Reverse Vaccinology: An Approach for Identifying Leptospiral Vaccine Candidates
Abstract
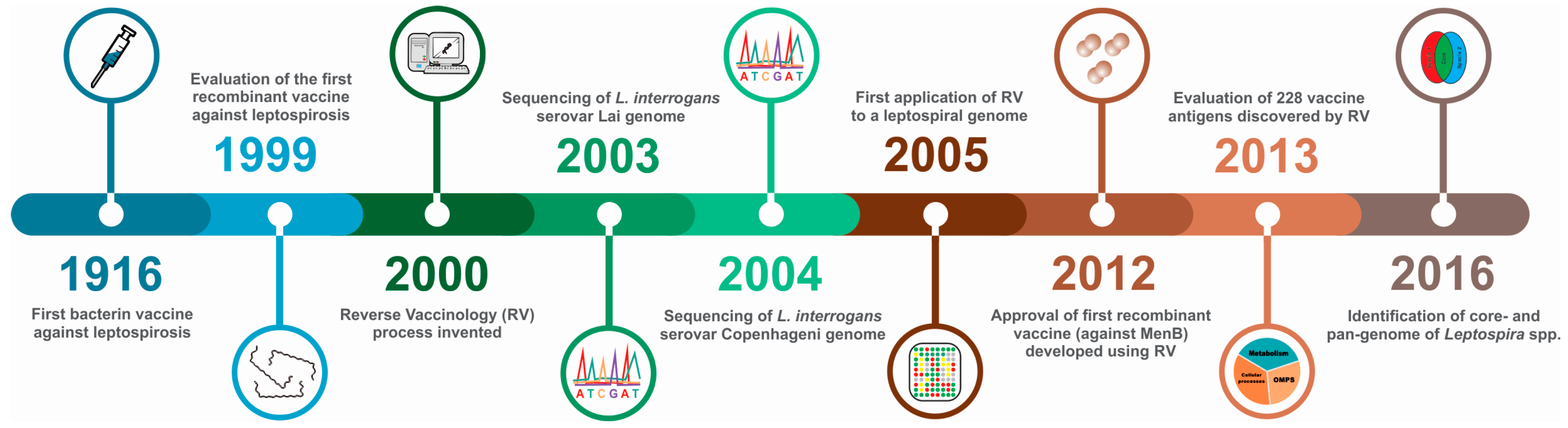
:1. Introduction
2. Leptospira and Leptospirosis
3. Conventional Vaccines Available against Leptospirosis
4. Recombinant Vaccines Based on Classically-Identified Antigens
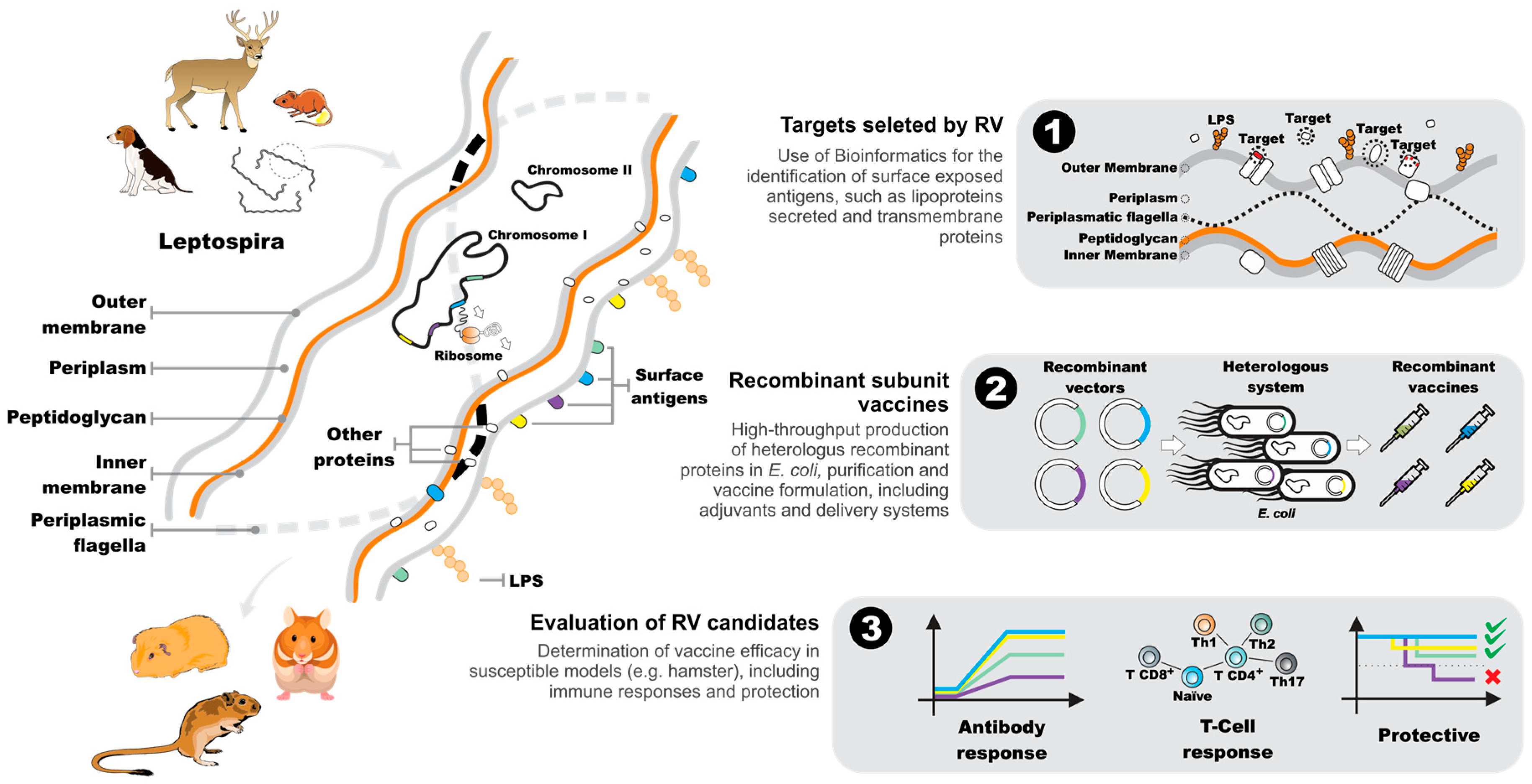
5. The Reverse Vaccinology Process
6. Leptospiral Genomes as a Source of Vaccine-Related Information
7. Downstream Analysis of Candidate Reverse Vaccinology Targets
8. Conclusions
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Leptospirosis worldwide, 1999. Wkly. Epidemiol. Rec. 1999, 74, 237–242. [Google Scholar]
- Costa, F.; Hagan, J.E.; Calcagno, J.; Kane, M.; Torgerson, P.; Martinez-Silveira, M.S.; Stein, C.; Abela-Ridder, B.; Ko, A.I. Global morbidity and mortality of leptospirosis: A systematic review. PLoS Negl. Trop. Dis. 2015, 9, e0003898. [Google Scholar] [CrossRef] [PubMed]
- Ellis, W.A. Animal leptospirosis. Curr. Top. Microbiol. Immunol. 2015, 387, 99–137. [Google Scholar] [PubMed]
- McBride, A.J.; Athanazio, D.A.; Reis, M.G.; Ko, A.I. Leptospirosis. Curr. Opin. Infect. Dis. 2005, 18, 376–386. [Google Scholar] [CrossRef] [PubMed]
- Dellagostin, O.A.; Grassmann, A.A.; Hartwig, D.D.; Felix, S.R.; da Silva, E.F.; McBride, A.J. Recombinant vaccines against leptospirosis. Hum. Vaccine 2011, 7, 1215–1224. [Google Scholar] [CrossRef] [PubMed]
- Rappuoli, R. Reverse vaccinology. Curr. Opin. Microbiol. 2000, 3, 445–450. [Google Scholar] [CrossRef]
- Rappuoli, R.; Pizza, M.; del Giudice, G.; de Gregorio, E. Vaccines, new opportunities for a new society. Proc. Natl. Acad. Sci. USA 2014, 111, 12288–12293. [Google Scholar] [CrossRef] [PubMed]
- Rappuoli, R. Vaccines, emerging viruses, and how to avoid disaster. BMC Biol. 2014, 12, 100. [Google Scholar] [CrossRef] [PubMed]
- Delany, I.; Rappuoli, R.; Seib, K.L. Vaccines, reverse vaccinology, and bacterial pathogenesis. Cold Spring Harb. Perspect. Med. 2013, 3, a012476. [Google Scholar] [CrossRef] [PubMed]
- Seib, K.L.; Zhao, X.; Rappuoli, R. Developing vaccines in the era of genomics: A decade of reverse vaccinology. Clin. Microbiol. Infect. 2012, 18, 109–116. [Google Scholar] [CrossRef] [PubMed]
- Michalik, M.; Djahanshiri, B.; Leo, J.C.; Linke, D. Reverse vaccinology: The pathway from genomes and epitope predictions to tailored recombinant vaccines. Methods Mol. Biol. 2016, 1403, 87–106. [Google Scholar] [PubMed]
- De la Fuente, J.; Kopacek, P.; Lew-Tabor, A.; Maritz-Olivier, C. Strategies for new and improved vaccines against ticks and tick-borne diseases. Parasite Immunol. 2016, 38, 754–769. [Google Scholar] [CrossRef] [PubMed]
- Adler, B. Leptospira and Leptospirosis; Springer: Berlin/Heidelberg, Germany, 2015. [Google Scholar]
- Levett, P.N. Systematics of leptospiraceae. Curr. Top. Microbiol. Immunol. 2015, 387, 11–20. [Google Scholar] [PubMed]
- Bourhy, P.; Collet, L.; Brisse, S.; Picardeau, M. Leptospira mayottensis sp. Nov., a pathogenic species of the genus Leptospira isolated from humans. Int. J. Syst. Evol. Microbiol. 2014, 64, 4061–4067. [Google Scholar] [CrossRef] [PubMed]
- Cameron, C.E. Leptospiral structure, physiology, and metabolism. Curr. Top. Microbiol. Immunol. 2015, 387, 21–41. [Google Scholar] [PubMed]
- Haake, D.A.; Zuckert, W.R. The leptospiral outer membrane. Curr. Top. Microbiol. Immunol. 2015, 387, 187–221. [Google Scholar] [PubMed]
- Picardeau, M. Genomics, proteomics, and genetics of leptospira. Curr. Top. Microbiol. Immunol. 2015, 387, 43–63. [Google Scholar] [PubMed]
- Murray, G.L. The molecular basis of leptospiral pathogenesis. Curr. Top. Microbiol. Immunol. 2015, 387, 139–185. [Google Scholar] [PubMed]
- Andre-Fontaine, G.; Aviat, F.; Thorin, C. Waterborne leptospirosis: Survival and preservation of the virulence of pathogenic Leptospira spp. In fresh water. Curr. Microbiol. 2015, 71, 136–142. [Google Scholar] [CrossRef] [PubMed]
