Divergent Expression Patterns and Function of Two cxcr4 Paralogs in Hermaphroditic Epinephelus coioides
Abstract
1. Introduction
2. Results
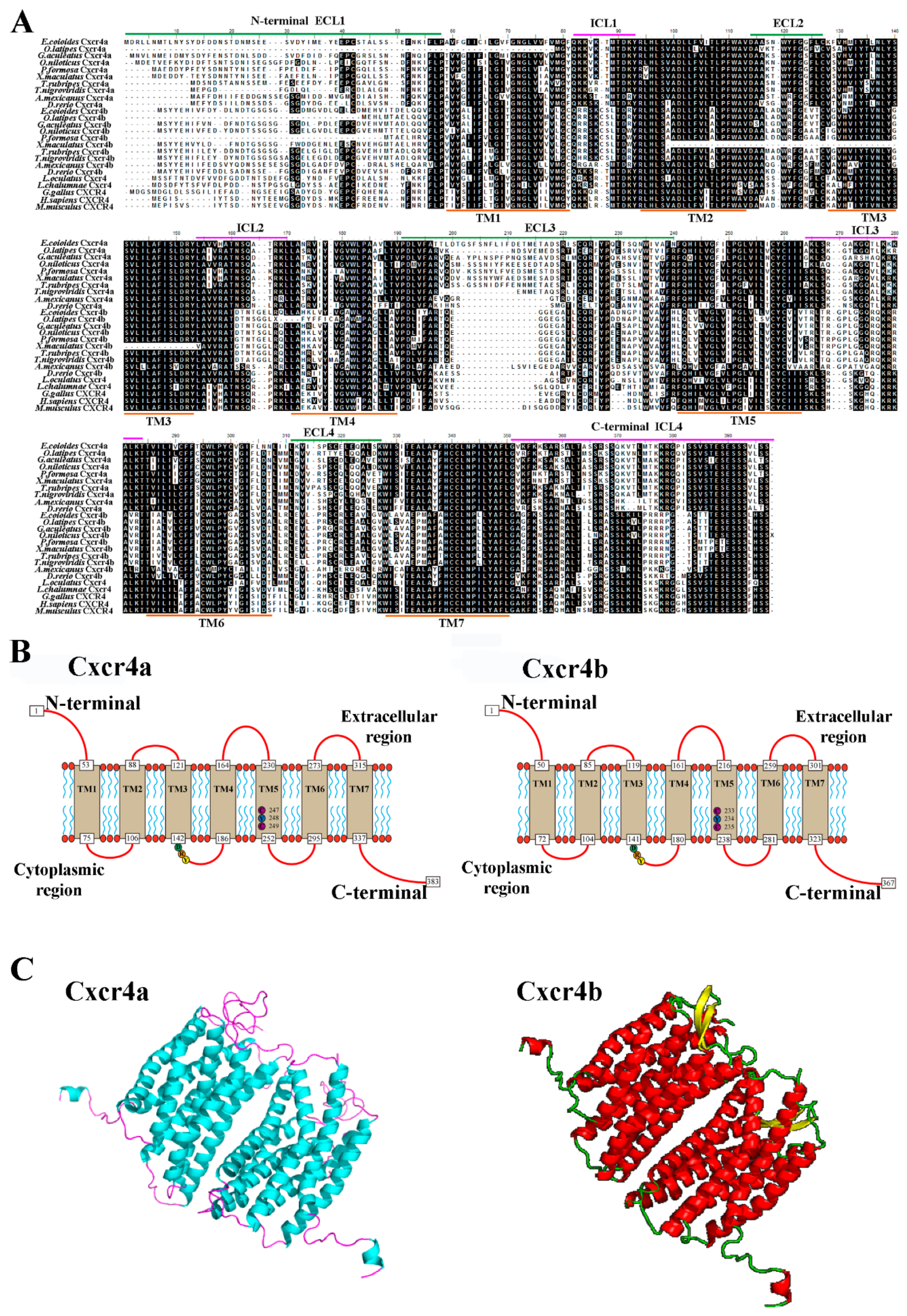
2.1. Molecular Characterization of Two cxcr4 Paralogs in Orange-Spotted Grouper
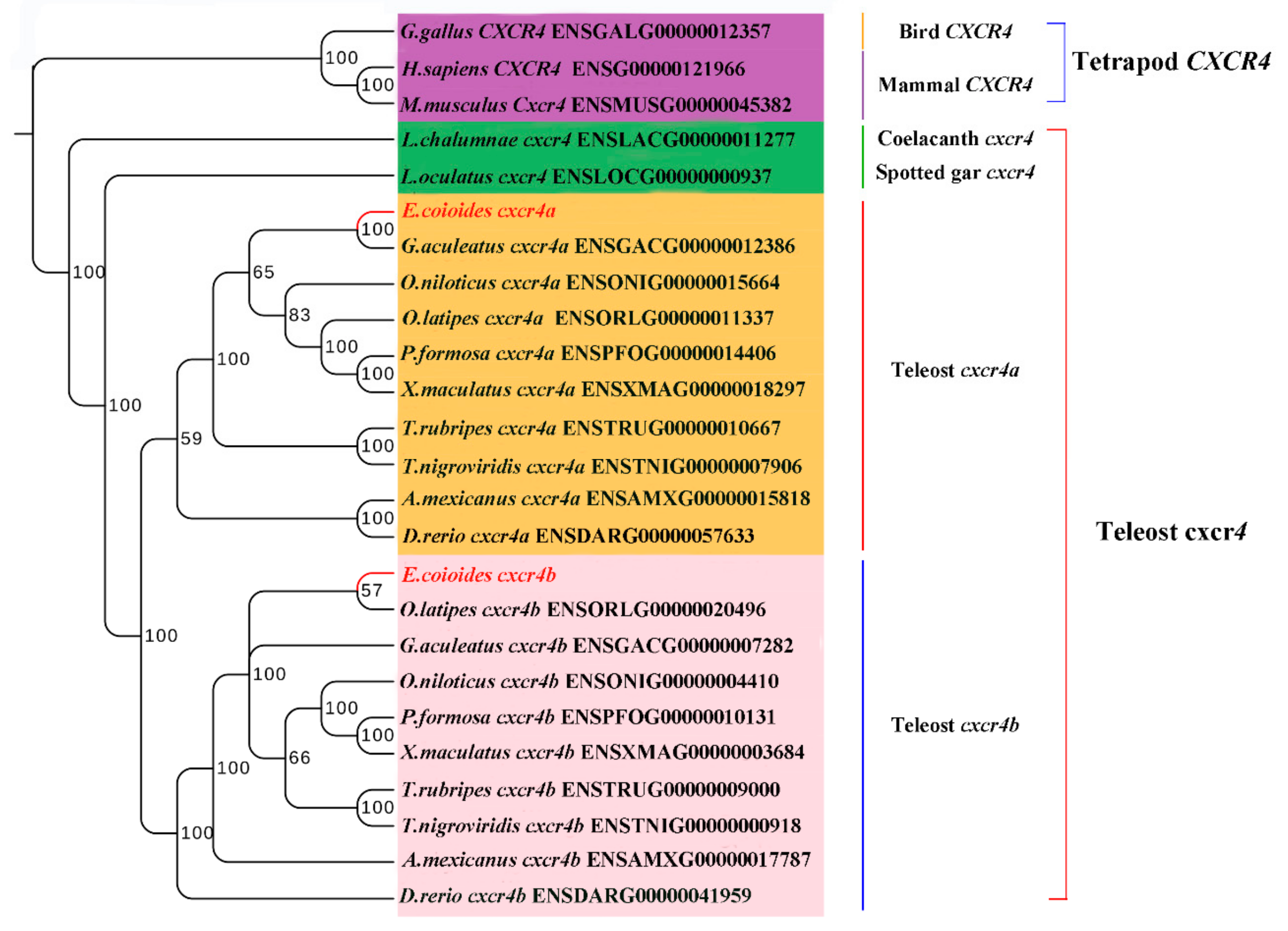
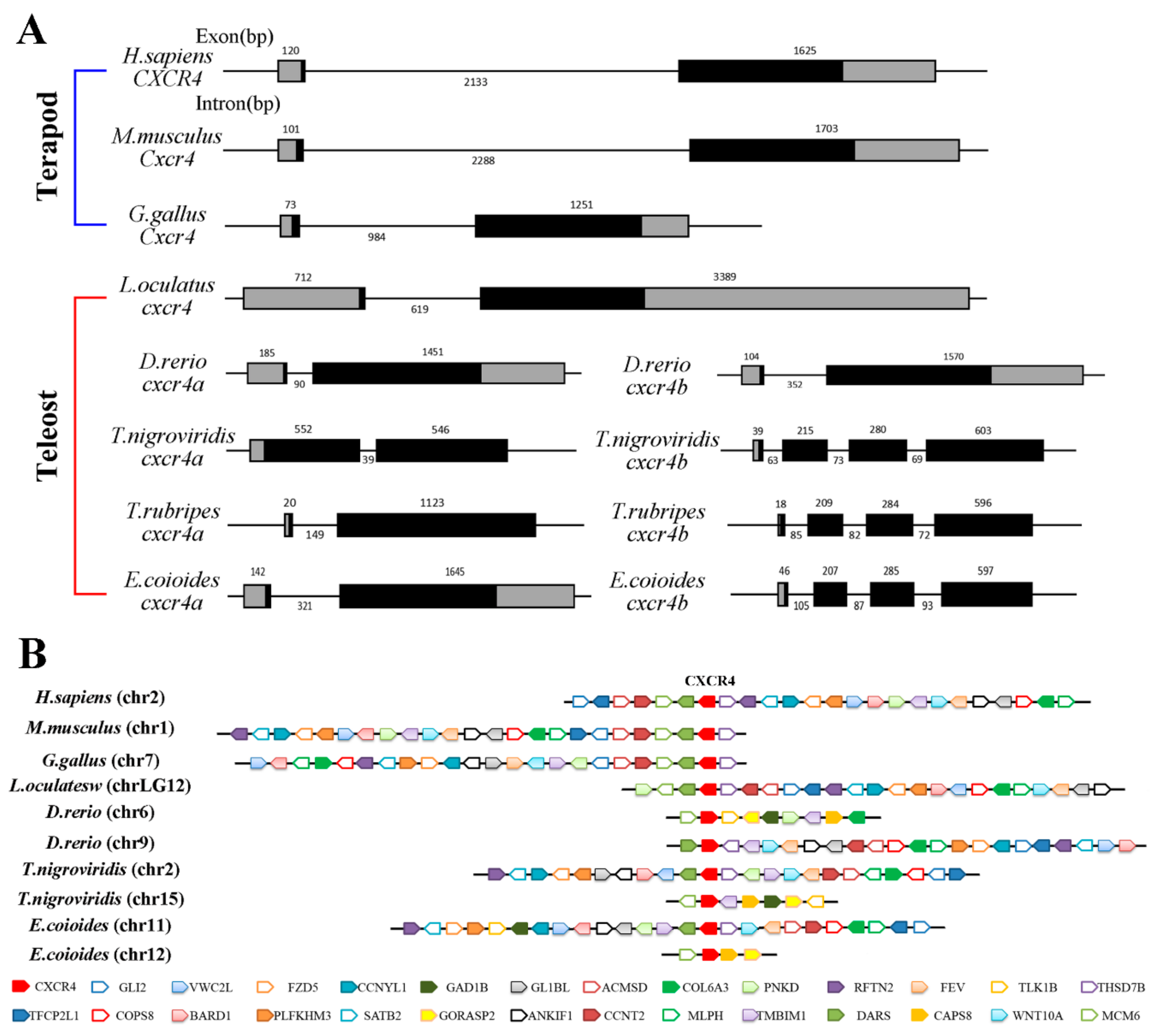
2.2. Phylogenetic Relationship, Gene Structure and Gene Synteny of Teleost and Tetrapod cxcr4s
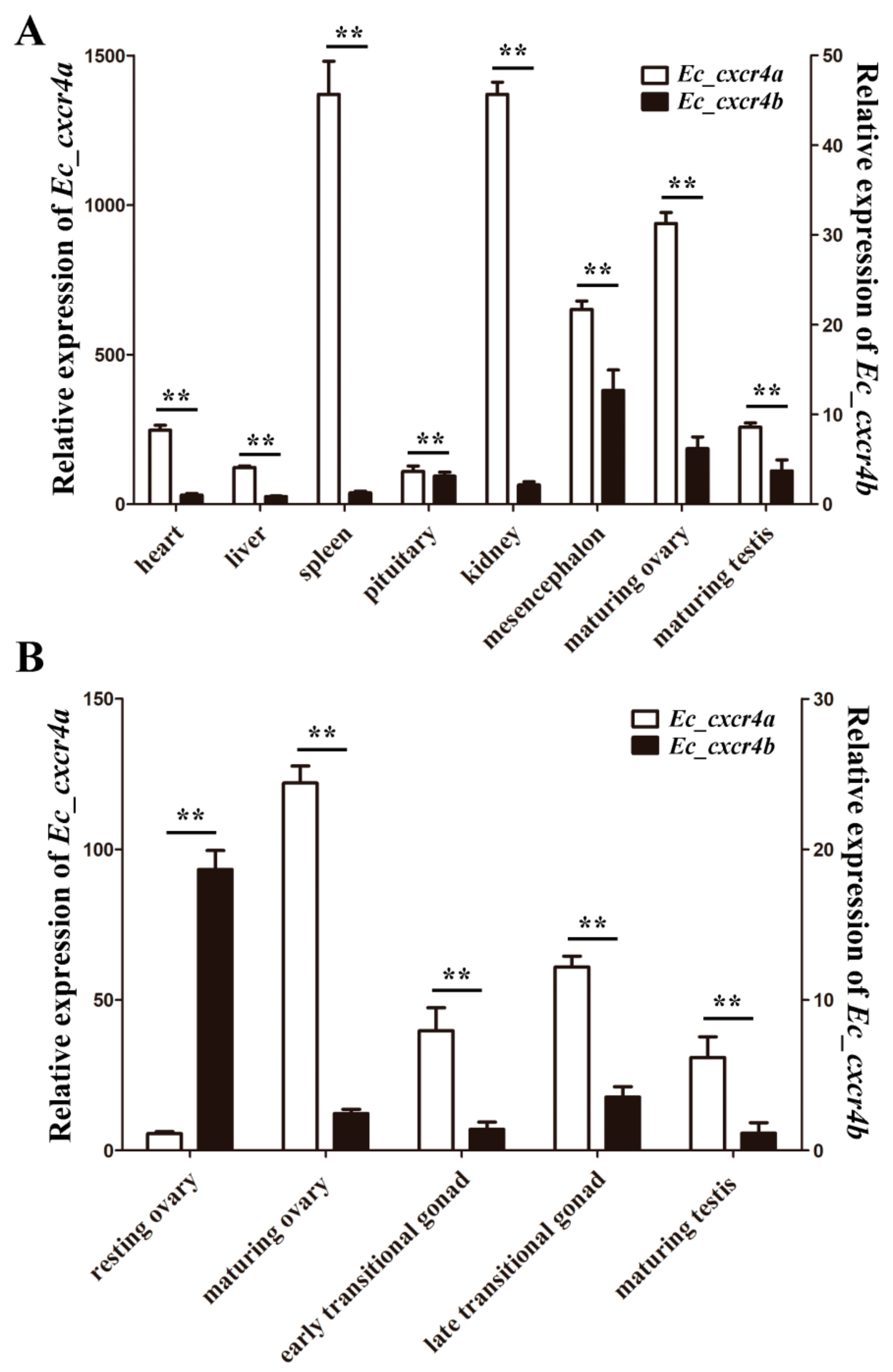
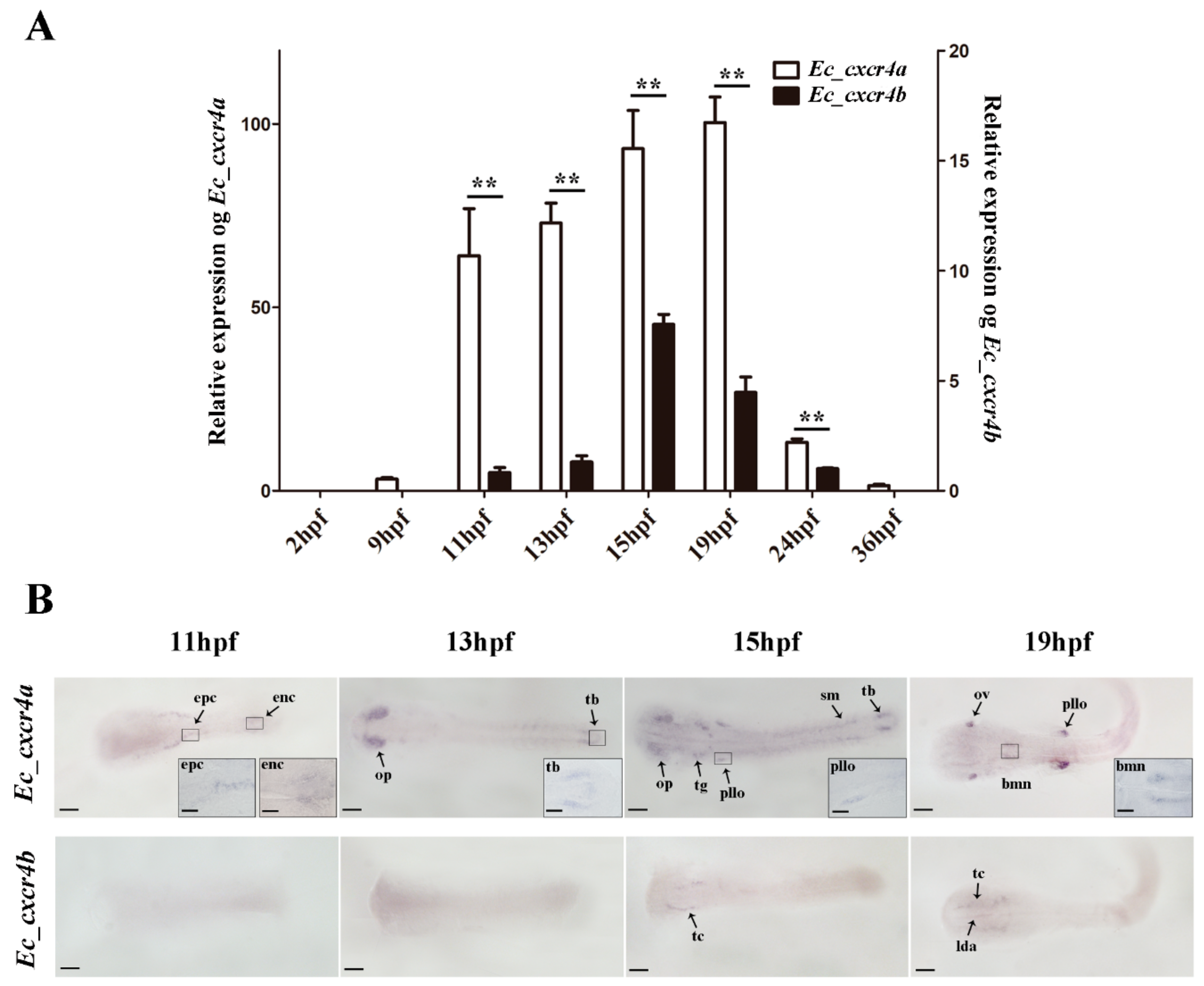
2.3. Biased and Differential Expression Patterns of Two cxcr4 Paralogs in Orange-Spotted Grouper
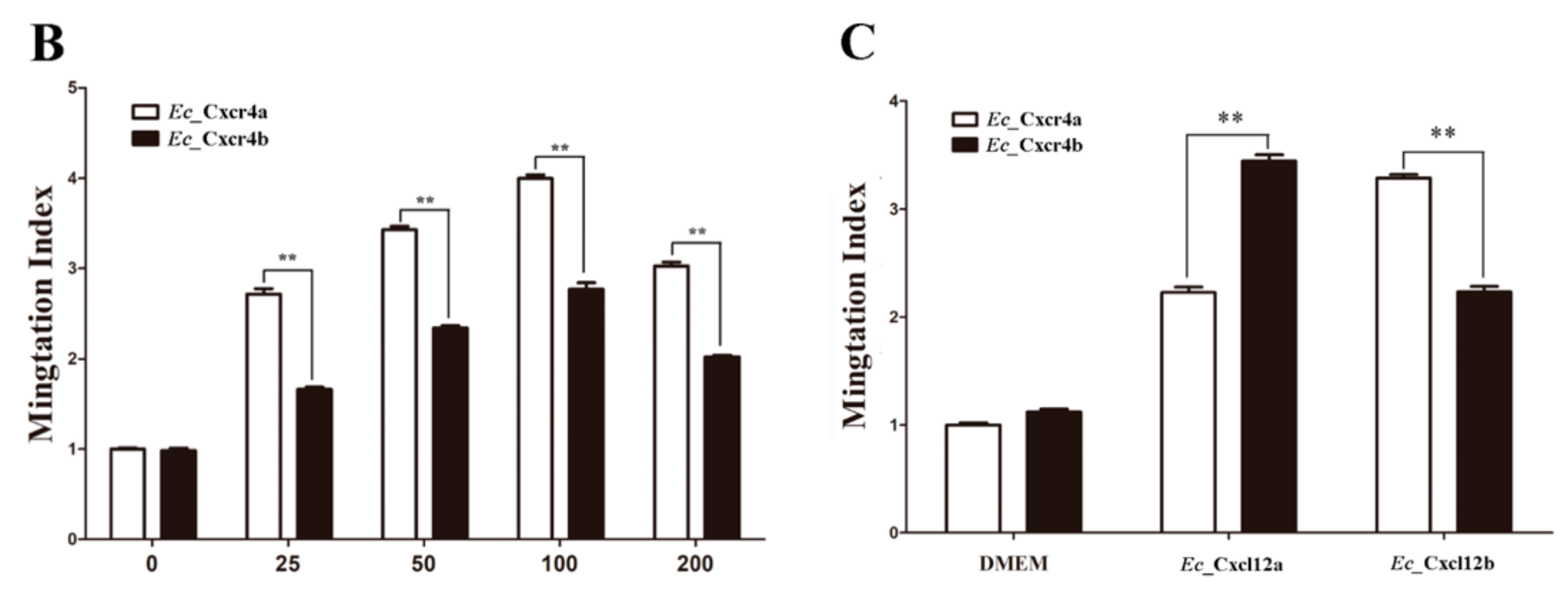
2.4. Differential Chemotactic Migratory Abilities of HEK293T Cells Transfected with Cxcr4a/b Induced by Cxcl12a/b
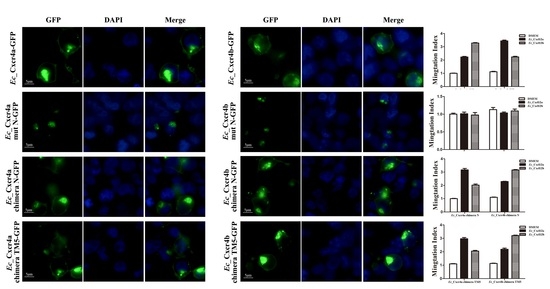
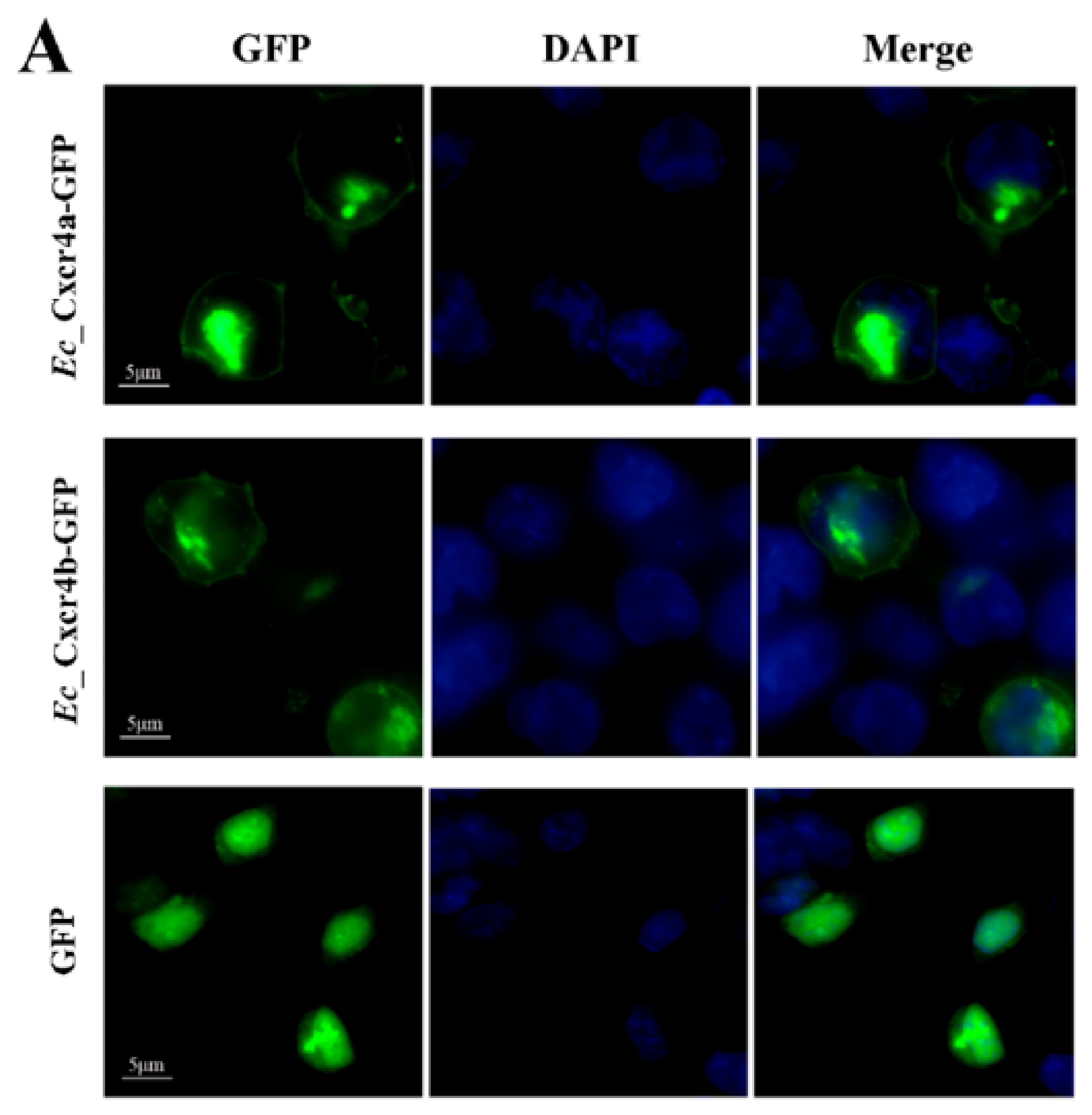
2.5. Specific Key Domains for Chemotactic Interaction of Ec_Cxcr4a/b-Ec_Cxcl12a/b
3. Discussion
4. Materials and Methods
4.1. Experimental Fish
4.2. RNA Extraction and qPCR
4.3. Sequence and Phylogenetic Analyses
4.4. Probe Synthesis and In Situ Hybridization
4.5. Plasmid Constructs
4.6. Subcellular Localization
4.7. Chemotaxis Assay
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Fredriksson, R.; Lagerstrom, M.C.; Lundin, L.G.; Schioth, H.B. The g-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol. Pharmacol. 2003, 63, 1256–1272. [Google Scholar] [CrossRef] [PubMed]
- Viola, A.; Luster, A.D. Chemokines and their receptors: Drug targets in immunity and inflammation. Annu. Rev. Pharmacol. 2008, 48, 171–197. [Google Scholar] [CrossRef] [PubMed]
- Bajoghli, B. Evolution and function of chemokine receptors in the immune system of lower vertebrates. Eur. J. Immunol. 2013, 43, 1686–1692. [Google Scholar] [CrossRef] [PubMed]
- Bussmann, J.; Raz, E. Chemokine-guided cell migration and motility in zebrafish development. EMBO J. 2015, 34, 1309–1318. [Google Scholar] [CrossRef] [PubMed]
- Calderon, L.; Boehm, T. Three chemokine receptors cooperatively regulate homing of hematopoietic progenitors to the embryonic mouse thymus. Proc. Natl. Acad. Sci. USA 2011, 108, 7517–7522. [Google Scholar] [CrossRef] [PubMed]
- Griffith, J.W.; Sokol, C.L.; Luster, A.D. Chemokines and chemokine receptors: Positioning cells for host defense and immunity. Annu. Rev. Immunol. 2014, 32, 659–702. [Google Scholar] [CrossRef] [PubMed]
- Nagarsheth, N.; Wicha, M.S.; Zou, W.P. Chemokines in the cancer microenvironment and their relevance in cancer immunotherapy. Nat. Rev. Immunol. 2017, 17, 559–572. [Google Scholar] [CrossRef] [PubMed]
- Schall, T.J.; Proudfoot, A.E.I. Overcoming hurdles in developing successful drugs targeting chemokine receptors. Nat. Rev. Immunol. 2011, 11, 355–363. [Google Scholar] [CrossRef] [PubMed]
- Zlotnik, A.; Yoshie, O. The chemokine superfamily revisited. Immunity 2012, 36, 705–716. [Google Scholar] [CrossRef] [PubMed]
- Bajoghli, B.; Aghaallaei, N.; Hess, I.; Rode, I.; Netuschil, N.; Tay, B.H.; Venkatesh, B.; Yu, J.K.; Kaltenbach, S.L.; Holland, N.D.; et al. Evolution of genetic networks underlying the emergence of thymopoiesis in vertebrates. Cell 2009, 138, 186–197. [Google Scholar] [CrossRef] [PubMed]
- Huising, M.O.; Stolte, E.; Flik, G.; Savelkoul, H.F.J.; Verburg-van Kemenade, B.M.L. Cxc chemokines and leukocyte chemotaxis in common carp (Cyprinus carpio L.). Dev. Comp. Immunol. 2003, 27, 875–888. [Google Scholar] [CrossRef]
