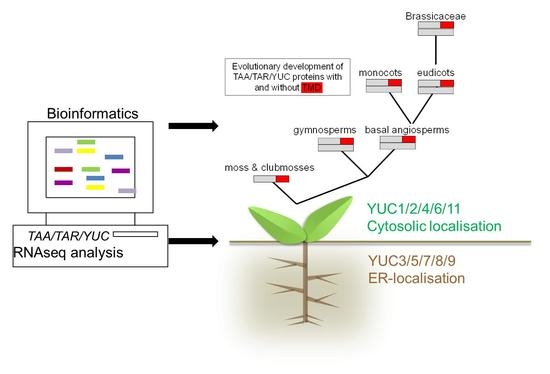
Bioinformatics Analysis of Phylogeny and Transcription of TAA/YUC Auxin Biosynthetic Genes
Abstract
1. Introduction
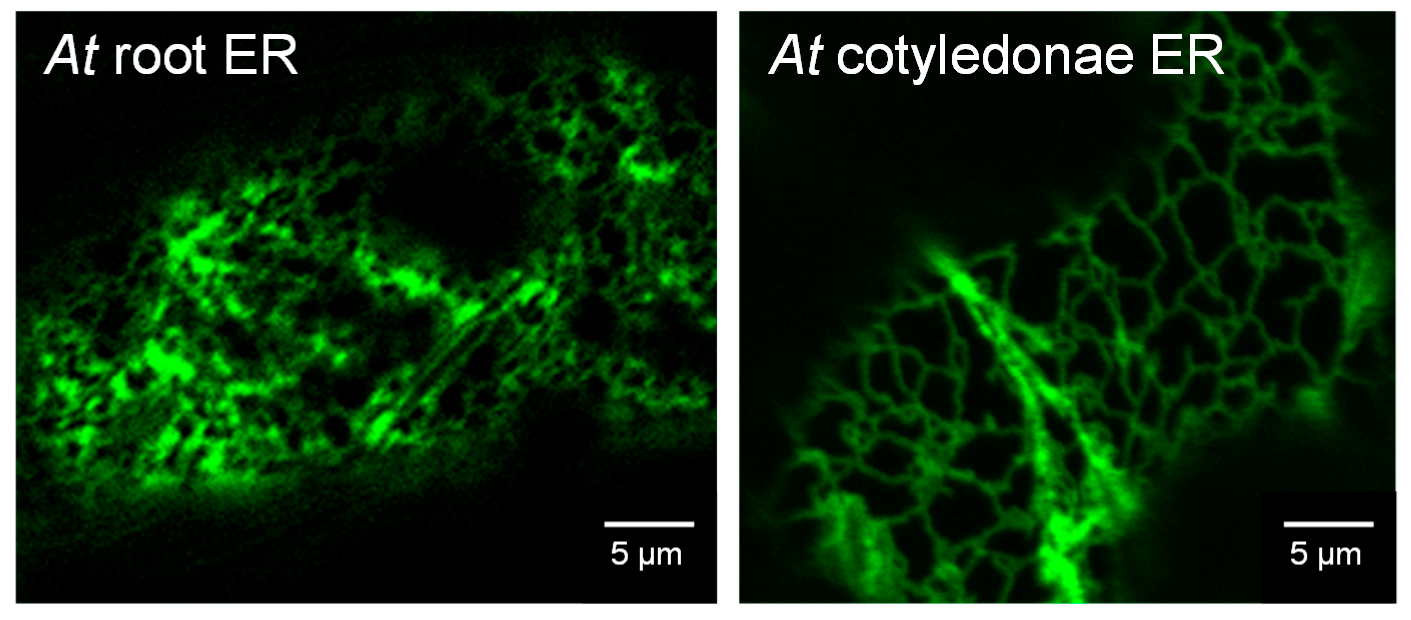
1.1. Subcellular Location of Auxin Biosynthetic Genes
2. Results
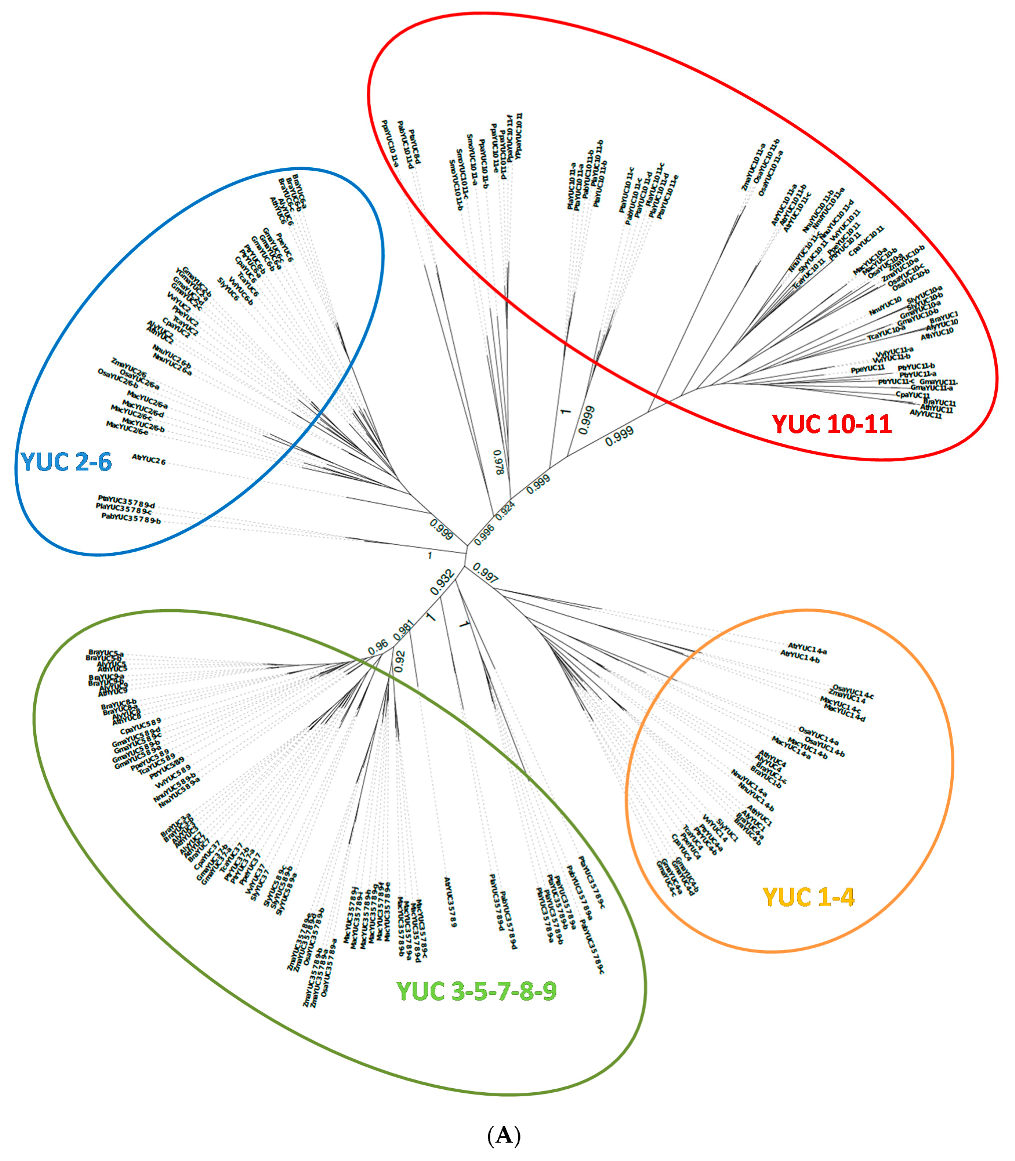
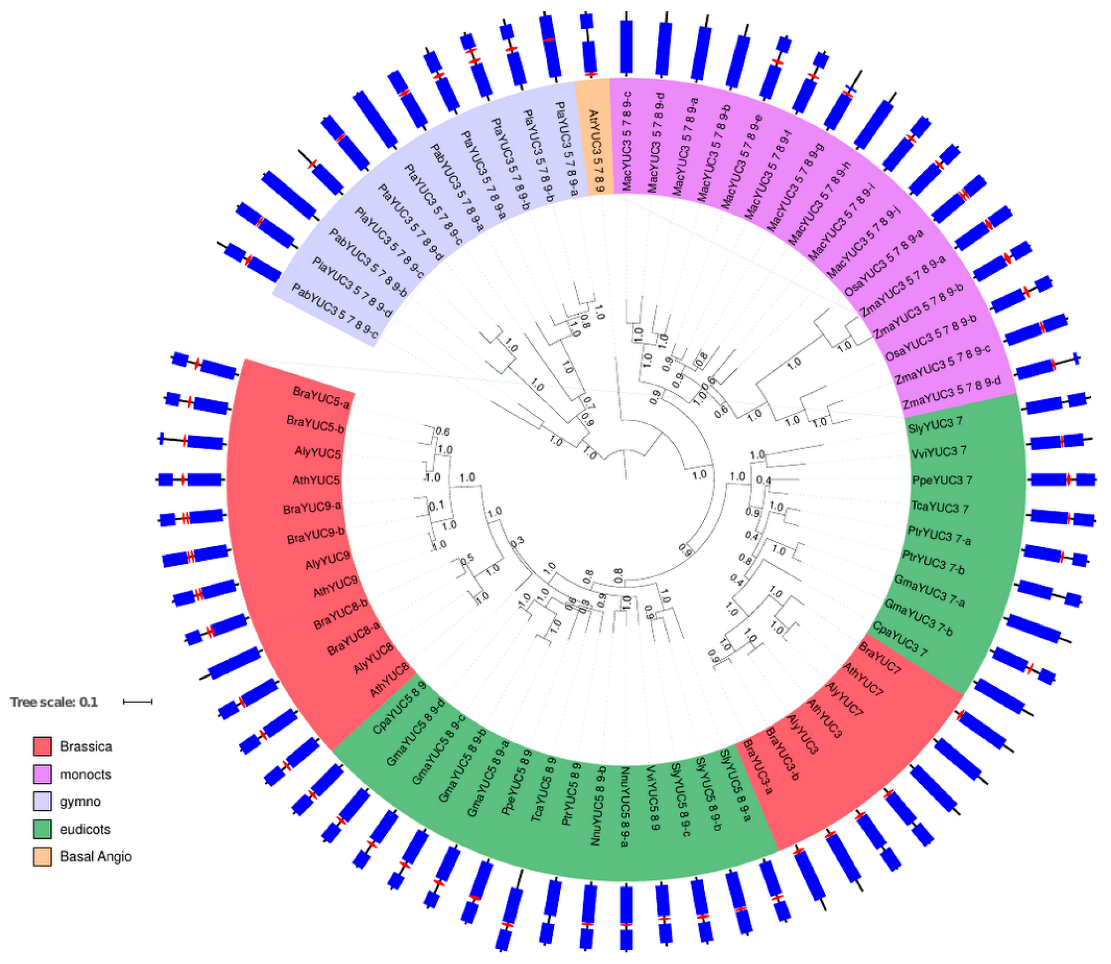
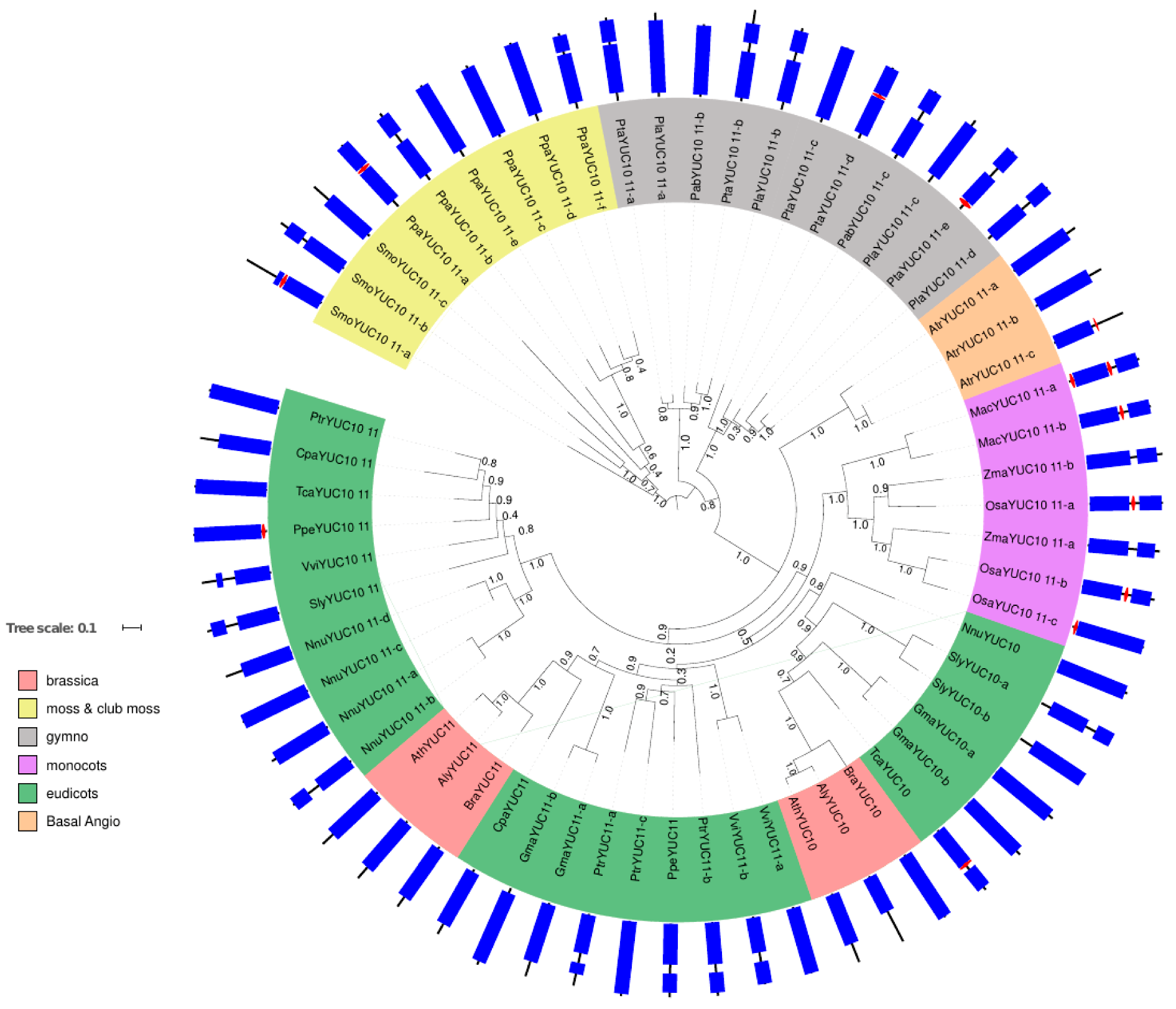
2.1. Phylogenetic Analysis of YUC Proteins
2.1.1. Phylogenetic Analysis for Shoot-Localised YUC Proteins
2.1.2. Phylogenetic Analysis for Root-Localised YUC Proteins
2.1.3. Phylogenetic Analysis for YUC Proteins Localised in Flowers and Siliques
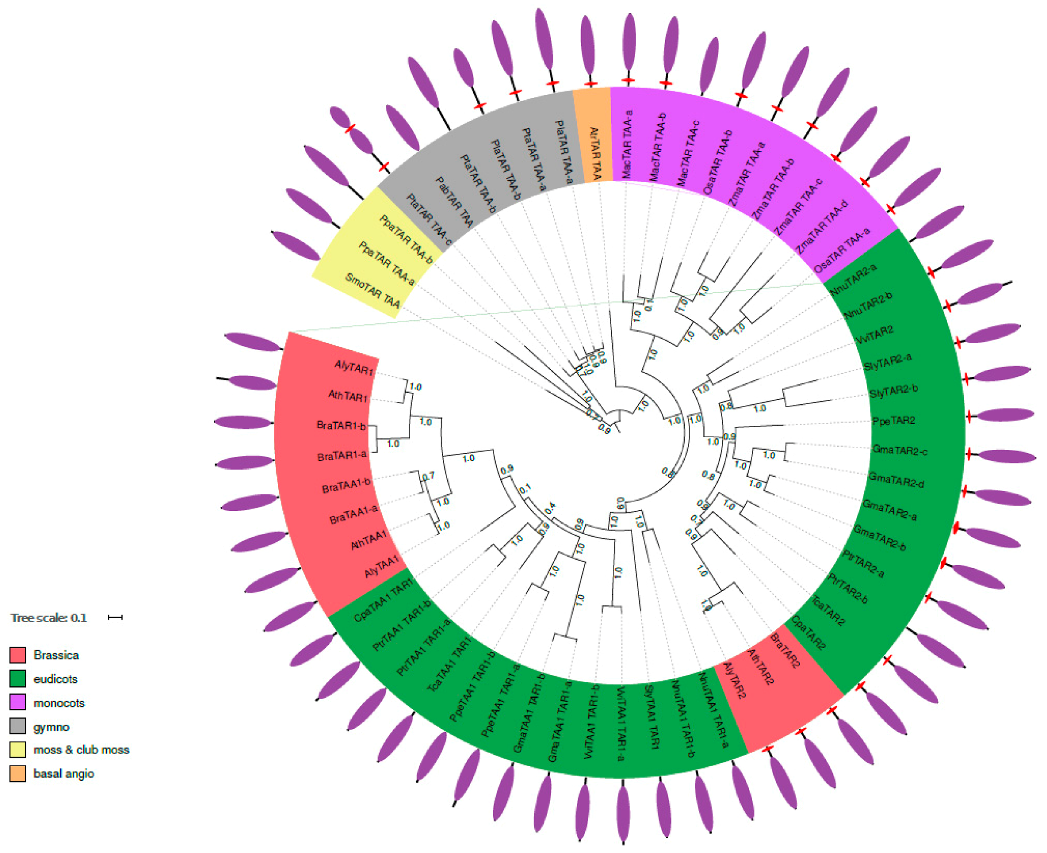
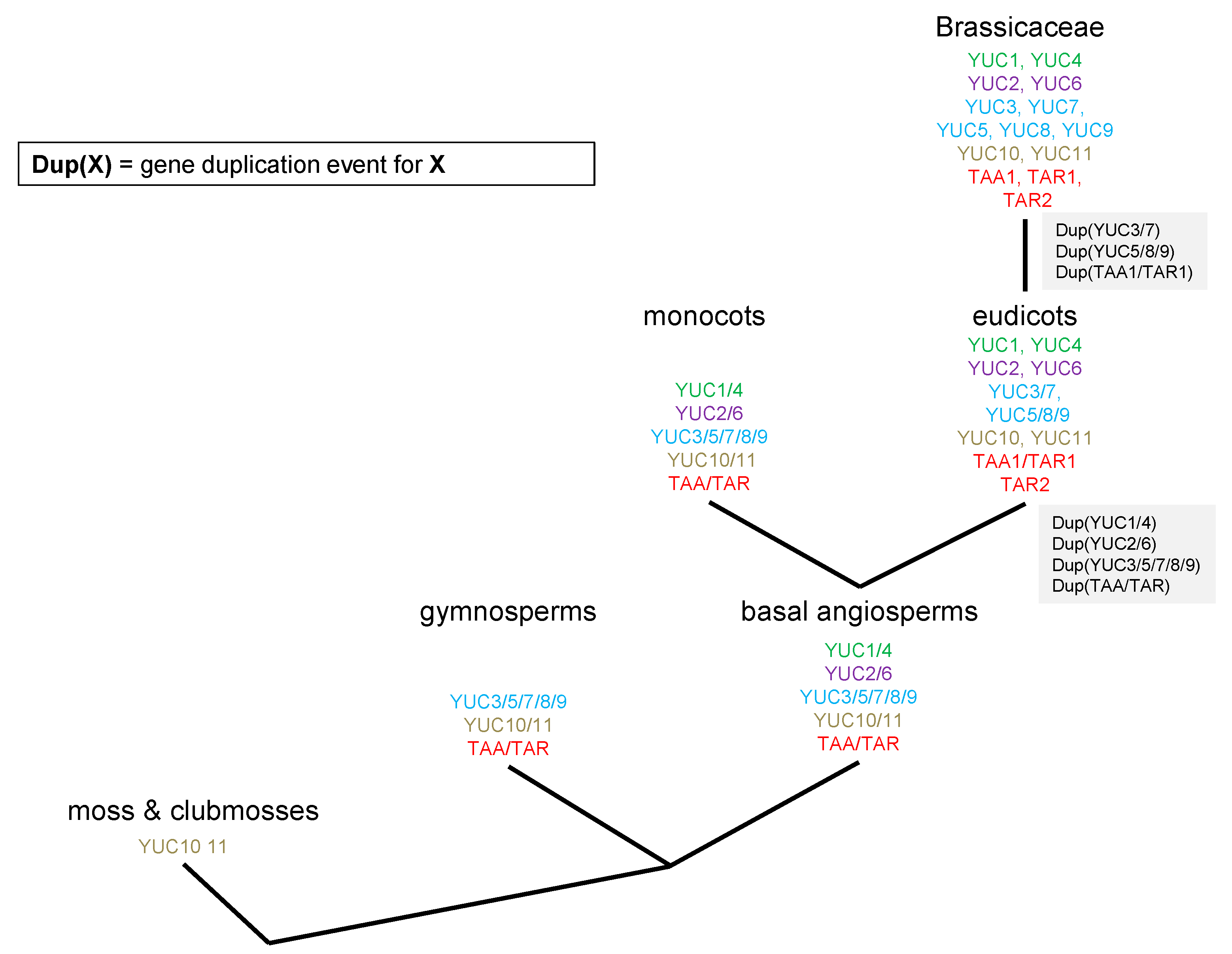
2.2. Phylogenetic Analysis of TAR and TAA Proteins
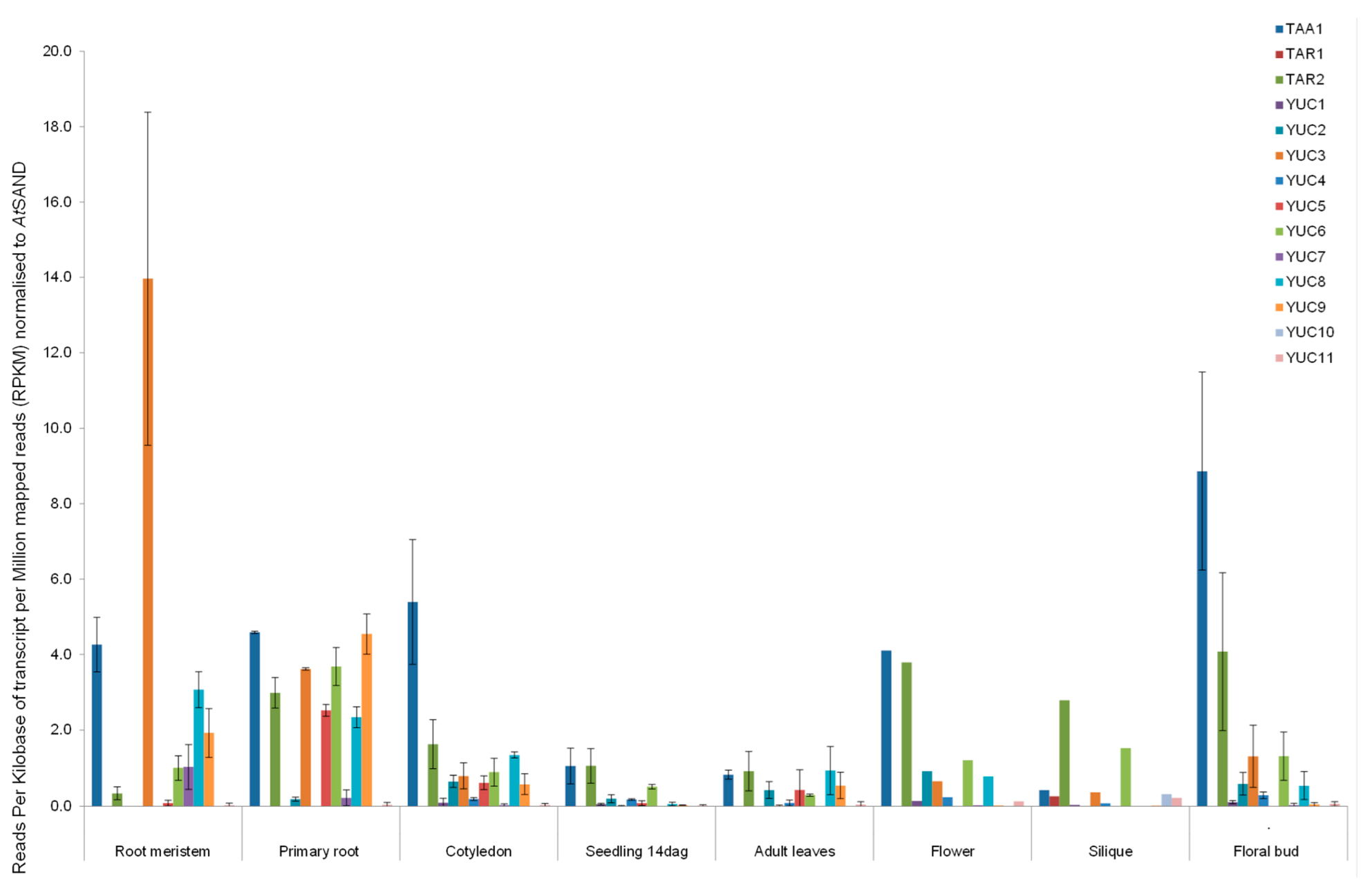
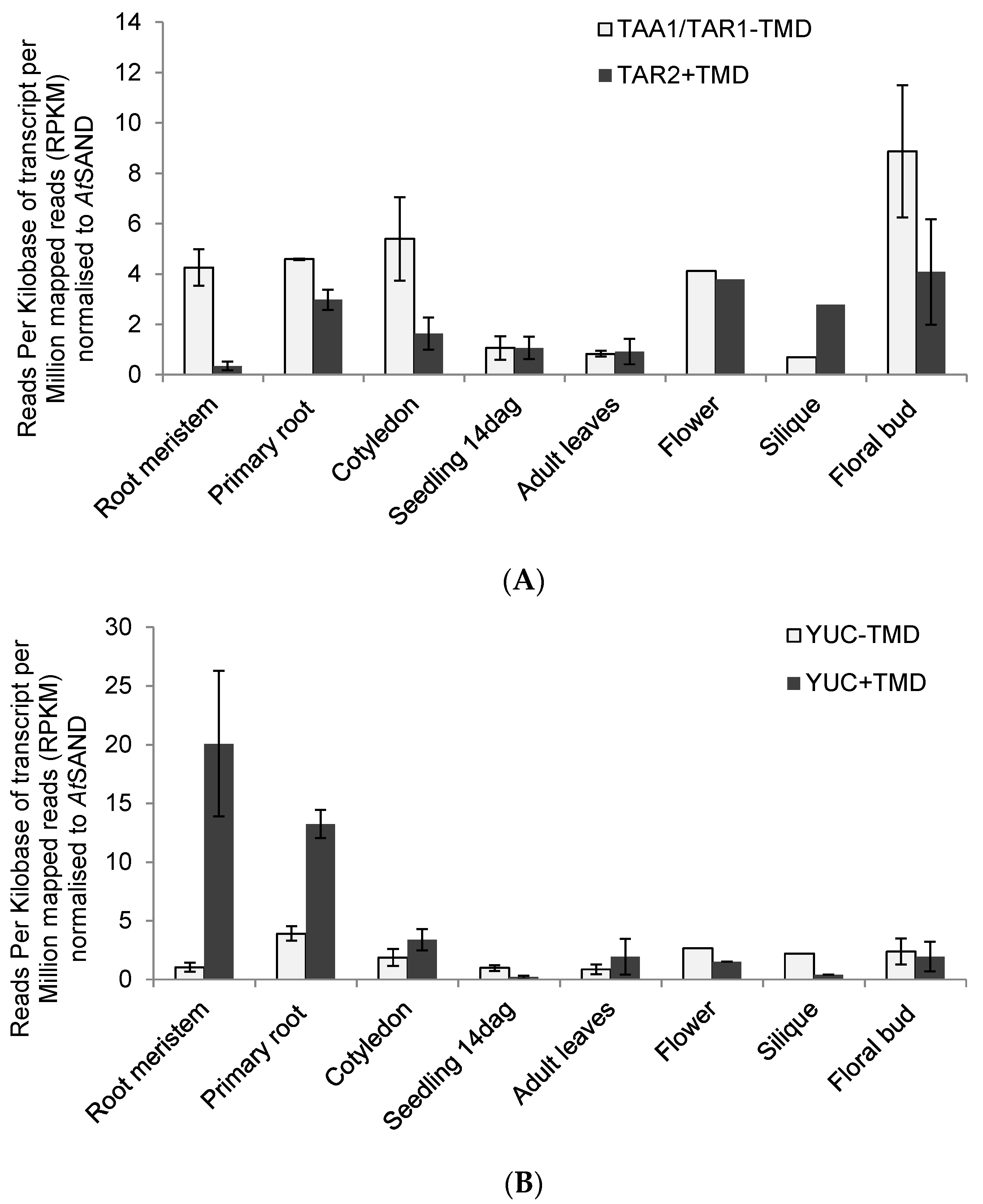
2.3. RNAseq Analysis of Auxin Biosynthetic Enzymes in the TAA/YUC Pathway
3. Material and Methods
3.1. Homologous Protein Detection
3.2. Phylogenetic Reconstruction
3.3. RNA Sequencing Data and Analysis
3.4. Confocal Imaging of Arabidopsis Seedlings