- Ganoza, C.A.; Matthias, M.A.; Saito, M.; Cespedes, M.; Gotuzzo, E.; Vinetz, J.M. Asymptomatic renal colonization of humans in the peruvian amazon by leptospira. PLoS Negl. Trop. Dis. 2010, 4, e612. [Google Scholar] [CrossRef] [PubMed]
- Gouveia, E.L.; Metcalfe, J.; de Carvalho, A.L.; Aires, T.S.; Villasboas-Bisneto, J.C.; Queirroz, A.; Santos, A.C.; Salgado, K.; Reis, M.G.; Ko, A.I. Leptospirosis-associated severe pulmonary hemorrhagic syndrome, salvador, brazil. Emerg. Infect. Dis. 2008, 14, 505–508. [Google Scholar] [CrossRef] [PubMed]
- Segura, E.R.; Ganoza, C.A.; Campos, K.; Ricaldi, J.N.; Torres, S.; Silva, H.; Cespedes, M.J.; Matthias, M.A.; Swancutt, M.A.; Lopez Linan, R.; et al. Clinical spectrum of pulmonary involvement in leptospirosis in a region of endemicity, with quantification of leptospiral burden. Clin. Infect. Dis. 2005, 40, 343–351. [Google Scholar] [CrossRef] [PubMed]
- Fraga, T.R.; Barbosa, A.S.; Isaac, L. Leptospirosis: Aspects of innate immunity, immunopathogenesis and immune evasion from the complement system. Scand. J. Immunol. 2011, 73, 408–419. [Google Scholar] [CrossRef] [PubMed]
- Haake, D.A.; Levett, P.N. Leptospirosis in humans. Curr. Top. Microbiol. Immunol. 2015, 387, 65–97. [Google Scholar] [PubMed]
- World Health Organization; International Leptospirosis Society. Human Leptospirosis: Guidance for Diagnosis, Surveillance and Control; World Health Organization: Geneva, Switzerland, 2003. [Google Scholar]
- Torgerson, P.R.; Hagan, J.E.; Costa, F.; Calcagno, J.; Kane, M.; Martinez-Silveira, M.S.; Goris, M.G.; Stein, C.; Ko, A.I.; Abela-Ridder, B. Global burden of leptospirosis: Estimated in terms of disability adjusted life years. PLoS Negl. Trop. Dis. 2015, 9, e0004122. [Google Scholar] [CrossRef] [PubMed]
- Ido, Y.; Hoki, R.; Ito, H.; Wani, H. The prophylaxis of weil’s disease (spirochaetosis icterohaemorrhagica). J. Exp. Med. 1916, 24, 471–483. [Google Scholar] [CrossRef] [PubMed]
- Adler, B. Vaccines against leptospirosis. Curr. Top. Microbiol. Immunol. 2015, 387, 251–272. [Google Scholar] [PubMed]
- Da Cunha, C.E.; Felix, S.R.; Neto, A.C.; Campello-Felix, A.; Kremer, F.S.; Monte, L.G.; Amaral, M.G.; de Oliveira Nobre, M.; da Silva, E.F.; Hartleben, C.P.; et al. Infection with Leptospira kirschneri serovar mozdok: First report from the southern hemisphere. Am. J. Trop. Med. Hyg. 2016, 94, 519–521. [Google Scholar] [CrossRef] [PubMed]
- Sykes, J.E.; Hartmann, K.; Lunn, K.F.; Moore, G.E.; Stoddard, R.A.; Goldstein, R.E. 2010 acvim small animal consensus statement on leptospirosis: Diagnosis, epidemiology, treatment, and prevention. J. Vet. Intern. Med. 2011, 25, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Faine, S.B.; Adler, B.; Bolin, C.; Perolat, P. Leptospira and Leptospirosis, 2nd ed.; MediSci: Melbourne, Australia, 1999. [Google Scholar]
- Haake, D.A.; Mazel, M.K.; McCoy, A.M.; Milward, F.; Chao, G.; Matsunaga, J.; Wagar, E.A. Leptospiral outer membrane proteins OmpL1 and LipL41 exhibit synergistic immunoprotection. Infect. Immun. 1999, 67, 6572–6582. [Google Scholar] [PubMed]
- Shang, E.S.; Summers, T.A.; Haake, D.A. Molecular cloning and sequence analysis of the gene encoding LipL41, a surface-exposed lipoprotein of pathogenic Leptospira species. Infect. Immun. 1996, 64, 2322–2330. [Google Scholar] [PubMed]
- Haake, D.A.; Walker, E.M.; Blanco, D.R.; Bolin, C.A.; Miller, M.N.; Lovett, M.A. Changes in the surface of Leptospira interrogans serovar grippotyphosa during in vitro cultivation. Infect. Immun. 1991, 59, 1131–1140. [Google Scholar] [PubMed]
- Lin, X.; Sun, A.; Ruan, P.; Zhang, Z.; Yan, J. Characterization of conserved combined t and b cell epitopes in Leptospira interrogans major outer membrane proteins OmpL1 and LipL41. BMC Microbiol. 2011, 11, 21. [Google Scholar] [CrossRef] [PubMed]
