthracnose resistance to Colletotrichum capsici in chili pepper (Capsicum annuum L.). Breed. Sci. 2008, 58, 137–141. [Google Scholar] [CrossRef]
- Lin, S.W.; Gniffke, P.A.; Wang, T.C. Inheritance of resistance to pepper anthracnose by Colletotrichum acutatum. Acta Hortic. 2007, 760, 329–334. [Google Scholar] [CrossRef]
- Lin, Q.; Kanchana-Udomkan, C.; Jaunet, T.; Mongkolporn, O. Genetic analysis of resistance to pepper anthracnose caused by Colletotrichum capsici. Thail. J. Agric. Sci. 2002, 35, 259–264. [Google Scholar]
- Mahasuk, P.; Taylor, P.W.J.; Mongkolporn, O. Identification of two new genes conferring resistance to Colletotrichum acutatum in Capsicum baccatum. Phytopathology 2009, 99, 1100–1104. [Google Scholar] [CrossRef] [PubMed]
- Flor, H.H. Current status of the gene-for-gene concept. Annu. Rev. Phytopathol. 1971, 9, 275–296. [Google Scholar] [CrossRef]
- Cheema, D.S.; Singh, D.P.; Rawal, R.D.; Deshpande, A.A. Inheritance of resistance to anthracnose disease in chillies. Capsicum Eggplant Newsl. 1984, 3, 44. [Google Scholar]
- Ahmed, N.; Dey, S.K.; Hundal, J.S. Inheritance of resistance to anthracnose in chilli. Indian Phytopathol. 1991, 44, 402–403. [Google Scholar]
- Qing, L.; Kanchana-Udomkarn, K.; Jaunet, T.; Mongkolporn, O. Inheritance of resistance to pepper anthracnose caused by Colletotrichum capsici. Capsicum Eggplant Newsl. 2002, 21, 85–88. [Google Scholar]
- Nanda, C.; Rao, M.A.; Ramesh, S.; Hittalmani, S.; Prathibha, V.H. Tagging SSR Markers associated with genomic regions controlling anthracnose resistance in chilli (Capsicum baccatum L.). Vegetos 2016, 29, 130–134. [Google Scholar] [CrossRef]
- Neetha, J.; Rao, A.M.; Ramesh, S.; Manjunath, B.; Mangalagowri, N.; Ashwini, M. Tagging SSR markers to genomic regions associated with anthracnose disease resistance and productivity per se traits in hot pepper (Capsicum annuum L.). Environ. Ecol. 2016, 34, 1440–1446. [Google Scholar]
