Analytic and Diagnostic Performances of Human Papillomavirus E6/E7 mRNA Test on up-to 11-Year-Old Liquid-Based Cervical Samples. A Biobank-Based Longitudinal Study
Abstract
:1. Introduction
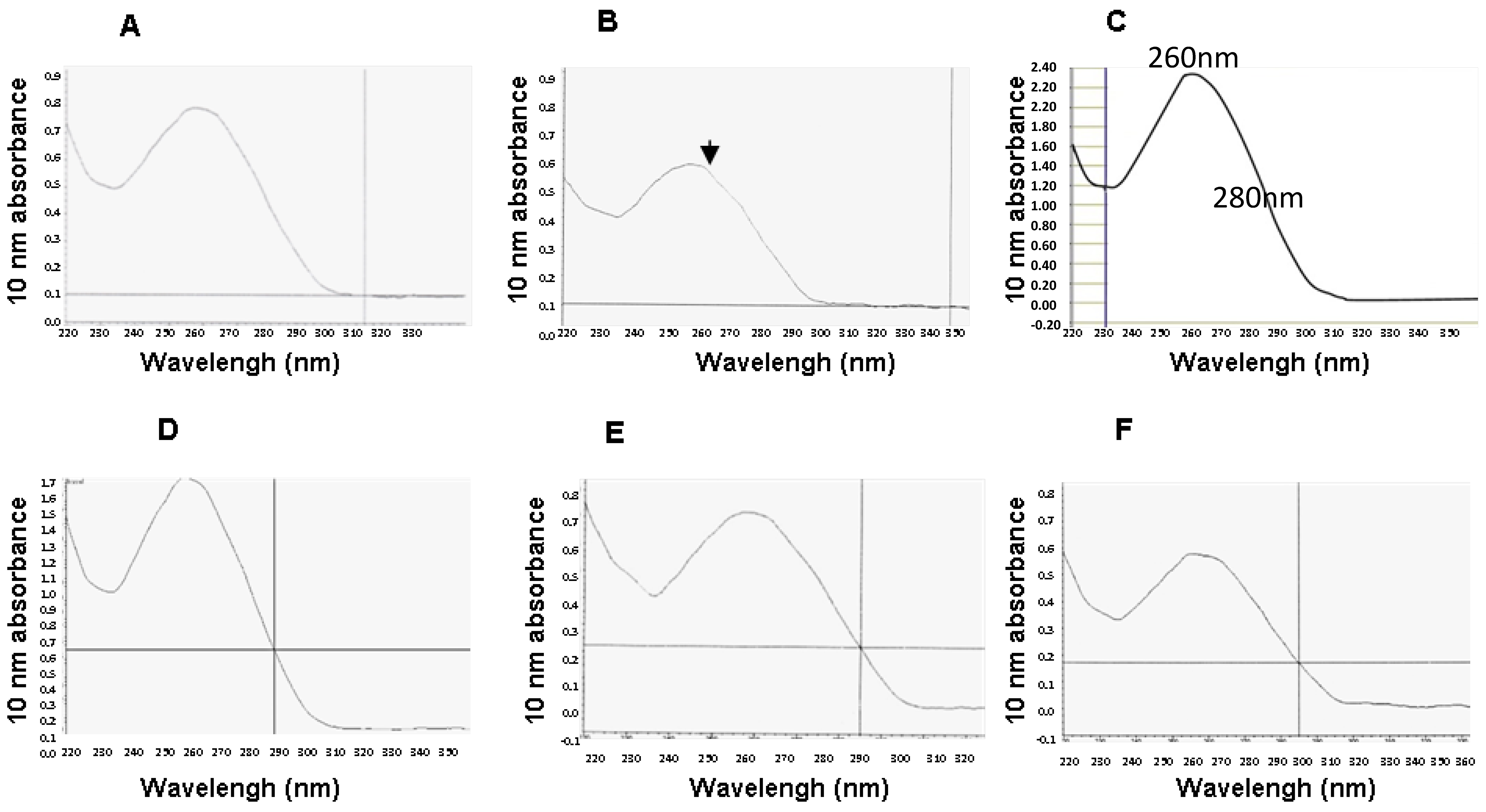
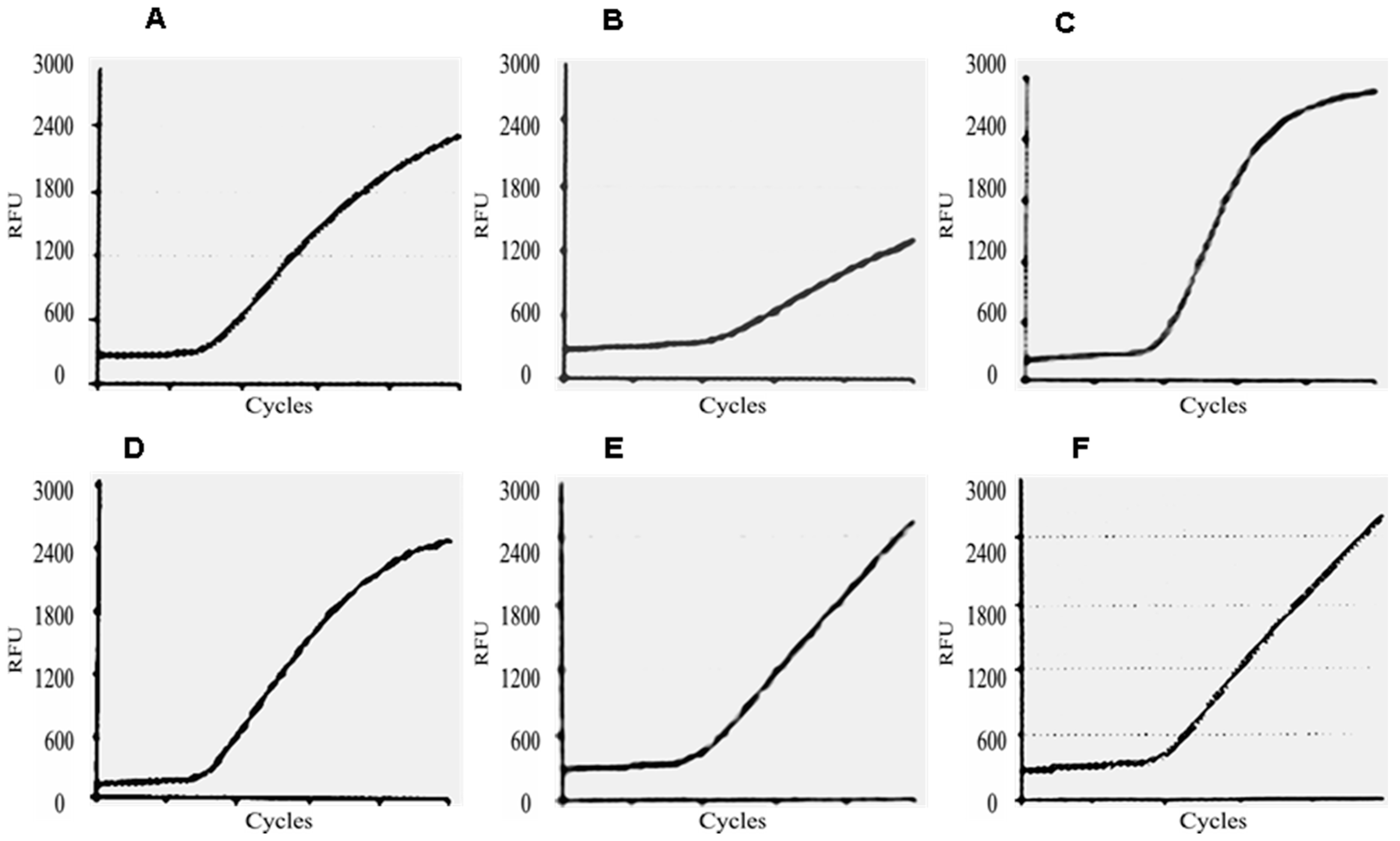
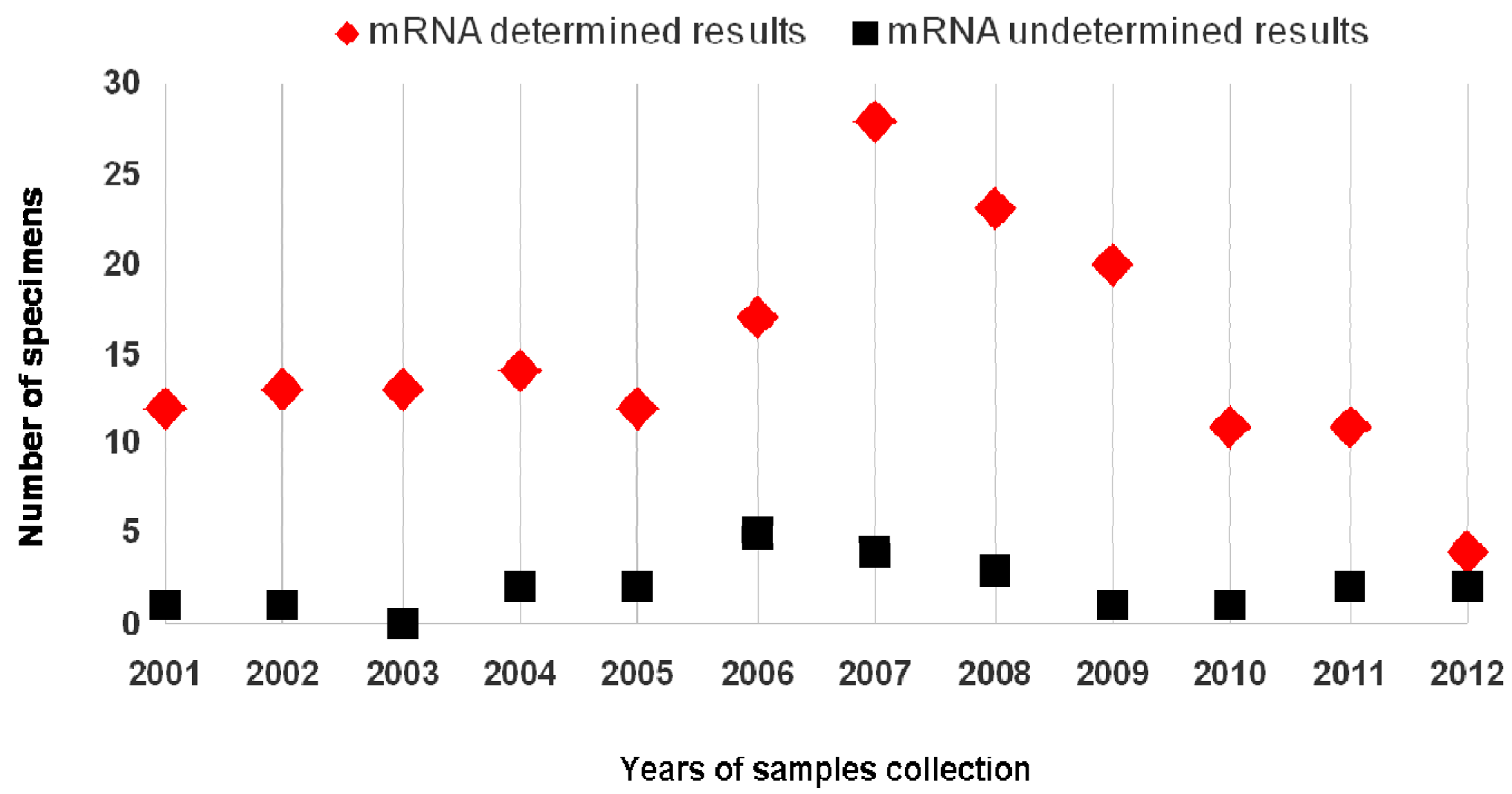
2. Results
3. Discussion
4. Materials and Methods
4.1. Setting
4.2. Cytological and Histological Diagnosis
4.3. HPV-DNA Detection
4.4. RNA Isolation
4.5. Quantification of RNA Yield and Purity
4.6. E6/E7 mRNA Test
4.7. Statistical Analyses
4.8. Ethical Aspects
5. Conclusions
Author Contributions
Conflicts of Interest
References
- Walboomers, J.M.; Jacobs, M.V.; Manos, M.M.; Bosch, F.X.; Kummer, J.A.; Shah, K.V.; Snijders, P.J.F.; Peto, J.; Meijer, C.J.L.M.; Munoz, N. Human Papillomavirus is a necessary cause of invasive cervical cancer worldwide. J. Pathol. 1999, 189, 12–19. [Google Scholar] [CrossRef]
- Munoz, N.; Castelsague, X.; Gonzalez, A.B.; Gissman, L. Chapter I: HPV in the etiology of human cancer. Vaccine 2006, 24, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Zappacosta, R.; Rosini, S. Cervical cancer screening: From molecular basis to diagnostic practice, going through new technologies. Technol. Cancer. Res. Treat 2008, 7, 161–174. [Google Scholar] [CrossRef] [PubMed]
- Ronco, G.; Dillner, J.; Elfström, K.M.; Tunesi, S.; Snijders, P.J.F.; Arbyn, M.; Kitchener, H.; Segnan, N.; Gilham, C. Efficacy of HPV-based screening for prevention of invasive cervical cancer. Follow-up of four European randomized controlled trials. Lancet 2014, 383, 524–532. [Google Scholar] [CrossRef]
- Sankaranarayanan, R.; Nene, B.M.; Shastri, S.S.; Jayant, K.; Muwonge, R.; Budukh, A.M.; Hingmire, S.; Malvi, S.G.; Thorat, R.; Kothari, A.; et al. HPV screening for cervical cancer in rural India. N. Engl. J. Med. 2009, 360, 1385–1394. [Google Scholar] [CrossRef] [PubMed]