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Zhao, Y. Arabidopsis Book; The American Society of Plant Biologists: Rockville, MD, USA, 2014; Volume 12. [Google Scholar]
- Tivendale, N.D.; Ross, J.J.; Cohen, J.D. The shifting paradigm of auxin biosynthesis. Trends Plant Sci. 2014, 19, 44–51. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Christensen, S.K.; Fankhauser, C.; Cashman, J.R.; Cohen, J.D.; Weigel, D.; Chory, J. A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 2001, 291, 306–309. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Dai, X.; Zhao, Y. Auxin biosynthesis by the YUCCA Flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev. 2006, 20, 1790–1799. [Google Scholar] [CrossRef] [PubMed]
- Stepanova, A.N.; Robertson-Hoyt, J.; Yun, J.; Benavente, L.M.; Xie, D.Y.; Dolezal, K.; Schlereth, A.; Jürgens, G.; Alonso, J.M. TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 2008, 133, 177–191. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Dai, X.; De-Paoli, H.; Cheng, Y.; Takebayashi, Y.; Kasahara, H.; Kamiya, Y.; Zhao, Y. Auxin overproduction in shoots cannot rescue auxin deficiencies in Arabidopsis roots. Plant Cell Physiol. 2014, 55, 1072–1079. [Google Scholar] [CrossRef] [PubMed]