- Cencioni, C.; Capogrossi, M.C.; Napolitano, M. The sdf-1/cxcr4 axis in stem cell preconditioning. Cardiovasc. Res. 2012, 94, 400–407. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.B.; Liu, S.B.; Li, Y.; Wang, X.H.; Xue, W.J.; Ge, G.Q.; Luo, X.H. The role of sdf-1-cxcr4/cxcr7 axis in the therapeutic effects of hypoxia-preconditioned mesenchymal stem cells for renal ischemia/reperfusion injury. PLoS ONE 2012. [Google Scholar] [CrossRef] [PubMed]
- Nagasawa, T.; Hirota, S.; Tachibana, K.; Takakura, N.; Nishikawa, S.; Kitamura, Y.; Yoshida, N.; Kikutani, H.; Kishimoto, T. Defects of b-cell lymphopoiesis and bone-marrow myelopoiesis in mice lacking the cxc chemokine pbsf/sdf-1. Nature 1996, 382, 635–638. [Google Scholar] [CrossRef] [PubMed]
- Tachibana, K.; Hirota, S.; Iizasa, H.; Yoshida, H.; Kawabata, K.; Kataoka, Y.; Kitamura, Y.; Matsushima, K.; Yoshida, N.; Nishikawa, S.; et al. The chemokine receptor cxcr4 is essential for vascularization of the gastrointestinal tract. Nature 1998, 393, 591–594. [Google Scholar] [CrossRef] [PubMed]
- Zou, Y.R.; Kottmann, A.H.; Kuroda, M.; Taniuchi, I.; Littman, D.R. Function of the chemokine receptor cxcr4 in haematopoiesis and in cerebellar development. Nature 1998, 393, 595–599. [Google Scholar] [CrossRef] [PubMed]
- Knaut, H.; Werz, C.; Geisler, R.; Nusslein-Volhard, C. A zebrafish homologue of the chemokine receptor cxcr4 is a germ-cell guidance receptor. Nature 2003, 421, 279–282. [Google Scholar] [CrossRef] [PubMed]
- Katsumoto, K.; Kume, S. The role of cxcl12-cxcr4 signaling pathway in pancreatic development. Theranostics 2013, 3, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Alejo, A.; Tafalla, C. Chemokines in teleost fish species. Dev. Comp. Immunol. 2011, 35, 1215–1222. [Google Scholar] [CrossRef] [PubMed]
- Bird, S.; Tafalla, C. Teleost chemokines and their receptors. Biology 2015, 4, 756–784. [Google Scholar] [CrossRef] [PubMed]
- Peatman, E.; Liu, Z.J. Evolution of cc chemokines in teleost fish: A case study in gene duplication and implications for immune diversity. Immunogenetics 2007, 59, 613–623. [Google Scholar] [CrossRef] [PubMed]
- Zou, W.P.; Max, S.W. Chemokines and cellular plasticity of ovarian cancer stem cells. Oncoscience 2015. [Google Scholar] [CrossRef] [PubMed]
- Amores, A.; Force, A.; Yan, Y.L.; Joly, L.; Amemiya, C.; Fritz, A.; Ho, R.K.; Langeland, J.; Prince, V.; Wang, Y.L.; et al. Zebrafish hox clusters and vertebrate genome evolution. Science 1998, 282, 1711–1714. [Google Scholar] [CrossRef] [PubMed]
- Postlethwait, J.H.; Yan, Y.L.; Gates, M.A.; Horne, S.; Amores, A.; Brownlie, A.; Donovan, A.; Egan, E.S.; Force, A.; Gong, Z.Y.; et al. Vertebrate genome evolution and the zebrafish gene map. Nat. Genet. 1998, 18, 345–349. [Google Scholar] [CrossRef] [PubMed]
- Chong, S.W.; Emelyanov, A.; Gong, Z.Y.; Korzh, V. Expression pattern of two zebrafish genes, cxcr4a and cxcr4b. Mech. Dev. 2001, 109, 347–354. [Google Scholar] [CrossRef]
- Doitsidou, M.; Reichman-Fried, M.; Stebler, J.; Koprunner, M.; Dorries, J.; Meyer, D.; Esguerra, C.V.; Leung, T.; Raz, E. Guidance of primordial germ cell migration by the chemokine sdf-1. Cell 2002, 111, 647–659. [Google Scholar] [CrossRef]