- Maggino, T.; Sciarrone, R.; Murer, B.; dei Rossi, M.R.; Fedato, C.; Maran, M.; Lorio, M.; Soldà, M.; Zago, F.; Rossi, P.G.; Zorzi, M. Screening women for cervical cancer carcinoma with a HPV mRNA test: First results from the Venice pilot program. Br. J. Cancer 2016, 23, 525–532. [Google Scholar] [CrossRef] [PubMed]
- Arbyn, M.; Ronco, G.; Cuzich, J.; Wentzensen, N.; Castle, P.E. How to evaluate emerging technologies in cervical cancer screening? Int. J. Cancer 2009, 125, 2489–2496. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Yang, B.; Zhang, A.; Zhou, A.; Yuan, J.; Wang, Y.; Sun, L.; Cao, H.; Wang, J.; Zheng, W. Association between human papillomavirus type 16 E6 and E7 variants with subsequent persistent infection and recurrence of cervical high-grade squamous intraepithelial lesion after conization. J. Med. Virol. 2016, 88, 1982–1988. [Google Scholar] [CrossRef] [PubMed]
- Doorbar, J.; Egawa, N.; Griffin, H.; Kranjec, C.; Murakami, I. Human papillomavirus molecular biology and disease association. Rev. Med. Virol. 2015, 25, 2–23. [Google Scholar] [CrossRef] [PubMed]
- Nor Rashid, N.; Yusof, R.; Watson, R.J. Disruption of pocket protein dream complexes by E7 proteins of different types of human papillomaviruses. Acta Virol. 2013, 57, 447–451. [Google Scholar] [CrossRef] [PubMed]
- Park, J.W.; Shin, M.K.; Pitot, H.C.; Lambert, P.F. High incidence of HPV-associated head and neck cancers in FA deficient mice is associated with E7’s induction of DNA damage through its inactivation of pocket proteins. PLoS ONE 2013, 8, e75056. [Google Scholar] [CrossRef] [PubMed]
- Park, J.W.; Shin, M.K.; Lambert, P.F. High incidence of female reproductive tract cancers in FA-deficient HPV16-transgenic mice correlates with E7’s induction of DNA damage response, an activity mediated by E7’s inactivation of pocket proteins. Oncogene 2013, 9, 54–67. [Google Scholar] [CrossRef] [PubMed]