- Kriechbaumer, V.; Seo, H.; Park, W.J.; Hawes, C. Endoplasmic reticulum localization and activity of maize auxin biosynthetic enzymes. Plant Physiol. 2015, 169, 1933–1945. [Google Scholar] [CrossRef] [PubMed]
- Kriechbaumer, V.; Botchway, S.W.; Hawes, C. Localization and interactions between Arabidopsis auxin biosynthetic enzymes in the TAA/YUC-dependent pathway. J. Exp. Bot. 2016, 67, 4195–4207. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.; Ferrer, J.L.; Ljung, K.; Pojer, F.; Hong, F.; Long, J.A.; Li, L.; Moreno, J.E.; Bowman, M.E.; Ivans, L.J.; et al. Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 2008, 133, 164–176. [Google Scholar] [CrossRef] [PubMed]
- Mravec, J.; Skupa, P.; Bailly, A.; Hoyerová, K.; Krecek, P.; Bielach, A.; Petrásek, J.; Zhang, J.; Gaykova, V.; Stierhof, Y.D.; et al. Subcellular homeostasis of phytohormone auxin is mediated by the ER localized PIN5 transporter. Nature 2009, 459, 1136–1140. [Google Scholar] [CrossRef] [PubMed]
- Dal Bosco, C.D.; Dovzhenko, A.; Liu, X.; Woerner, N.; Rensch, T.; Eismann, M.; Eimer, S.; Hegermann, J.; Paponov, I.A.; Ruperti, B.; et al. The endoplasmic reticulum localized PIN8 is a pollen-specific auxin carrier involved in intracellular auxin homeostasis. Plant J. 2012, 71, 860–870. [Google Scholar] [CrossRef] [PubMed]