- Gao, W.H.; Fang, L.; Yang, D.Q.; Ai, K.T.; Luo, K.; Tian, G.M.; Zhou, J.W.; Hu, W.; Yuan, H.W.; Xu, Q.Q. Cloning and expression of Asian swamp eel (Monopterus albus) cxcr4 paralogues, and their modulation by pathogen infection. Aquaculture 2016, 457, 50–60. [Google Scholar] [CrossRef]
- Fu, Q.; Yang, Y.J.; Li, C.; Zeng, Q.F.; Zhou, T.; Li, N.; Liu, Y.; Liu, S.K.; Liu, Z.J. The cc and cxc chemokine receptors in channel catfish (Ictalurus punctatus) and their involvement in disease and hypoxia responses. Dev. Comp. Immunol. 2017, 77, 241–251. [Google Scholar] [CrossRef] [PubMed]
- Kurokawa, H.; Saito, D.; Nakamura, S.; Katoh-Fukui, Y.; Ohta, K.; Baba, T.; Morohashi, K.I.; Tanaka, M. Germ cells are essential for sexual dimorphism in the medaka gonad. Proc. Natl. Acad. Sci. USA 2007, 104, 16958–16963. [Google Scholar] [CrossRef] [PubMed]
- Jia, A.R.; Zhang, X.H. Molecular cloning, characterization, and expression analysis of the cxcr4 gene from turbot: Scophthalmus maximus. J. Biomed. Biotechnol. 2009, 2009. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.Y.; Chen, Y.M.; Hsu, H.H.; Shiu, C.T.; Kuo, H.C.; Chen, T.Y. Grouper (Epinephelus coioides) cxcr4 is expressed in response to pathogens infection and early stage of development. Dev. Comp. Immunol. 2012, 36, 112–120. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.X.; Kang, L.S.; Liu, W.; Lou, B.; Wu, C.W.; Jiang, L.H. Molecular characterization and expression analysis of the large yellow croaker (Larimichthys crocea) chemokine receptors cxcr2, cxcr3, and cxcr4 after bacterial and poly I:C challenge. Fish Shellfish Immun. 2017, 70, 228–239. [Google Scholar] [CrossRef] [PubMed]
- Priyathilaka, T.T.; Oh, M.; Bathige, S.D.N.K.; De Zoysa, M.; Lee, J. Two distinct cxc chemokine receptors (cxcr3 and cxcr4) from the big-belly seahorse hippocampus abdominalis: Molecular perspectives and immune defensive role upon pathogenic stress. Fish Shellfish Immun. 2017, 65, 59–70. [Google Scholar] [CrossRef] [PubMed]
- Thulasitha, W.S.; Umasuthan, N.; Revathy, K.S.; Whang, I.; Lee, J. Molecular characterization, genomic structure and expressional profiles of a cxc chemokine receptor 4 (cxcr4) from rock bream oplegnathus fasciatus. Fish Shellfish Immun. 2015, 44, 471–477. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Luo, H.J.; Yin, G.B.; Dong, C.R.; Xu, M.; Chen, G.G.; Liu, Z.M. Overexpression of hif-2 alpha, twist, and cxcr4 is associated with lymph node metastasis in papillary thyroid carcinoma. Clin. Dev. Immunol. 2013, 2013. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.J.; Zhu, Z.H.; Sun, Y.N.; Ren, L.P.; Wang, R.X. Characterization and expression of the cxcr1 and cxcr4 in miiuy croaker and evolutionary analysis shows the strong positive selection pressures imposed in mammal cxcr1. Dev. Comp. Immunol. 2014, 44, 133–144. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.Y.; Zhang, B.; Lin, Y.; Yang, Y.; Liu, X.Q.; Lu, F. Breast cancer metastasis suppressor 1 inhibits sdf-1 alpha-induced migration of non-small cell lung cancer by decreasing cxcr4 expression. Cancer Lett. 2008, 269, 46–56. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wei, X.L.; Chen, L.P.; Chen, N.; Li, Y.H.; Wang, W.M.; Wang, H.L. Sequence analysis and expression differentiation of chemokine receptor cxcr4b among three populations of megalobrama amblycephala. Dev. Comp. Immunol. 2013, 40, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Alabyev, B.Y.; Najakshin, A.M.; Mechetina, L.V.; Taranin, A.V. Cloning of a cxcr4 homolog in chondrostean fish and characterization of the cxcr4-specific structural features. Dev. Comp. Immunol. 2000, 24, 765–770. [Google Scholar] [CrossRef]