- Kraus, I.; Molden, T.; Holm, R.; Lie, A.K.; Karlsen, F.; Kristensen, G.B. Presence of E6 and E7 mRNA from human Papillomavirus types 16, 18, 31, 33, and 45 in the majority of cervical carcinomas. J. Clin. Microbiol. 2006, 44, 1310–1317. [Google Scholar] [CrossRef] [PubMed]
- Guess, J.C.; McCance, D.J. Decreased migration of Langerhans precursor-like cells in response to human keratinocytes expressing human papillomavirus type 16 E6/E7 is related to reduced macrophage inflammatory protein-3 production. J. Virol. 2005, 79, 14852–14857. [Google Scholar] [CrossRef] [PubMed]
- Biswas, S.K.; Sodhi, A. Effect of monocyte chemoattractant protein-1 on murine bone marrow cells: Proliferation, colony-forming ability and signal transduction pathway involved. Int. J. Immunopathol. Pharmacol. 2002, 15, 183–187. [Google Scholar] [CrossRef] [PubMed]
- Scott, M.; Nakagawa, M.; Moscicki, A. Cell-mediated immune response to human papillomavirus infection. Clin. Diagn. Lab. Immunol. 2001, 8, 209–213. [Google Scholar] [CrossRef] [PubMed]
- Denton, K.J.; Bergeron, C.; Klement, P.; Trunk, M.J.; Keller, T.; Ridder, R.; European CINtec Cytology Study Group. European CINtec Cytology Study Group. The sensitivity and specificity of p16 (INK4a) cytology vs. HPV testing for detecting high-grade cervical disease in the triage of ASC-US and LSIL pap cytology results. Am. J. Clin. Pathol. 2010, 134, 12–21. [Google Scholar] [CrossRef] [PubMed]
- Giorgi Rossi, P.; Benevolo, M.; Vocaturo, A.; Caraceni, D.; Ciccocioppo, L.; Frega, A.; Terrenato, I.; Zappacosta, R.; French, D.; Rosini, S. Prognostic value of HPV E6/E7 mRNA assay in women with negative colposcopy or CIN1 histology result: A follow-up study. PLoS ONE 2013, 8, e57600. [Google Scholar] [CrossRef] [PubMed]