- Ding, Z.; Wang, B.; Moreno, I.; Dupláková, N.; Simon, S.; Carraro, N.; Reemmer, J.; Pěnčík, A.; Chen, X.; Tejos, R.; et al. ER-localized auxin transporter PIN8 regulates auxin homeostasis and male gametophyte development in Arabidopsis. Nat. Commun. 2012, 3, 941. [Google Scholar] [CrossRef] [PubMed]
- Barbez, E.; Kubes, M.; Rolcik, J.; Béziat, C.; Pěnčík, A.; Wang, B.; Rosquete, M.R.; Zhu, J.; Dobrev, P.I.; Lee, Y.; et al. A novel putative auxin carrier family regulates intracellular auxin homeostasis in plants. Nature 2012, 485, 119–122. [Google Scholar] [CrossRef] [PubMed]
- Bender, R.L.; Fekete, M.L.; Klinkenberg, P.M.; Hampton, M.; Bauer, B.; Malecha, M.; Lindgren, K.; A Maki, J.; Perera, M.A.; Nikolau, B.J.; et al. PIN6 is required for nectary auxin response and short stamen development. Plant J. 2013, 74, 893–904. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Attwood, T.K.; Babbitt, P.C.; Bateman, A.; Bork, P.; Bridge, A.J.; Chang, H.