- Ko, C.F.; Chiou, T.T.; Chen, T.T.; Wu, J.L.; Chen, J.C.; Lu, J.K. Molecular cloning of myostatin gene and characterization of tissue-specific and developmental stage-specific expression of the gene in orange spotted grouper, epinephelus coioides. Mar. Biotechnol. 2007, 9, 20–32. [Google Scholar] [CrossRef] [PubMed]
- Lo, W.Y.; Chang, C.F.; Song, Y.L. Evaluation of dorsal aorta cannulation for immunological studies of grouper (Epinephelus malabaricus). Fish Shellfish Immun. 2003, 14, 289–303. [Google Scholar] [CrossRef]
- Yao, B.; Zhou, L.; Wang, Y.; Xia, W.; Gui, J.F. Differential expression and dynamic changes of sox3 during gametogenesis and sex reversal in protogynous hermaphroditic fish. J. Exp. Zool. Part A 2007, 307, 207–219. [Google Scholar] [CrossRef] [PubMed]
- Yeh, S.L.; Dai, Q.C.; Chu, Y.T.; Kuo, C.M.; Ting, Y.Y.; Chang, C.F. Induced sex change, spawning and larviculture of potato grouper, epinephelus tukula. Aquaculture 2003, 228, 371–381. [Google Scholar] [CrossRef]
- Zhou, L.; Gui, J.F. Molecular mechanisms underlying sex change in hermaphroditic groupers. Fish Physiol. Biochem. 2010, 36, 181–193. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.H.; Zhou, L.; Li, Z.; Liu, X.C.; Li, S.S.; Wang, Y.; Gui, J.F. Sexual dimorphic expression of dnd in germ cells during sex reversal and its requirement for primordial germ cell survival in protogynous hermaphroditic grouper. Comp. Biochem. Phys. B 2017, 208, 47–57. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.H.; Wang, Y.; Lu, W.J.; Li, Z.; Liu, X.C.; Li, S.S.; Zhou, L.; Gui, J.F. Divergent expression patterns and function implications of four nanos genes in a hermaphroditic fish, epinephelus coioides. Int. J. Mol. Sci. 2017, 18, 685. [Google Scholar] [CrossRef] [PubMed]
- Son, B.R.; Marquez-Curtis, L.A.; Kucia, M.; Wysoczynski, M.; Turner, A.R.; Ratajczak, J.; Ratajczak, M.Z.; Janowska-Wieczorek, A. Migration of bone marrow and cord blood mesenchymal stem cells in vitro is regulated by stromal-derived factor-1-cxcr4 and hepatocyte growth factor-c-met axes and involves matrix metalloproteinases. Stem Cells 2006, 24, 1254–1264. [Google Scholar] [CrossRef] [PubMed]
- Doranz, B.J.; Orsini, M.J.; Turner, J.D.; Hoffman, T.L.; Berson, J.F.; Hoxie, J.A.; Peiper, S.C.; Brass, L.F.; Doms, R.W. Identification of cxcr4 domains that support coreceptor and chemokine receptor functions. J. Virol. 1999, 73, 2752–2761. [Google Scholar] [PubMed]
- Roland, J.; Murphy, B.J.; Ahr, B.; Robert-Hebmann, W.; Delauzun, V.; Nye, K.E.; Devaux, C.; Biard-Piechaczyk, M. Role of the intracellular domains of cxcr4 in sdf-1-mediated signaling. Blood 2003, 101, 399–406. [Google Scholar] [CrossRef] [PubMed]
- Amores, A.; Catchen, J.; Ferrara, A.; Fontenot, Q.; Postlethwait, J.H. Genome evolution and meiotic maps by massively parallel DNA sequencing: Spotted gar, an outgroup for the teleost genome duplication. Genetics 2011, 188, 799–808. [Google Scholar] [CrossRef] [PubMed]
- Braasch, I.; Gehrke, A.R.; Smith, J.J.; Kawasaki, K.; Manousaki, T.; Pasquier, J.; Amores, A.; Desvignes, T.; Batzel, P.; Catchen, J.; et al. The spotted gar genome illuminates vertebrate evolution and facilitates human-teleost comparisons. Nat. Genet. 2016, 48, 427–437. [Google Scholar] [CrossRef] [PubMed]