- Pepe, M.S.; Etzioni, R.; Feng, Z.; Potter, J.D.; Thompson, M.L.; Thornquist, M.; Winget, M.; Yasui, Y. Phases of biomarker development for early detection of cancer. J. Natl. Cancer Inst. 2001, 93, 1054–1061. [Google Scholar] [CrossRef] [PubMed]
- Pepe, M.S.; Feng, Z.; Janes, H.; Bossuyt, P.M.; Potter, J.D. Pivotal evaluation of the accuracy of a biomarker used for classification or prediction: Standards for study design. J. Natl. Cancer Inst. 2008, 100, 1432–1438. [Google Scholar] [CrossRef] [PubMed]
- Kinkorovaà, J. Biobanks in the era of personalized medicine: Objectives, challenges, and innovations. EPMA J. 2015, 7, 4–9. [Google Scholar] [CrossRef] [PubMed]
- Castle, P.E.; Solomon, D.; Hildesheim, A.; Herrero, R.; Bratti, M.C.; Sherman, M.E.; Rodriguez, A.C.; Alfaro, M.; Hutchinson, M.L.; Dunn, S.T.; et al. Stability of archived liquid-based cervical cytologic specimens. Cancer 2003, 99, 89–96. [Google Scholar] [CrossRef] [PubMed]
- Waldstrøm, M.; Ørnskov, D. Clinical Performance of a Human Papillomavirus messenger RNA test (Aptima HPV Assay) on residual material from archived 3-year-old PreservCyt samples with low-grade squamous intraepithelial lesion. Arch. Pathol. Lab. Med. 2011, 135, 1052–1056. [Google Scholar] [CrossRef] [PubMed]
- Desjardins, P.; Conklin, D. NanoDrop microvolume quantitation of nucleic acids. J. Vis. Exp. 2010, 45, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Hewitt, R.E. Biobanking: The foundation of personalized medicine. Curr. Opin. Oncol. 2011, 23, 112–119. [Google Scholar] [CrossRef] [PubMed]
- Park, A. 10 Ideas Changing the World Right Now. Available online: http://content.time.com/time/specials/packages/article/0,28804,1884779_1884782_1884766,00.html (accessed on 15 December 2015).