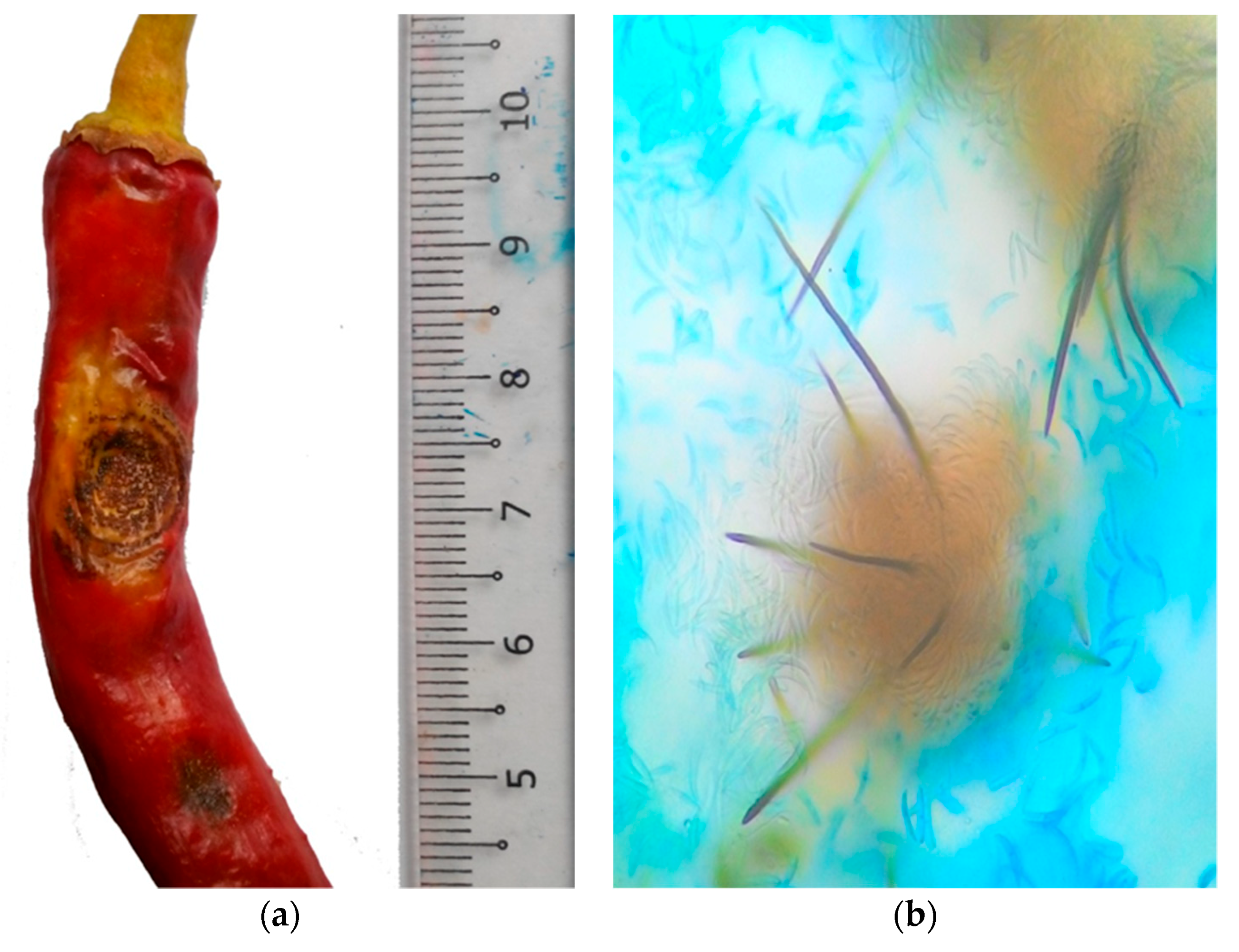
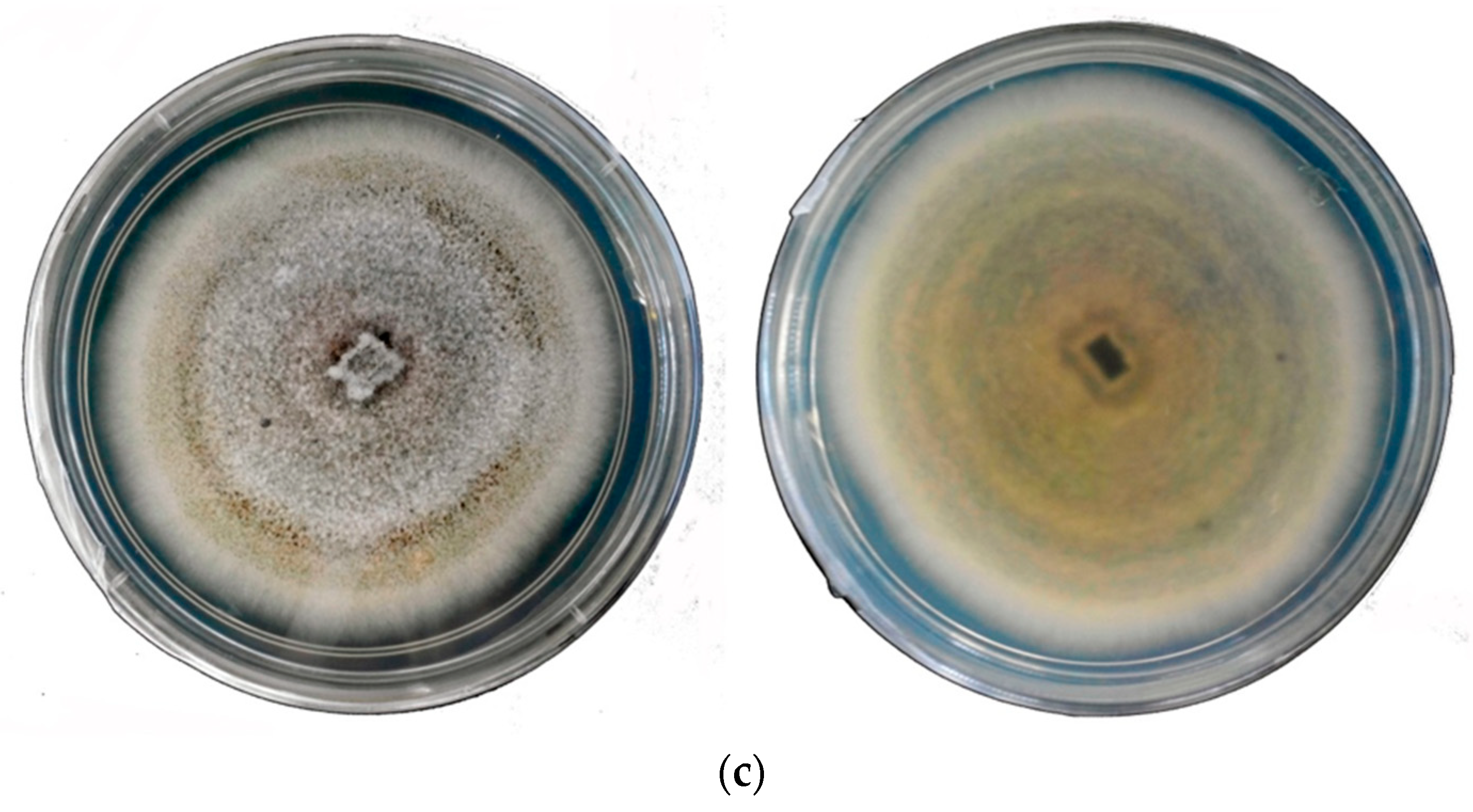
| Region/Country | Pathogen | Source |
|---|---|---|
| Malaysia | C. truncatum | Sariah [9]; Mahmodi et al. [54] |
| Thailand | C. acutatum, C. truncatum and C. gloeosporioides | Than et al. [53]; Montri [18] |
| Indonesia | C. acutatum, C. truncatum and C. gloeosporioides | Voorrips et al. [55] |
| India | C. truncatum, C. dematium, C. gloeosporioides, C. graminicola, C. acutatum, C. piperatum, C. atramentaum, C. truncatum, C. fructicola and C. siamense | Thind and Jhooty [56]; Hedge et al. [57]; Paul and Behl [58]; Susheela [59]; Selvakumar [60]; Kaur and Singh [61]; Ramachandran and Rathnamma [62]; Sharma et al. [33]; Sharma and Shenoy [63] |
| Korea | C. acutatum, C. gloeosporioides, C. coccodes and C. dematium | Park and Kim [64] |
| Papua New Guinea | C. truncatum and C. gloeosporioides | Pearson et al. [65] |
| New Zealand | C. coccodes | Johnston and Jones [66] |
| Taiwan | C. acutatum, C. truncatum and C. gloeosporioides | Manandhar et al. [47] |
| Australia | C. acutatum, C. atramentarium, C. dematium, C. gloeosporioides var. minor and C. gloeosporioides var. gloeosporioides | Simmonds [67] |
| United Kingdom | C. acutatum and Glomerella cingulata | Adikaram et al. [68] |
| USA | C. gloeosporioides, C. acutatum | Harp et al. [13]; Roberts et al. [45] |
| Vietnam | C. acutatum, C. truncatum, C. gloeosporioides and C. nigrum | Don et al. [69] |
| Sri Lanka | C. truncatum | Rajapakse and Ranasinghe [48] |
| Capsicum Species | Colletotrichum Species | Marker Type | Marker Name | Reference |
|---|---|---|---|---|