- Jaillon, O.; Aury, J.M.; Brunet, F.; Petit, J.L.; Stange-Thomann, N.; Mauceli, E.; Bouneau, L.; Fischer, C.; Ozouf-Costaz, C.; Bernot, A.; et al. Genome duplication in the teleost fish tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature 2004, 431, 946–957. [Google Scholar] [CrossRef] [PubMed]
- Kasahara, M.; Naruse, K.; Sasaki, S.; Nakatani, Y.; Qu, W.; Ahsan, B.; Yamada, T.; Nagayasu, Y.; Doi, K.; Kasai, Y.; et al. The medaka draft genome and insights into vertebrate genome evolution. Nature 2007, 447, 714–719. [Google Scholar] [CrossRef] [PubMed]
- Meyer, A.; Van de Peer, Y. From 2r to 3r: Evidence for a fish-specific genome duplication (fsgd). In BioEssays: News and Reviews in Molecular, Cellular and Developmental Biology; Wiley: Hoboken, NJ, USA, 2005; Volume 27, pp. 937–945. [Google Scholar]
- Zhou, L.; Gui, J. Natural and artificial polyploids in aquaculture. Aquac. Fish. 2017, 2, 103–111. [Google Scholar] [CrossRef]
- Amemiya, C.T.; Alfoldi, J.; Lee, A.P.; Fan, S.H.; Philippe, H.; MacCallum, I.; Braasch, I.; Manousaki, T.; Schneider, I.; Rohner, N.; et al. The african coelacanth genome provides insights into tetrapod evolution. Nature 2013, 496, 311–316. [Google Scholar] [CrossRef] [PubMed]
- Wapinski, I.; Pfeffer, A.; Friedman, N.; Regev, A. Natural history and evolutionary principles of gene duplication in fungi. Nature 2007, 449, 54–61. [Google Scholar] [CrossRef] [PubMed]
- Scannell, D.R.; Byrne, K.P.; Gordon, J.L.; Wong, S.; Wolfe, K.H. Multiple rounds of speciation associated with reciprocal gene loss in polyploid yeasts. Nature 2006, 440, 341–345. [Google Scholar] [CrossRef] [PubMed]
- Markov, A.V.; Kaznacheev, I.S. Evolutionary consequences of polyploidy in prokaryotes and the origin of mitosis and meiosis. Biol. Direct 2016. [Google Scholar] [CrossRef] [PubMed]
- Otto, S.P. The evolutionary consequences of polyploidy. Cell 2007, 131, 452–462. [Google Scholar] [CrossRef] [PubMed]
- Lien, S.; Koop, B.F.; Sandve, S.R.; Miller, J.R.; Kent, M.P.; Nome, T.; Hvidsten, T.R.; Leong, J.S.; Minkley, D.R.; Zimin, A.; et al. The atlantic salmon genome provides insights into rediploidization. Nature 2016, 533, 200–205. [Google Scholar] [CrossRef] [PubMed]
- Xiong, J.; Lu, X.Y.; Lu, Y.M.; Zeng, H.H.; Yuan, D.X.; Feng, L.F.; Chang, Y.; Bowen, J.; Gorovsky, M.; Fu, C.J.; et al. Tetrahymena gene expression database (tged): A resource of microarray data and co-expression analyses for tetrahymena. Sci. China Life Sci. 2011, 54, 65–67. [Google Scholar] [CrossRef] [PubMed]
- Comai, L. The advantages and disadvantages of being polyploid. Nat. Rev. Genet. 2005, 6, 836–846. [Google Scholar] [CrossRef] [PubMed]
- Buggs, R.J.A.; Chamala, S.; Wu, W.; Tate, J.A.; Schnable, P.S.; Soltis, D.E.; Soltis, P.S.; Barbazuk, W.B. Rapid, repeated, and clustered loss of duplicate genes in allopolyploid plant populations of independent origin. Curr. Biol. 2012, 22, 248–252. [Google Scholar] [CrossRef] [PubMed]
- Doyle, J.J.; Flagel, L.E.; Paterson, A.H.; Rapp, R.A.; Soltis, D.E.; Soltis, P.S.; Wendel, J.F. Evolutionary genetics of genome merger and doubling in plants. Annu. Rev. Genet. 2008, 42, 443–461. [Google Scholar] [CrossRef] [PubMed]