- Arbyn, M.; van Veen, E.B.; Andersson, K.; Bogers, J.; Boulet, G.; Bergeron, C.; von Knebel-Doeberitz, M.; Dillner, J. Cervical cytology biobanking in Europe. Int. J. Biol. Markers 2010, 25, 117–125. [Google Scholar] [PubMed]
- Arbyn, M.; Herbert, A.; Martin-Hirsch, P.; Schenck, U.; Baldauf, J.-J.; da Silva, D.; Anttila, A.; Nieminen, P.; Prendiville, W. European guidelines for quality assurance in cervical cancer screening: Recommendations for collecting samples for conventional and liquid-based cytology. Cytopathology 2007, 18, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Sobradillo, P.; Pozo, F.; Agustí, A. P4 medicine: The future around the corner. Arch. Bronconeumol. 2011, 47, 35–40. [Google Scholar] [CrossRef] [PubMed]
- Larsson, G.L.; Karlsson, M.G.; Helenius, G. HPV testing of biobanked liquid-based cytology-a validation study. Int. Biol. Markers 2016, 31, 218–223. [Google Scholar] [CrossRef] [PubMed]
- Fujii, T.; Asano, A.; Shimada, K.; Tatsumi, Y.; Obayashi, C.; Konishi, N. Evaluation of RNA and DNA extraction from liquid-based cytology specimens. Diagn. Cytopathol. 2016, 44, 833–840. [Google Scholar] [CrossRef] [PubMed]
- Hogarth, S.; Hopkins, M.; Rotolo, D. Technological Accretion in Diagnostics: HPV Testing and Cytology in Cervical Cancer Screening; Routledge: New York, NY, USA, 2015. [Google Scholar]
- Fabre, A.L.; Colotte, M.; Luis, A.; Tuffet, S.; Bonnet, J. An efficient method for long-term room temperature storage of RNA. Eur. J. Hum. Genet. 2014, 22, 379–385. [Google Scholar] [CrossRef] [PubMed]
- Vincek, V.; Nassiri, M.; Nadji, M.; Morales, A.R. A tissue fixative that protects macromolecules (DNA, RNA, and protein) and histomorphology in clinical samples. Lab. Investig. 2003, 83, 1427–1435. [Google Scholar] [CrossRef] [PubMed]
- Micke, P.; Ohshima, M.; Tahmasebpoor, S.; Ren, Z.; Östman, A.; Pontén, F.; Botling, J. Biobanking of fresh frozen tissue: RNA is stable in nonfixed surgical specimens. Lab. Investig. 2006, 86, 202–211. [Google Scholar] [CrossRef] [PubMed]
- Rossi, E.D.; Bizzarro, T.; Longatto-Filho, A.; Gerhard, R.; Schmitt, F. The diagnostic and prognostic role of liquid-based cytology: Are we ready to monitor therapy and resistance? Expert Rev. Anticancer Ther. 2015, 15, 911–921. [Google Scholar] [CrossRef] [PubMed]
- Hoda, R.S.; Loukeris, K.; Abdul-Karim, F.W. Gynaecologic cytology on conventional and liquid-based preparations: A comprehensive review of similarities and differences. Diagn. Cytopathol. 2013, 41, 257–278. [Google Scholar] [CrossRef] [PubMed]
- Boulet, C.A.; Horvath, C.A.; Berghmans, S.; Moeneclaey, L.M.; Duys, I.S.M.; Arbyn, M.; Depuydt, C.E.; Vereecken, A.J.; Sahebali, S.; Bogers, J.J. Cervical cytology biobanking: Quality of DNA from archivial cervical pap-stained smears. J. Clin. Pathol. 2008, 61, 637–641. [Google Scholar] [CrossRef] [PubMed]
- Habis, A.H.; Vermon, S.D.; Lee, D.R.; Verma, M.; Unger, E.R. Molecular quality of exfoliated cervical cells: Implications for molecular epidemiology and biomarker discovery. Cancer Epidemiol. Biomarker Prev. 2004, 13, 492–496. [Google Scholar]