- Feng, C.Y.; Li, Q.T.; Zhang, X.Y.; Dong, K.; Hu, B.Y.; Guo, X.K. Immune strategies using single-component LipL32 and multi-component recombinant LipL32–41-OmpL1 vaccines against leptospira. Braz. J. Med. Biol. Res. 2009, 42, 796–803. [Google Scholar] [CrossRef] [PubMed]
- Maneewatch, S.; Tapchaisri, P.; Sakolvaree, Y.; Klaysing, B.; Tongtawe, P.; Chaisri, U.; Songserm, T.; Wongratanacheewin, S.; Srimanote, P.; Chongsa-nguanz, M.; et al. OmpL1 DNA vaccine cross-protects against heterologous Leptospira spp. Challenge. Asian Pac. J. Allergy Immunol. 2007, 25, 75–82. [Google Scholar] [PubMed]
- Haake, D.A.; Chao, G.; Zuerner, R.L.; Barnett, J.K.; Barnett, D.; Mazel, M.; Matsunaga, J.; Levett, P.N.; Bolin, C.A. The leptospiral major outer membrane protein LipL32 is a lipoprotein expressed during mammalian infection. Infect. Immun. 2000, 68, 2276–2285. [Google Scholar] [CrossRef] [PubMed]
- Haake, D.A.; Suchard, M.A.; Kelley, M.M.; Dundoo, M.; Alt, D.P.; Zuerner, R.L. Molecular evolution and mosaicism of leptospiral outer membrane proteins involves horizontal DNA transfer. J. Bacteriol. 2004, 186, 2818–2828. [Google Scholar] [CrossRef] [PubMed]
- Malmstrom, J.; Beck, M.; Schmidt, A.; Lange, V.; Deutsch, E.W.; Aebersold, R. Proteome-wide cellular protein concentrations of the human pathogen Leptospira interrogans. Nature 2009, 460, 762–765. [Google Scholar] [CrossRef] [PubMed]
- Caimano, M.J.; Sivasankaran, S.K.; Allard, A.; Hurley, D.; Hokamp, K.; Grassmann, A.A.; Hinton, J.C.; Nally, J.E. A model system for studying the transcriptomic and physiological changes associated with mammalian host-adaptation by Leptospira interrogans serovar copenhageni. PLoS Pathog. 2014, 10, e1004004. [Google Scholar] [CrossRef] [PubMed]
- Hoke, D.E.; Egan, S.; Cullen, P.A.; Adler, B. LipL32 is an extracellular matrix-interacting protein of Leptospira spp. And pseudoalteromonas tunicata. Infect. Immun. 2008, 76, 2063–2069. [Google Scholar] [CrossRef] [PubMed]
- Hauk, P.; Macedo, F.; Romero, E.C.; Vasconcellos, S.A.; de Morais, Z.M.; Barbosa, A.S.; Ho, P.L. In LipL32, the major leptospiral lipoprotein, the c terminus is the primary immunogenic domain and mediates interaction with collagen IV and plasma fibronectin. Infect Immun. 2008, 76, 2642–2650. [Google Scholar] [CrossRef] [PubMed]
- Guerreiro, H.; Croda, J.; Flannery, B.; Mazel, M.; Matsunaga, J.; Galvao, R.M.; Levett, P.N.; Ko, A.I.; Haake, D.A. Leptospiral proteins recognized during the humoral immune response to leptospirosis in humans. Infect. Immun. 2001, 69, 4958–4968. [Google Scholar] [CrossRef] [PubMed]
- Pinne, M.; Haake, D.A. LipL32 is a subsurface lipoprotein of Leptospira interrogans: Presentation of new data and reevaluation of previous studies. PLoS ONE 2013, 8, e51025. [Google Scholar] [CrossRef] [PubMed]
- Murray, G.L.; Srikram, A.; Hoke, D.E.; Wunder, E.A., Jr.; Henry, R.; Lo, M.; Zhang, K.; Sermswan, R.W.; Ko, A.I.; Adler, B. Major surface protein LipL32 is not required for either acute or chronic infection with Leptospira interrogans. Infect. Immun. 2009, 77, 952–958. [Google Scholar] [CrossRef] [PubMed]
- Lucas, D.S.; Cullen, P.A.; Lo, M.; Srikram, A.; Sermswan, R.W.; Adler, B. Recombinant LipL32 and LigA from Leptospira are unable to stimulate protective immunity against leptospirosis in the hamster model. Vaccine 2011, 29, 3413–3418. [Google Scholar] [CrossRef] [PubMed]
- Branger, C.; Chatrenet, B.; Gauvrit, A.; Aviat, F.; Aubert, A.; Bach, J.M.; Andre-Fontaine, G. Protection against Leptospira interrogans sensu lato challenge by DNA immunization with the gene encoding hemolysin-associated protein 1. Infect. Immun. 2005, 73, 4062–4069. [Google Scholar] [CrossRef] [PubMed]
- Seixas, F.K.; da Silva, E.F.; Hartwig, D.D.; Cerqueira, G.M.; Amaral, M.; Fagundes, M.Q.; Dossa, R.G.; Dellagostin, O.A. Recombinant mycobacterium bovis bcg expressing the LipL32 antigen of Leptospira interrogans protects hamsters from challenge. Vaccine 2007, 26, 88–95. [Google Scholar] [CrossRef] [PubMed]