- Grover, C.E.; Gallagher, J.P.; Szadkowski, E.P.; Yoo, M.J.; Flagel, L.E.; Wendel, J.F. Homoeolog expression bias and expression level dominance in allopolyploids. New Phytol. 2012, 196, 966–971. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.J.; Koh, J.; Yoo, M.J.; Chen, S.X.; Wendel, J.F. Gene-expression novelty in allopolyploid cotton: A proteomic perspective. Genetics 2015, 200, 91–104. [Google Scholar] [CrossRef] [PubMed]
- Jackson, S.; Chen, Z.J. Genomic and expression plasticity of polyploidy. Curr. Opin. Plant Biol. 2010, 13, 153–159. [Google Scholar] [CrossRef] [PubMed]
- Koh, J.; Chen, S.X.; Zhu, N.; Yu, F.H.; Soltis, P.S.; Soltis, D.E. Comparative proteomics of the recently and recurrently formed natural allopolyploid Tragopogon mirus (asteraceae) and its parents. New Phytol. 2012, 196, 292–305. [Google Scholar] [CrossRef] [PubMed]
- Lashermes, P.; Hueber, Y.; Combes, M.C.; Severac, D.; Dereeper, A. Inter-genomic DNA exchanges and homeologous gene silencing shaped the nascent allopolyploid coffee genome (Coffea arabica L.). G3-Genes Genom. Genet. 2016, 6, 2937–2948. [Google Scholar] [CrossRef] [PubMed]
- Page, J.T.; Liechty, Z.S.; Alexander, R.H.; Clemons, K.; Hulse-Kemp, A.M.; Ashrafi, H.; Van Deynze, A.; Stelly, D.M.; Udall, J.A. DNA sequence evolution and rare homoeologous conversion in tetraploid cotton. PLoS Genet. 2016. [Google Scholar] [CrossRef]
- Pala, I.; Coelho, M.M.; Schartl, M. Dosage compensation by gene-copy silencing in a triploid hybrid fish. Curr. Biol. 2008, 18, 1344–1348. [Google Scholar] [CrossRef] [PubMed]
- Salmon, A.; Flagel, L.; Ying, B.; Udall, J.A.; Wendel, J.F. Homoeologous nonreciprocal recombination in polyploid cotton. New Phytol. 2010, 186, 123–134. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.F.; Dong, Q.L.; Li, X.C.; Yuliang, A.Z.; Yu, Y.N.; Li, N.; Liu, B.; Gong, L. Cytonuclear variation of rubisco in synthesized rice hybrids and allotetraploids. Plant Genome 2017. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.H.; Liu, D.Y.; Wang, X.W.; Ji, C.M.; Cheng, F.; Liu, B.N.; Hu, Z.Y.; Chen, S.; Pental, D.; Ju, Y.H.; et al. The genome sequence of allopolyploid brassica juncea and analysis of differential homoeolog gene expression influencing selection. Nat. Genet. 2016, 48, 1225–1232. [Google Scholar] [CrossRef] [PubMed]
- Yoo, M.J.; Szadkowski, E.; Wendel, J.F. Homoeolog expression bias and expression level dominance in allopolyploid cotton. Heredity 2013, 110, 171–180. [Google Scholar] [CrossRef] [PubMed]
- Amat-Fernandez, J.; Hammond, M.J.; Liang, D.; Wang, T.F.; Ventura, T.; Elizur, A.; Cummins, S.F. Molecular characterization of sdf1 and cxcr4 in the mozambique tilapia, oreochromis mossambicus. Anim. Reprod. Sci. 2017, 176, 51–63. [Google Scholar] [CrossRef] [PubMed]
- Paredes-Zuniga, S.; Morales, R.A.; Munoz-Sanchez, S.; Munoz-Montecinos, C.; Parada, M.; Tapia, K.; Rubilar, C.; Allende, M.L.; Pena, O.A. Cxcl12a/cxcr4b acts to retain neutrophils in caudal hematopoietic tissue and to antagonize recruitment to an injury site in the zebrafish larva. Immunogenetics 2017, 69, 341–349. [Google Scholar] [CrossRef] [PubMed]