- Shabihkhani, M.; Lucey, G.M.; Wei, B.; Mareninov, S.; Lou, J.J.; Vinters, H.V.; Singer, E.J.; Cloughesy, T.F.; Yong, W.H. The procurement, storage, and quality assurance of frozen blood and tissue biospecimens in pathology, biorepository, and biobank settings. Clin. Biochem. 2014, 47, 258–266. [Google Scholar] [CrossRef] [PubMed]
- Agred, P.M.; Beitman, G.H.; Gutierrez, E.C. Long-term stability of human genomic and human papillomavirus DNA stored in BD SurePath and Hologic PreservCyt liquid-based cytology media. J. Clin. Microbiol. 2013, 51, 2702–2706. [Google Scholar] [CrossRef] [PubMed]
- Tarkowsky, T.A.; Rajeevan, M.S.; Lee, D.R.; Unger, E.R. Improved detection of viral from liquid-based cytology samples. Mol. Diagn. 2001, 6, 125–130. [Google Scholar] [CrossRef]
- Boom, R.; Salimans, M.M.; Jansen, C.L.; Wertheim-van Dillen, P.M.; van der Noordaa, J. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 1990, 28, 495–503. [Google Scholar] [PubMed]
- Cheung, R.C.; Matsui, S.M.; Greenberg, H.B. Rapid and sensitive method for detection of hepatitis C virus RNA by using silica particles. J. Clin. Microbiol. 1994, 32, 2593–2597. [Google Scholar] [PubMed]
- Santella, R.M. Approaches to DNA/RNA extraction and whole genome amplification. Cancer Epidemiol. Biomarkers Prev. 2006, 15, 1585–1587. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, Y.; Yamaguchi, H.; Einaga, N.; Esumi, M. Pitfalls of DNA quantification using DNA-binding fluorescent dyes and suggested solutions. PLoS ONE 2016, 11, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Jenatet, D.; Schwarzmann, F.; Tromp, J.; Melchers, W.J.G.; van der Wurff, A.A.M.; Oosterlaken, T.; Jacobs, M.; Troesch, A. Nuclisens EasyQ HPV v1 test–Testing for oncogenic activity of human papillomaviruses. J. Clin. Virol. 2009, 45, 529–537. [Google Scholar] [CrossRef]
- Cuschieri, K.S.; Beattie, G.; Hassan, S.; Robertson, K.; Cubie, H. Assessment of human papillomavirus mRNA detection over time in cervical specimens collected in liquid based cytology medium. J. Clin. Virol. Methods 2005, 124, 211–215. [Google Scholar] [CrossRef] [PubMed]
- Burger, E.A.; Kornør, H.; Klemp, M.; Lauvrak, V.; Kristiansen, I.S. HPV mRNA tests for the detection of cervical intraepithelial neoplasia: A systematic review. Gynecol. Oncol. 2011, 120, 430–438. [Google Scholar] [CrossRef] [PubMed]
- Westre, B.; Giske, A.; Guttomsen, H.; Sørbye, S.W.; Skjeldestad, F.E. 5-tipe HPV mRNA versus 14-type HPV DNA test: Test performance, over-diagnosis and overtreatment in triage of women with minor cervical lesions. BMC Clin. Pathol. 2016, 16, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Jordan, J.; Arbyn, M.; Hirsch, P.M.; Schenck, U.; Baldauf, J.J.; da Silva, D.; Anttila, A.; Nieminen, P.; Prendiville, W. European guidelines for quality assurance in cervical screening: Recommendations for clinical management of abnormal cervical cytology. Cytopathology 2008, 19, 342–354. [Google Scholar] [CrossRef] [PubMed]
- Guidelines of the Italian Society of Cervical Pathology and Colposcopy (SICPVCP). Available online: www.colposcopiaitaliana.it/pdf07/Linee-Guida-2006.pdf (accessed on 27 March 2017).