- Branger, C.; Sonrier, C.; Chatrenet, B.; Klonjkowski, B.; Ruvoen-Clouet, N.; Aubert, A.; Andre-Fontaine, G.; Eloit, M. Identification of the hemolysis-associated protein 1 as a cross-protective immunogen of Leptospira interrogans by adenovirus-mediated vaccination. Infect. Immun. 2001, 69, 6831–6838. [Google Scholar] [CrossRef] [PubMed]
- Grassmann, A.A.; Felix, S.R.; dos Santos, C.X.; Amaral, M.G.; Seixas Neto, A.C.; Fagundes, M.Q.; Seixas, F.K.; da Silva, E.F.; Conceicao, F.R.; Dellagostin, O.A. Protection against lethal leptospirosis after vaccination with LipL32 coupled or coadministered with the B subunit of Escherichia coli heat-labile enterotoxin. Clin. Vaccine Immunol. 2012, 19, 740–745. [Google Scholar] [CrossRef] [PubMed]
- Habarta, A.; Abreu, P.A.; Olivera, N.; Hauk, P.; Cedola, M.T.; Ferrer, M.F.; Ho, P.L.; Gomez, R.M. Increased immunogenicity to LipL32 of Leptospira interrogans when expressed as a fusion protein with the cholera toxin B subunit. Curr. Microbiol. 2011, 62, 526–531. [Google Scholar] [CrossRef] [PubMed]
- Matsunaga, J.; Barocchi, M.A.; Croda, J.; Young, T.A.; Sanchez, Y.; Siqueira, I.; Bolin, C.A.; Reis, M.G.; Riley, L.W.; Haake, D.A.; et al. Pathogenic Leptospira species express surface-exposed proteins belonging to the bacterial immunoglobulin superfamily. Mol. Microbiol. 2003, 49, 929–945. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Faisal, S.M.; Yan, W.; Chang, Y.C.; McDonough, S.P.; Zhang, N.; Akey, B.L.; Chang, Y.F. Evaluation of novel fusion proteins derived from extracellular matrix binding domains of LigB as vaccine candidates against leptospirosis in a hamster model. Vaccine 2011, 29, 7379–7386. [Google Scholar] [CrossRef] [PubMed]
- Yan, W.; Faisal, S.M.; McDonough, S.P.; Divers, T.J.; Barr, S.C.; Chang, C.F.; Pan, M.J.; Chang, Y.F. Immunogenicity and protective efficacy of recombinant Leptospira immunoglobulin-like protein B (rLigB) in a hamster challenge model. Microbes Infect. 2009, 11, 230–237. [Google Scholar] [CrossRef] [PubMed]
- Coutinho, M.L.; Choy, H.A.; Kelley, M.M.; Matsunaga, J.; Babbitt, J.T.; Lewis, M.S.; Aleixo, J.A.; Haake, D.A. A LigA three-domain region protects hamsters from lethal infection by Leptospira interrogans. PLoS Negl. Trop. Dis. 2011, 5, e1422. [Google Scholar] [CrossRef] [PubMed]
- Raja, V.; Natarajaseenivasan, K. Pathogenic, diagnostic and vaccine potential of leptospiral outer membrane proteins (OMPs). Crit. Rev. Microbiol. 2015, 41, 1–17. [Google Scholar] [CrossRef] [PubMed]
- He, H.J.; Wang, W.Y.; Wu, Z.D.; Lv, Z.Y.; Li, J.; Tan, L.Z. Protection of guinea pigs against Leptospira interrogans serovar Lai by LipL21 DNA vaccine. Cell. Mol. Immunol. 2008, 5, 385–391. [Google Scholar] [CrossRef] [PubMed]
- Nascimento, A.L.; Ko, A.I.; Martins, E.A.; Monteiro-Vitorello, C.B.; Ho, P.L.; Haake, D.A.; Verjovski-Almeida, S.; Hartskeerl, R.A.; Marques, M.V.; Oliveira, M.C.; et al. Comparative genomics of two Leptospira interrogans serovars reveals novel insights into physiology and pathogenesis. J. Bacteriol. 2004, 186, 2164–2172. [Google Scholar] [CrossRef] [PubMed]
- Ren, S.X.; Fu, G.; Jiang, X.G.; Zeng, R.; Miao, Y.G.; Xu, H.; Zhang, Y.X.; Xiong, H.; Lu, G.; Lu, L.F.; et al. Unique physiological and pathogenic features of Leptospira interrogans revealed by whole-genome sequencing. Nature 2003, 422, 888–893. [Google Scholar] [CrossRef] [PubMed]
- Gamberini, M.; Gomez, R.M.; Atzingen, M.V.; Martins, E.A.; Vasconcellos, S.A.; Romero, E.C.; Leite, L.C.; Ho, P.L.; Nascimento, A.L. Whole-genome analysis of Leptospira interrogans to identify potential vaccine candidates against leptospirosis. FEMS Microbiol. Lett. 2005, 244, 305–313. [Google Scholar] [CrossRef] [PubMed]
- Serruto, D.; Bottomley, M.J.; Ram, S.; Giuliani, M.M.; Rappuoli, R. The new multicomponent vaccine against meningococcal serogroup B, 4CMenB: Immunological, functional and structural characterization of the antigens. Vaccine 2012, 30, B87–B97. [Google Scholar] [CrossRef] [PubMed]