-C.; Dosztányi, Z.; El-Gebali, S.; Fraser, M.; et al. InterPro in 2017—beyond protein family and domain annotations. Nucleic Acids Res. 2017, 45, D190–D199. [Google Scholar] [CrossRef] [PubMed]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. Fasttree 2 approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef] [PubMed]
- Kriechbaumer, V.; Wang, P.; Hawes, C.; Abell, B.M. Alternative splicing of the auxin biosynthesis gene YUCCA4 determines its subcellular compartmentation. Plant J. 2012, 70, 292–302. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinformat. 2004, 5, 113. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life v2: Online annotation and display of phylogenetic trees made easy. Nucleic Acids Res. 2011, 39, 475–478. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 25, R36. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeqa Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Czechowski, T.; Stitt, M.; Altmann, T.; Udvardi, M.K.; Scheible, W.R. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 2005, 139, 5–17. [Google Scholar] [CrossRef] [PubMed]

| Species | Abbreviation | Group | Genome | Chromosome No. |
|---|---|---|---|---|
| Arabidopsis thaliana | Ath | eudicots | 150 Mb | 5 |
| Arabidopsis lyrata | Aly | eudicots | 206.7 Mb | 8 |
| Brassica rapa | Bra | eudicots | 283.8 Mb | 10 |
| Prunus persica | Ppe | eudicots | 224.6 Mb | 8 |
| Glycine max | Gma | eudicots | 950 Mb | 20 |
| Carica papaya | Cpa | eudicots | 372 Mb | 9 |
| Theobroma cacao | Tca | eudicots | 326.9 Mb | 10 |
| Nelumbo nucifera | Nnu | eudicots | 804 Mb | 8 |
| Vitis cinifera | Vvi | eudicots | 487 Mb | 19 |
| Populus trichocarpa | Ptr | eudicots | 485 Mb | 19 |
| Solanum lycopersicum | Sly | eudicots | 900 Mb | 12 |
| Zea mays | Zma | monocots | 2.3 Gb | 10 |
| Oryza sativa | Osa | monocots | 372 Mb | 12 |
| Musa acuminata | Mac | monocots | 472 Mb | 8 |
| Amborella trichopoda | Atr | basal angiosperms | 706 Mb | 13 |
| Pinus abies | Pab | gymnosperms | 19.6 Gb | 12 |
| Pinus lambertiana | Pil | gymnosperms | 31 Gb | 12 |
| Pinus taeda | Pta | gymnosperms | 20.15 Gb | 12 |
| Physcomitrella patens | Ppa | moss | 473 Mb | 27 |
| Selaginella moellendorffii | Smo | lycophytes | 212.5 Mb | 27 |
| Gene Name | Gene ID | Protein Length | Localisation | Flavin- Dependent Monooxygenase Domain | Tryptophan Amino- Transferase Domain | Predicted TMD |