- Zappacosta, R.; Gatta, D.M.P.; Marinucci, P.; Capanna, S.; Lattanzio, G.; Caraceni, D.; Rosini, S. Role of E6/E7 mRNA test in the diagnostic algorithm of HPV-positive patients showing ASCUS and LSIL: Clinical and economic implications in a publicly financed healthcare system. Expert Rev. Mol. Diagn. 2015, 15, 137–150. [Google Scholar] [CrossRef] [PubMed]
- Molden, T.; Nygard, J.F.; Kraus, I.; Karlsen, F.; Nygård, M.; Skare, G.B.; Skomedal, H.; Steinar, Ø.T.; Hagmar, B. Predicting CIN2+ when detecting HPV mRNA and DNA by PreTect HPV-proofer and consensus PCR: A 2-year follow-up of women with ASCUS or LSIL Pap smear. Int. J. Cancer 2005, 10, 973–977. [Google Scholar] [CrossRef] [PubMed]
- Arbyn, M.; Ronco, G.; Anttila, A.; Meijer, C.J.L.M.; Poljak, M.; Ogilvie, G.; Koliopoulos, G.; Naucler, P.; Sankaranarayanan, R.; Peto, J. Evidence regarding human papillomavirus testing in secondary prevention of cervical cancer. Vaccine 2012, 30, 88–99. [Google Scholar] [CrossRef] [PubMed]
- Benevolo, M.; Vocaturo, A.; Caraceni, D.; French, D.; Rosini, S.; Zappacosta, R.; Terrenato, I.; Ciccocioppo, L.; Frega, A.; Giorgi Rossi, P. Sensitivity, specificity, and clinical value of human papillomavirus (HPV) E6/E7 mRNA assay as a triage test for cervical cytology and HPV DNA test. J. Clin. Microbiol. 2011, 49, 2643–2650. [Google Scholar] [CrossRef] [PubMed]
- Clifford, G.M.; Smith, J.S.; Plummer, M.; Munõz, N.; Franceschi, S. Human papillomavirus types in invasive cervical ancer worldwide: A meta-analysis. Br. J. Cancer 2003, 88, 53–73. [Google Scholar] [CrossRef] [PubMed]
- Von Knebel Doeberitz, M. Biomarkers in screening of cervical cancer. In Emerging Issues on HPV Infections: From Science to Practice; Monsonego, J., Ed.; Karger: Basel, Switzerland, 2006; pp. 1–19. [Google Scholar]
- Skomedal, H.; Kraus, I.; Molden, T.; Molden, T.; Hovland, S.; Morland, G.; Morland, E.; Karlsen, F. Direct detection of cervical carcinogenesis through mRNA. In Emerging Issues on HPV Infections: From Science to Practice; Monsonego, J., Ed.; Karger: Basel, Switzerland, 2006; pp. 82–102. [Google Scholar]
- Castle, P.E.D.; Solomon, C.M.; Wheeler, P.E.; Gravitt, P.E.; Wacholder, S.; Schiffman, M. Human papillomavirus genotype specificity of Hybrid Capture 2. J. Clin. Microbiol. 2003, 89, 101–105. [Google Scholar] [CrossRef] [PubMed]
- Solomon, D.; Davey, D.; Kurman, R.; Moriarty, A.; O’Connor, D.; Prey, M.; Raab, S.; Sherman, M.; Wilbur, D.; Wright, T.; et al. Bethesda 2001 Workshop. The 2001 Bethesda System: Terminology for reporting results of cervical cytology. JAMA 2002, 287, 2114–2119. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization: International Agency for Research on Cancer. DA cervix cancer screening. In IARC Handbooks of Cancer Prevention; IARC Press: Lyon, France, 2005; Volume 10. [Google Scholar]
- Landis, J.R.; Koch, G.G. The measurement of observer agreement for categorical data. Biometrics 1977, 1, 159–174. [Google Scholar] [CrossRef]
- Dillner, J. Methods in Biobanking; Springer: New York, NY, USA, 2011. [Google Scholar]
- Perskvist, N.; Norlin, L.; Dillner, J. The process of moving from a regional based cervical cytology biobank to a national infrastructure. Biopreserv. Biobank 2015, 13, 94–97. [Google Scholar] [CrossRef] [PubMed]
- Perskvist, N.; Björklund, C.; Dillner, J. A complex intervention for workflow enhancement at the Swedish cervical cytology biobank. Biopreserv. Biobank 2014, 12, 69–73. [Google Scholar] [CrossRef] [PubMed]
- Perskvist, N.; Norman, I.; Eklund, C.; Litton, J.E.; Dillner, J. The Swedish cervical cytology biobank: Sample handling and storage process. Biopreserv. Biobank 2013, 11, 19–24. [Google Scholar] [CrossRef] [PubMed]