- Murray, G.L.; Lo, M.; Bulach, D.M.; Srikram, A.; Seemann, T.; Quinsey, N.S.; Sermswan, R.W.; Allen, A.; Adler, B. Evaluation of 238 antigens of Leptospira borgpetersenii serovar Hardjo for protection against kidney colonisation. Vaccine 2013, 31, 495–499. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Zhu, Y.; Wang, Y.; Chang, Y.F.; Zhang, Y.; Jiang, X.; Zhuang, X.; Zhu, Y.; Zhang, J.; Zeng, L.; et al. Whole genome sequencing revealed host adaptation-focused genomic plasticity of pathogenic Leptospira. Sci. Rep. 2016, 6, 20020. [Google Scholar] [CrossRef] [PubMed]
- Fouts, D.E.; Matthias, M.A.; Adhikarla, H.; Adler, B.; Amorim-Santos, L.; Berg, D.E.; Bulach, D.; Buschiazzo, A.; Chang, Y.F.; Galloway, R.L.; et al. What makes a bacterial species pathogenic? Comparative genomic analysis of the genus Leptospira. PLoS Negl. Trop. Dis. 2016, 10, e0004403. [Google Scholar] [CrossRef] [PubMed]
- Bowman, B.N.; McAdam, P.R.; Vivona, S.; Zhang, J.X.; Luong, T.; Belew, R.K.; Sahota, H.; Guiney, D.; Valafar, F.; Fierer, J.; et al. Improving reverse vaccinology with a machine learning approach. Vaccine 2011, 29, 8156–8164. [Google Scholar] [CrossRef] [PubMed]
- Heinson, A.I.; Woelk, C.H.; Newell, M.L. The promise of reverse vaccinology. Int. Health 2015, 7, 85–89. [Google Scholar] [CrossRef] [PubMed]
- Picardeau, M.; Bulach, D.M.; Bouchier, C.; Zuerner, R.L.; Zidane, N.; Wilson, P.J.; Creno, S.; Kuczek, E.S.; Bommezzadri, S.; Davis, J.C.; et al. Genome sequence of the saprophyte Leptospira biflexa provides insights into the evolution of Leptospira and the pathogenesis of leptospirosis. PLoS ONE 2008, 3, e1607. [Google Scholar] [CrossRef] [PubMed]
- Ricaldi, J.N.; Fouts, D.E.; Selengut, J.D.; Harkins, D.M.; Patra, K.P.; Moreno, A.; Lehmann, J.S.; Purushe, J.; Sanka, R.; Torres, M.; et al. Whole genome analysis of Leptospira licerasiae provides insight into leptospiral evolution and pathogenicity. PLoS Negl. Trop. Dis. 2012, 6, e1853. [Google Scholar] [CrossRef] [PubMed]
- Bulach, D.M.; Zuerner, R.L.; Wilson, P.; Seemann, T.; McGrath, A.; Cullen, P.A.; Davis, J.; Johnson, M.; Kuczek, E.; Alt, D.P.; et al. Genome reduction in Leptospira borgpetersenii reflects limited transmission potential. Proc. Natl. Acad. Sci. USA 2006, 103, 14560–14565. [Google Scholar] [CrossRef] [PubMed]
- Chou, L.F.; Chen, Y.T.; Lu, C.W.; Ko, Y.C.; Tang, C.Y.; Pan, M.J.; Tian, Y.C.; Chiu, C.H.; Hung, C.C.; Yang, C.W. Sequence of Leptospira santarosai serovar shermani genome and prediction of virulence-associated genes. Gene 2012, 511, 364–370. [Google Scholar] [CrossRef] [PubMed]
- Kremer, F.S.; Eslabao, M.R.; Jorge, S.; Oliveira, N.R.; Labonde, J.; Santos, M.N.; Monte, L.G.; Grassmann, A.A.; Cunha, C.E.; Forster, K.M.; et al. Draft genome of the Leptospira interrogans strains, Acegua, RCA, Prea, and Capivara, obtained from wildlife maintenance hosts and infected domestic animals. Mem. Inst. Oswaldo Cruz 2016, 111, 280–283. [Google Scholar] [CrossRef] [PubMed]
- Barragan, V.; Sahl, J.W.; Wiggins, K.; Chiriboga, J.; Salinas, A.; Cantos, N.E.; Loor, M.N.; Intriago, B.I.; Morales, M.; Trueba, G.; et al. Draft genome sequence of the first pathogenic Leptospira isolates from ecuador. Genome Announc. 2016. [Google Scholar] [CrossRef] [PubMed]
- Slamti, L.; Picardeau, M. Construction of a library of random mutants in the spirochete Leptospira biflexa using a mariner transposon. Methods Mol. Biol. 2012, 859, 169–176. [Google Scholar] [PubMed]
- Figueira, C.P.; Croda, J.; Choy, H.A.; Haake, D.A.; Reis, M.G.; Ko, A.I.; Picardeau, M. Heterologous expression of pathogen-specific genes LigA and LigB in the saprophyte Leptospira biflexa confers enhanced adhesion to cultured cells and fibronectin. BMC Microbiol. 2011, 11, 129. [Google Scholar] [CrossRef] [PubMed]