|---|---|---|---|---|---|---|
| YUC1 | AT4G32540 | 414 | shoot + cytosolic | yes | no | No |
| YUC2 | AT4G13260 | 415 | shoot + cytosolic | yes | no | No |
| YUC3 | AT1G04610 | 437 | root + cytosolic | yes | no | Yes |
| YUC4 | AT5G11320 | 410 | shoot + cytosolic | yes | no | No |
| YUC5 | AT5G43890 | 424 | root + ER | yes | no | Yes |
| YUC6 | AT5G25620 | 426 | shoot + cytosolic | yes | no | No |
| YUC7 | AT2G33230 | 431 | root and ER | yes | no | No |
| YUC8 | AT4G28720 | 426 | root + ER | yes | no | Yes |
| YUC9 | AT1G04180 | 421 | root + ER | yes | no | Yes |
| YUC10 | AT1G48910 | 383 | flower + cytosolic | yes | no | No |
| YUC11 | AT1G21430 | 391 | flower + cytosolic | yes | no | No |
| TAA1 | AT1G70560 | 391 | whole plant cytosolic | no | yes | No |
| TAR1 | AT1G23320 | 388 | silique + cytosolic | no | yes | No |
| TAR2 | AT4G24670 | 440 | whole plant and ER | no | yes | Yes |
| Tissue | SRA File Number for RNAseq Analysis |
|---|---|
| Root meristematic zone | SRR1740401 |
| SRR1740402 | |
| SRR1740403 | |
| Primary root | SRR1042766 |
| SRR1042767 | |
| Cotyledon | SRR1292207 |
| SRR605657 | |
| SRR605658 | |
| Seedling 14 dag | SRR346552 |