| C. annuum × C. chinense C. annuum × C. baccatum | C. truncatum C. acutatum | SSR, SCAR | HpmsE032, HpmsE143, InDel | Lee et al. [155]; Wang [156]; Suwor et al. [142] |
| C. annuum × C. chinense | C. acutatum | SSR, SCAR, CAPS | HpmsE057, HpmsE116, HpmsE126, ES382, Gpms161, Gp20068, ES64, Epms745, Gp20068, ES118, ES181, InDel, C2_At4g03400 | Sun et al. [141] |
| C. annuum × C. baccatum | C. truncatum C. acutatum | SSR, CAPS | HpmsE032, HpmsE143, CaR12.2M1, CcR9M1 | Lee et al. [155] |
| C. annuum × C. baccatum | C. truncatum C. acutatum | SSR | Hpms2-24, HpmsE143, HpmsE092, HpmsE032, HpmsE063 | Lee et al. [143] |
| C. annuum × C. chinense | C. gloeosporioides C. truncatum | AFLP, SSR | B1, B2, D1, G1, H1, CA-MS6, CA-MS12, CA-MS22, CA-MS23, CA-MS25 | Voorrips et al. [55] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ridzuan, R.; Rafii, M.Y.; Ismail, S.I.; Mohammad Yusoff, M.; Miah, G.; Usman, M. Breeding for Anthracnose Disease Resistance in Chili: Progress and Prospects. Int. J. Mol. Sci. 2018, 19, 3122. https://doi.org/10.3390/ijms19103122
Ridzuan R, Rafii MY, Ismail SI, Mohammad Yusoff M, Miah G, Usman M. Breeding for Anthracnose Disease Resistance in Chili: Progress and Prospects. International Journal of Molecular Sciences. 2018; 19(10):3122. https://doi.org/10.3390/ijms19103122
Chicago/Turabian StyleRidzuan, Raihana, Mohd Y. Rafii, Siti Izera Ismail, Martini Mohammad Yusoff, Gous Miah, and Magaji Usman. 2018. "Breeding for Anthracnose Disease Resistance in Chili: Progress and Prospects" International Journal of Molecular Sciences 19, no. 10: 3122. https://doi.org/10.3390/ijms19103122
APA StyleRidzuan, R., Rafii, M. Y., Ismail, S. I., Mohammad Yusoff, M., Miah, G., & Usman, M. (2018). Breeding for Anthracnose Disease Resistance in Chili: Progress and Prospects. International Journal of Molecular Sciences, 19(10), 3122. https://doi.org/10.3390/ijms19103122