- Girons, I.S.; Bourhy, P.; Ottone, C.; Picardeau, M.; Yelton, D.; Hendrix, R.W.; Glaser, P.; Charon, N. The LE1 bacteriophage replicates as a plasmid within Leptospira biflexa: Construction of an L. Biflexa–Escherichia coli shuttle vector. J. Bacteriol 2000, 182, 5700–5705. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhuang, X.; Zhong, Y.; Zhang, C.; Zhang, Y.; Zeng, L.; Zhu, Y.; He, P.; Dong, K.; Pal, U.; et al. Distribution of plasmids in distinct Leptospira pathogenic species. PLoS Negl. Trop. Dis. 2015, 9, e0004220. [Google Scholar] [CrossRef] [PubMed]
- Umamaheswari, A.; Pradhan, D.; Hemanthkumar, M. Computer aided subunit vaccine design against pathogenic Leptospira serovars. Interdiscip. Sci. 2012, 4, 38–45. [Google Scholar] [CrossRef] [PubMed]
- Hartwig, D.D.; Seixas, F.K.; Cerqueira, G.M.; McBride, A.J.; Dellagostin, O.A. Characterization of the immunogenic and antigenic potential of putative lipoproteins from Leptospira interrogans. Curr. Microbiol. 2011, 62, 1337–1341. [Google Scholar] [CrossRef] [PubMed]
- Yan, W.; Faisal, S.M.; McDonough, S.P.; Chang, C.F.; Pan, M.J.; Akey, B.; Chang, Y.F. Identification and characterization of OmpA-like proteins as novel vaccine candidates for leptospirosis. Vaccine 2010, 28, 2277–2283. [Google Scholar] [CrossRef] [PubMed]
- Atzingen, M.V.; Goncales, A.P.; de Morais, Z.M.; Araujo, E.R.; de Brito, T.; Vasconcellos, S.A.; Nascimento, A.L. Characterization of leptospiral proteins that afford partial protection in hamsters against lethal challenge with Leptospira interrogans. J. Med. Microbiol. 2010, 59, 1005–1015. [Google Scholar] [CrossRef] [PubMed]
- Viratyosin, W.; Ingsriswang, S.; Pacharawongsakda, E.; Palittapongarnpim, P. Genome-wide subcellular localization of putative outer membrane and extracellular proteins in Leptospira interrogans serovar Lai genome using bioinformatics approaches. BMC Genom. 2008, 9, 181. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.F.; Chen, C.S.; Palaniappan, R.U.; He, H.; McDonough, S.P.; Barr, S.C.; Yan, W.; Faisal, S.M.; Pan, M.J.; Chang, C.F. Immunogenicity of the recombinant leptospiral putative outer membrane proteins as vaccine candidates. Vaccine 2007, 25, 8190–8197. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.L.; Zhu, Y.Z.; Qin, J.H.; He, P.; Jiang, X.C.; Zhao, G.P.; Guo, X.K. In silico and microarray-based genomic approaches to identifying potential vaccine candidates against Leptospira interrogans. BMC Genom. 2006, 7, 293. [Google Scholar] [CrossRef] [PubMed]
- Hartwig, D.D.; Forster, K.M.; Oliveira, T.L.; Amaral, M.; McBride, A.J.; Dellagostin, O.A. A prime-boost strategy using the novel vaccine candidate, lema, protects hamsters against leptospirosis. Clin. Vaccine Immunol. 2013, 20, 747–752. [Google Scholar] [CrossRef] [PubMed]
- De Alvarenga Mudadu, M.; Carvalho, V.; Leclercq, S.Y. Nonclassically secreted proteins as possible antigens for vaccine development: A reverse vaccinology approach. Appl. Biochem. Biotechnol. 2015, 175, 3360–3370. [Google Scholar] [CrossRef] [PubMed]
- Orenstein, W.A.; Bernier, R.H.; Dondero, T.J.; Hinman, A.R.; Marks, J.S.; Bart, K.J.; Sirotkin, B. Field evaluation of vaccine efficacy. Bull. World Health Organ. 1985, 63, 1055–1068. [Google Scholar] [PubMed]
- Buyuktimkin, B.; Saier, M.H., Jr. Comparative analyses of transport proteins encoded within the genomes of Leptospira species. Microb. Pathog. 2016, 98, 118–131. [Google Scholar] [CrossRef] [PubMed]
- Diaz-Mejia, J.J.; Babu, M.; Emili, A. Computational and experimental approaches to chart the Escherichia coli cell-envelope-associated proteome and interactome. FEMS Microbiol. Rev. 2009, 33, 66–97. [Google Scholar] [CrossRef] [PubMed]
- Lo, M.; Cordwell, S.J.; Bulach, D.M.; Adler, B. Comparative transcriptional and translational analysis of leptospiral outer membrane protein expression in response to temperature. PLoS Negl. Trop. Dis. 2009, 3, e560. [Google Scholar] [CrossRef] [PubMed]