| SRR346553 | |
| Adult Leaves | SRR1159821 |
| SRR1159827 | |
| SRR1159831 | |
| SRR1159837 | |
| Flower | SRR656217 |
| Silique | SRR656218 |
| Flower bud | SRR800753 |
| SRR800754 | |
| SRR314815 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Poulet, A.; Kriechbaumer, V. Bioinformatics Analysis of Phylogeny and Transcription of TAA/YUC Auxin Biosynthetic Genes. Int. J. Mol. Sci. 2017, 18, 1791. https://doi.org/10.3390/ijms18081791
Poulet A, Kriechbaumer V. Bioinformatics Analysis of Phylogeny and Transcription of TAA/YUC Auxin Biosynthetic Genes. International Journal of Molecular Sciences. 2017; 18(8):1791. https://doi.org/10.3390/ijms18081791
Chicago/Turabian StylePoulet, Axel, and Verena Kriechbaumer. 2017. "Bioinformatics Analysis of Phylogeny and Transcription of TAA/YUC Auxin Biosynthetic Genes" International Journal of Molecular Sciences 18, no. 8: 1791. https://doi.org/10.3390/ijms18081791
APA StylePoulet, A., & Kriechbaumer, V. (2017). Bioinformatics Analysis of Phylogeny and Transcription of TAA/YUC Auxin Biosynthetic Genes. International Journal of Molecular Sciences, 18(8), 1791. https://doi.org/10.3390/ijms18081791