- Hubel, A.; Spindler, R.; Skubitz, A. Storage of human biospecimens: Selection of the optimal storage temperature. Biopreserv. Biobank 2014, 12, 165–175. [Google Scholar] [CrossRef] [PubMed]
- Pukkala, E.; Andersen, A.; Berglund, G.; Gislefoss, R.; Gudnason, V.; Hallmans, G.; Jellum, E.; Jousilahti, P.; Knekt, P.; Koskela, P.; et al. Nordic biological specimen banks as basis for studies of cancer causes and control-more than 2 million sample donors, 25 million person years and 100,000 prospective cancers. Acta Oncol. 2007, 46, 286–307. [Google Scholar] [CrossRef] [PubMed]
- Mirghani, H.; Casiraghi, O.; Amen, F.; He, M.; Ma, X.J.; Saulnier, P.; Lacroix, L.; Drusch, F.; Lakdhar, B.A.; Saint Guily, J.L.; et al. Diagnosis of HPV-driven head and neck cancer with a single test in routine clinical practice. Mod. Pathol. 2015, 28, 1518–1527. [Google Scholar] [CrossRef] [PubMed]

| Years of Sample Collection | E6/E7 mRNA Test Result (%) | ||
|---|---|---|---|
| Determined ° | Undetermined * | Total (%) | |
| 2001–2004 | 52 (29.2) | 4 (16.7) | 56 (27.7) |
| 2005–2008 | 81 (45.5) | 14 (58.3) | 95 (47) |
| 2009–2012 | 45 (25.3) | 6 (25) | 51 (25.3) |
| Total (%) | 178 (88,1) | 24 (11.9) | 202 |
| Hystological Diagnosis (%) | Cytological Diagnosis (%) | HPV-DNA Test (%) | E6/E7 mRNA Test (%) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| ASCUS | LSIL | ASC-H/HSIL | SCC | Positive | Negative | Positive | Negative | Undetermined * | |
| CIN2− | 24 (54.5) | 23 (43.4) | 9 (20.9) | 0 | 44 (32.6) | 12 (85.7) | 10 (10.8) | 31 (97) | 15 (62.5) |
| CIN2+ | 20 (45.5) | 30 (56.6) | 34 (79.1) | 9 (100) | 91 (67.4) | 2 (14.3) | 83 (89.2) | 1 (3) | 9 (37.5) |
| Total | 44 (29.5) | 53 (35.6) | 43 (28.9) | 9 (6) | 135 (90.6) | 14 (9.4) | 93 (62.4) | 32 (21.5) | 24 (11.9) |
| Age, Years | Clinical Performances of E6/E7 mRNA Test, % (95% CI) | |||
|---|---|---|---|---|
| Sensitivity | Specificity | PPV | NPV | |
| <30 (n = 38) | 96.3 (81.7–99.3) | 63.6 (35.3–84.8) | 86.7 (74.5–98.8) | 87.5 (64.6–100) |
| ≥30 (n = 87) | 100 (92.3–100) | 80 (62.2–90.7) | 90.5 (83.2–97.7) | 100 (100–100) |
| Molecular Testing | Clinical Performances, % (95% CI) | |||
|---|---|---|---|---|
| Sensitivity | Specificity | PPV | NPV | |
| HPV-DNA | 97.6 (90.8–99.5) | 17 (7–32.6) | 70.7 (61.4–78.6) | 77.8 (40.2–96) |
| HPV E6/E7 mRNA | 98.8 (92.6–100) | 75.6 (59.4–87) | 89.4 (80.7–94.4) | 97 (82–100) |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zappacosta, R.; Sablone, F.; Pansa, L.; Buca, D.; Buca, D.; Rosini, S. Analytic and Diagnostic Performances of Human Papillomavirus E6/E7 mRNA Test on up-to 11-Year-Old Liquid-Based Cervical Samples. A Biobank-Based Longitudinal Study. Int. J. Mol. Sci. 2017, 18, 1480. https://doi.org/10.3390/ijms18071480
Zappacosta R, Sablone F, Pansa L, Buca D, Buca D, Rosini S. Analytic and Diagnostic Performances of Human Papillomavirus E6/E7 mRNA Test on up-to 11-Year-Old Liquid-Based Cervical Samples. A Biobank-Based Longitudinal Study. International Journal of Molecular Sciences. 2017; 18(7):1480. https://doi.org/10.3390/ijms18071480
Chicago/Turabian StyleZappacosta, Roberta, Francesca Sablone, Lucia Pansa, Davide Buca, Danilo Buca, and Sandra Rosini. 2017. "Analytic and Diagnostic Performances of Human Papillomavirus E6/E7 mRNA Test on up-to 11-Year-Old Liquid-Based Cervical Samples. A Biobank-Based Longitudinal Study" International Journal of Molecular Sciences 18, no. 7: 1480. https://doi.org/10.3390/ijms18071480
APA StyleZappacosta, R., Sablone, F., Pansa, L., Buca, D., Buca, D., & Rosini, S. (2017). Analytic and Diagnostic Performances of Human Papillomavirus E6/E7 mRNA Test on up-to 11-Year-Old Liquid-Based Cervical Samples. A Biobank-Based Longitudinal Study. International Journal of Molecular Sciences, 18(7), 1480. https://doi.org/10.3390/ijms18071480