- Atzingen, M.V.; Barbosa, A.S.; de Brito, T.; Vasconcellos, S.A.; de Morais, Z.M.; Lima, D.M.; Abreu, P.A.; Nascimento, A.L. Lsa21, a novel leptospiral protein binding adhesive matrix molecules and present during human infection. BMC Microbiol. 2008, 8, 70. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.H.; Sheng, Y.Y.; Zhang, Z.M.; Shi, Y.Z.; He, P.; Hu, B.Y.; Yang, Y.; Liu, S.G.; Zhao, G.P.; Guo, X.K. Genome-wide transcriptional analysis of temperature shift in L. interrogans serovar Lai strain 56601. BMC Microbiol. 2006, 6, 51. [Google Scholar]
- Lo, M.; Bulach, D.M.; Powell, D.R.; Haake, D.A.; Matsunaga, J.; Paustian, M.L.; Zuerner, R.L.; Adler, B. Effects of temperature on gene expression patterns in Leptospira interrogans serovar Lai as assessed by whole-genome microarrays. Infect. Immun. 2006, 74, 5848–5859. [Google Scholar] [CrossRef] [PubMed]
- Eshghi, A.; Becam, J.; Lambert, A.; Sismeiro, O.; Dillies, M.A.; Jagla, B.; Wunder, E.A., Jr.; Ko, A.I.; Coppee, J.Y.; Goarant, C.; et al. A putative regulatory genetic locus modulates virulence in the pathogen Leptospira interrogans. Infect. Immun. 2014, 82, 2542–2552. [Google Scholar] [CrossRef] [PubMed]
- Matsunaga, J.; Medeiros, M.A.; Sanchez, Y.; Werneid, K.F.; Ko, A.I. Osmotic regulation of expression of two extracellular matrix-binding proteins and a haemolysin of Leptospira interrogans: Differential effects on LigA and Sph2 extracellular release. Microbiology 2007, 153, 3390–3398. [Google Scholar] [CrossRef] [PubMed]
- Matsunaga, J.; Sanchez, Y.; Xu, X.; Haake, D.A. Osmolarity, a key environmental signal controlling expression of leptospiral proteins LigA and LigB and the extracellular release of LigA. Infect. Immun. 2005, 73, 70–78. [Google Scholar] [CrossRef] [PubMed]
- Lo, M.; Murray, G.L.; Khoo, C.A.; Haake, D.A.; Zuerner, R.L.; Adler, B. Transcriptional response of Leptospira interrogans to iron limitation and characterization of a PerR homolog. Infect. Immun. 2010, 78, 4850–4859. [Google Scholar] [CrossRef] [PubMed]
- Patarakul, K.; Lo, M.; Adler, B. Global transcriptomic response of Leptospira interrogans serovar copenhageni upon exposure to serum. BMC Microbiol. 2010, 10, 31. [Google Scholar] [CrossRef] [PubMed]
- Eshghi, A.; Cullen, P.A.; Cowen, L.; Zuerner, R.L.; Cameron, C.E. Global proteome analysis of Leptospira interrogans. J. Proteome Res. 2009, 8, 4564–4578. [Google Scholar] [CrossRef] [PubMed]
- Vieira, M.L.; Pimenta, D.C.; de Morais, Z.M.; Vasconcellos, S.A.; Nascimento, A.L. Proteome analysis of Leptospira interrogans virulent strain. Open Microbiol. J. 2009, 3, 69–74. [Google Scholar] [CrossRef] [PubMed]
- Beck, M.; Malmstrom, J.A.; Lange, V.; Schmidt, A.; Deutsch, E.W.; Aebersold, R. Visual proteomics of the human pathogen Leptospira interrogans. Nat. Methods 2009, 6, 817–823. [Google Scholar] [CrossRef] [PubMed]
- Thongboonkerd, V.; Chiangjong, W.; Saetun, P.; Sinchaikul, S.; Chen, S.T.; Kositanont, U. Analysis of differential proteomes in pathogenic and non-pathogenic Leptospira: Potential pathogenic and virulence factors. Proteomics 2009, 9, 3522–3534. [Google Scholar] [CrossRef] [PubMed]
- Zeng, L.B.; Zhuang, X.R.; Huang, L.L.; Zhang, Y.Y.; Chen, C.Y.; Dong, K.; Zhang, Y.; Cui, Z.L.; Ding, X.L.; Chang, Y.F.; et al. Comparative subproteome analysis of three representative Leptospira interrogans vaccine strains reveals cross-reactive antigens and novel virulence determinants. J. Proteom. 2015, 112, 27–37. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.; Chang, X.; Cao, X.J.; Zhang, Y.; Zheng, H.; Zhu, Y.; Cai, C.; Cui, Z.; Li, Y.Y.; Jiang, X.G.; et al. Comparative proteogenomic analysis of the Leptospira interrogans virulence-attenuated strain IPAV against the pathogenic strain 56601. Cell Res. 2011, 21, 1210–1229. [Google Scholar] [CrossRef] [PubMed]
- Cullen, P.A.; Cordwell, S.J.; Bulach, D.M.; Haake, D.A.; Adler, B. Global analysis of outer membrane proteins from Leptospira interrogans serovar Lai. Infect. Immun. 2002, 70, 2311–2318. [Google Scholar] [CrossRef] [PubMed]