- Boldajipour, B.; Mahabaleshwar, H.; Kardash, E.; Reichman-Fried, M.; Blaser, H.; Minina, S.; Wilson, D.; Xu, Q.; Raz, E. Control of chemokine-guided cell migration by ligand sequestration. Cell 2008, 132, 463–473. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, J.A.; Bubner, E.J.; Takeuchi, Y.; Yoshizaki, G.; Wang, T.; Cummins, S.F.; Elizur, A. Primordial germ cell migration in the yellowtail kingfish (Seriola lalandi) and identification of stromal cell-derived factor 1. Gen. Comp. Endocrinol. 2015, 213, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Li, H.T.; Liang, R.; Lu, Y.N.; Wang, M.X.; Li, Z.D. Rtn3 regulates the expression level of chemokine receptor cxcr4 and is required for migration of primordial germ cells. Int. J. Mol. Sci. 2016. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Jones, D.; Borghesani, P.R.; Segal, R.A.; Nagasawa, T.; Kishimoto, T.; Bronson, R.T.; Springer, T.A. Impaired B-lymphopoiesis, myelopoiesis, and derailed cerebellar neuron migration in cxcr4- and sdf-1-deficient mice. Proc. Natl. Acad. Sci. USA 1998, 95, 9448–9453. [Google Scholar] [CrossRef] [PubMed]
- Power, C.A. Knock out models to dissect chemokine receptor function in vivo. J. Immunol. Methods 2003, 273, 73–82. [Google Scholar] [CrossRef]
- Mizoguchi, T.; Verkade, H.; Heath, J.K.; Kuroiwa, A.; Kikuchi, Y. Sdf1/cxcr4 signaling controls the dorsal migration of endodermal cells during zebrafish gastrulation. Development 2008, 135, 2521–2529. [Google Scholar] [CrossRef] [PubMed]
- Chong, S.W.; Nguyet, L.M.; Jiang, Y.J.; Korzh, V. The chemokine sdf-i and its receptor cxcr4 are required for formation of muscle in zebrafish. BMC Dev. Biol. 2007, 7. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Shirabe, K.; Kuwada, J.Y. Chemokine signaling regulates sensory cell migration in zebrafish. Dev. Biol. 2004, 269, 123–136. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Shirabe, K.; Thisse, C.; Thisse, B.; Okamoto, H.; Masai, I.; Kuwada, J.Y. Chemokine signaling guides axons within the retina in zebrafish. J. Neurosci. 2005, 25, 1711–1717. [Google Scholar] [CrossRef] [PubMed]
- Abe, P.; Molnar, Z.; Tzeng, Y.S.; Lai, D.M.; Arnold, S.J.; Stumm, R. Intermediate progenitors facilitate intracortical progression of thalamocortical axons and interneurons through cxcl12 chemokine signaling. J. Neurosci. 2015, 35, 13053–13063. [Google Scholar] [CrossRef] [PubMed]
- Mithal, D.S.; Banisadr, G.; Miller, R.J. Cxcl12 signaling in the development of the nervous system. J. Neuroimmune Pharm. 2012, 7, 820–834. [Google Scholar] [CrossRef] [PubMed]
- Nash, B.; Meucci, O. Functions of the chemokine receptor cxcr4 in the central nervous system and its regulation by mu-opioid receptors. Int. Rev. Neurobiol. 2014, 118, 105–128. [Google Scholar] [PubMed]
- Patel, J.R.; McCandless, E.E.; Dorsey, D.; Klein, R.S. Cxcr4 promotes differentiation of oligodendrocyte progenitors and remyelination. Proc. Natl. Acad. Sci. USA 2010, 107, 11062–11067. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Murakami, F. Chemokine cxcl12 and its receptors in the developing central nervous system: Emerging themes and future perspectives. Dev. Neurobiol. 2012, 72, 1349–1362. [Google Scholar] [CrossRef] [PubMed]
- Crump, M.P.; Spyracopoulos, L.; Lavigne, P.; Kim, K.S.; Clark-Lewis, I.; Sykes, B.D. Backbone dynamics of the human cc chemokine eotaxin: Fast motions, slow motions, and implications for receptor binding. Protein Sci. 1999, 8, 2041–2054. [Google Scholar] [CrossRef] [PubMed]
- Kofuku, Y.; Yoshiura, C.; Ueda, T.; Terasawa, H.; Hirai, T.; Tominaga, S.; Hirose, M.; Maeda, Y.; Takahashi, H.; Terashima, Y.; et al. Structural basis of the interaction between chemokine stromal cell-derived factor-1/cxcl12 and its g-protein-coupled receptor cxcr4. J. Biol. Chem. 2009, 284, 35240–35250. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.H.; Pan, C.H. Salvianolic acid b inhibits stromal cell-derived factor-1/cxcr4 axis and promotes apoptosis on vascular smooth muscle cells. FASEB J. 2010, 24. [Google Scholar]
- Boldajipour, B.; Doitsidou, M.; Tarbashevich, K.; Laguri, C.; Yu, S.R.; Ries, J.; Dumstrei, K.; Thelen, S.; Dorries, J.; Messerschmidt, E.M.; et al. Cxcl12 evolution—Subfunctionalization of a ligand through altered interaction with the chemokine receptor. Development 2011, 138, 2909–2914. [Google Scholar] [CrossRef] [PubMed]
- Szpakowska, M.; Bercoff, D.P.; Chevigne, A. Closing the ring: A fourth extracellular loop in chemokine receptors. Sci. Signal. 2014. [Google Scholar] [CrossRef] [PubMed]
- Vila-Coro, A.J.; Rodriguez-Frade, J.M.; De Ana, A.M.; Moreno-Ortiz, M.C.; Martinez, C.; Mellado, M. The chemokine sdf-1 alpha triggers cxcr4 receptor dimerization and activates the jak/stat pathway. FASEB J. 1999, 13, 1699–1710. [Google Scholar] [CrossRef] [PubMed]
- Pluhackova, K.; Gahbauer, S.; Kranz, F.; Wassenaar, T.A.; Bockmann, R.A. Dynamic cholesterol-conditioned dimerization of the g protein coupled chemokine receptor type 4. PLoS Comput. Biol. 2016. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Huelsenbeck, J.P. Mrbayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhou, L.; Li, Z.; Li, W.H.; Gui, J.F. Apolipoprotein c1 regulates epiboly during gastrulation in zebrafish. Sci. China Life Sci. 2013, 56, 975–984. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Sun, Z.H.; Zhou, L.; Li, Z.; Gui, J.F. Grouper tsh beta promoter-driven transgenic zebrafish marks proximal kidney tubule development. PLoS ONE 2014. [Google Scholar] [CrossRef]
- Shi, J.; Zhang, Y.B.; Liu, T.K.; Sun, F.; Gui, J.F. Subcellular localization and functional characterization of a fish irf9 from crucian carp carassius auratus. Fish Shellfish Immun. 2012, 33, 258–266. [Google Scholar] [CrossRef] [PubMed]

© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, W.-J.; Zhou, L.; Gao, F.-X.; Sun, Z.-H.; Li, Z.; Liu, X.-C.; Li, S.-S.; Wang, Y.; Gui, J.-F. Divergent Expression Patterns and Function of Two cxcr4 Paralogs in Hermaphroditic Epinephelus coioides. Int. J. Mol. Sci. 2018, 19, 2943. https://doi.org/10.3390/ijms19102943
Lu W-J, Zhou L, Gao F-X, Sun Z-H, Li Z, Liu X-C, Li S-S, Wang Y, Gui J-F. Divergent Expression Patterns and Function of Two cxcr4 Paralogs in Hermaphroditic Epinephelus coioides. International Journal of Molecular Sciences. 2018; 19(10):2943. https://doi.org/10.3390/ijms19102943
Chicago/Turabian StyleLu, Wei-Jia, Li Zhou, Fan-Xiang Gao, Zhi-Hui Sun, Zhi Li, Xiao-Chun Liu, Shui-Sheng Li, Yang Wang, and Jian-Fang Gui. 2018. "Divergent Expression Patterns and Function of Two cxcr4 Paralogs in Hermaphroditic Epinephelus coioides" International Journal of Molecular Sciences 19, no. 10: 2943. https://doi.org/10.3390/ijms19102943
APA StyleLu, W.-J., Zhou, L., Gao, F.-X., Sun, Z.-H., Li, Z., Liu, X.-C., Li, S.-S., Wang, Y., & Gui, J.-F. (2018). Divergent Expression Patterns and Function of Two cxcr4 Paralogs in Hermaphroditic Epinephelus coioides. International Journal of Molecular Sciences, 19(10), 2943. https://doi.org/10.3390/ijms19102943