- Monahan, A.M.; Callanan, J.J.; Nally, J.E. Proteomic analysis of Leptospira interrogans shed in urine of chronically infected hosts. Infect. Immun. 2008, 76, 4952–4958. [Google Scholar] [CrossRef] [PubMed]
- Nally, J.E.; Monahan, A.M.; Miller, I.S.; Bonilla-Santiago, R.; Souda, P.; Whitelegge, J.P. Comparative proteomic analysis of differentially expressed proteins in the urine of reservoir hosts of leptospirosis. PLoS ONE 2011, 6, e26046. [Google Scholar] [CrossRef] [PubMed]
- Witchell, T.D.; Eshghi, A.; Nally, J.E.; Hof, R.; Boulanger, M.J.; Wunder, E.A., Jr.; Ko, A.I.; Haake, D.A.; Cameron, C.E. Post-translational modification of LipL32 during Leptospira interrogans infection. PLoS Negl. Trop. Dis. 2014, 8, e3280. [Google Scholar] [CrossRef] [PubMed]
- Eshghi, A.; Pinne, M.; Haake, D.A.; Zuerner, R.L.; Frank, A.; Cameron, C.E. Methylation and in vivo expression of the surface-exposed Leptospira interrogans outer-membrane protein OmpL32. Microbiology 2012, 158, 622–635. [Google Scholar] [CrossRef] [PubMed]
- Hartwig, D.D.; Bacelo, K.L.; Oliveira, P.D.; Oliveira, T.L.; Seixas, F.K.; Amaral, M.G.; Hartleben, C.P.; McBride, A.J.; Dellagostin, O.A. Mannosylated ligani produced in pichia pastoris protects hamsters against leptospirosis. Curr. Microbiol. 2014, 68, 524–530. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, T.L.; Grassmann, A.A.; Schuch, R.A.; Seixas Neto, A.C.; Mendonca, M.; Hartwig, D.D.; McBride, A.J.; Dellagostin, O.A. Evaluation of the Leptospira interrogans outer membrane protein OmpL37 as a vaccine candidate. PLoS ONE 2015, 10, e0142821. [Google Scholar] [CrossRef] [PubMed]
- Pizza, M.; Scarlato, V.; Masignani, V.; Giuliani, M.M.; Arico, B.; Comanducci, M.; Jennings, G.T.; Baldi, L.; Bartolini, E.; Capecchi, B.; et al. Identification of vaccine candidates against serogroup b meningococcus by whole-genome sequencing. Science 2000, 287, 1816–1820. [Google Scholar] [CrossRef] [PubMed]
- Etz, H.; Minh, D.B.; Henics, T.; Dryla, A.; Winkler, B.; Triska, C.; Boyd, A.P.; Sollner, J.; Schmidt, W.; von Ahsen, U.; et al. Identification of in vivo expressed vaccine candidate antigens from Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 2002, 99, 6573–6578. [Google Scholar] [CrossRef] [PubMed]
- Lourdault, K.; Wang, L.C.; Vieira, A.; Matsunaga, J.; Melo, R.; Lewis, M.S.; Haake, D.A.; Gomes-Solecki, M. Oral immunization with Escherichia coli expressing a lipidated form of LigA protects hamsters against challenge with Leptospira interrogans serovar copenhageni. Infect. Immun. 2014, 82, 893–902. [Google Scholar] [CrossRef] [PubMed]
| Serovars a | No. CDS | RV Targets | Targets Screened | Localization | Expt. Data | % Efficacy d | Fisher (p) | Reference |
|---|---|---|---|---|---|---|---|---|
| Cop | 3737 | 206 | 16 | OMP/LIP | WB | ND | ND | [62] |
| Lai-1/Cop | 3672 b | 226 | NA | OMP/IM/PS/SEC | CGH/MA | ND | ND | [85] |
| Lai-1/Pom | 4727/3741 | NK | 12 | OMP | RV/HML | 51–100 | >0.05 | [84] |
| Lai-1/Cop | 4727 | 177 | ND | ND | ND | ND | ND | [83] |
| Cop | 3737 | 206 | 3 | OMP | WB/HML | 12–38 | >0.05 | [82] |
| Pom | NK | 6 | 6 | OmpA-like | ELISA/CR/HML | 43–80 | >0.05 | [81] |
| Cop | 3530 | 226 | 8 | LIP | ELISA/WB/HML | 88 | <0.01 | [80,86] * |
| Lai-1/Cop/Har-1 & 2 | 2689 b | 74 | 12 e (9) c | OMP | NA | ND | ND | [79] |
| Har-1 | 3412 | 262 | 238 (223) c | OMP/LIP/SEC | ELISA/HKCM | 0 | ND | [64] |
| Cop/Lai-1 & 2/ | 3667/4727 & 3711/ | 63 | 12 (26) c | SEC | NA | ND | ND | [87] |
| Har-1 & 2 | 3412 & 3277 |
© 2017 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dellagostin, O.A.; Grassmann, A.A.; Rizzi, C.; Schuch, R.A.; Jorge, S.; Oliveira, T.L.; McBride, A.J.A.; Hartwig, D.D. Reverse Vaccinology: An Approach for Identifying Leptospiral Vaccine Candidates. Int. J. Mol. Sci. 2017, 18, 158. https://doi.org/10.3390/ijms18010158
Dellagostin OA, Grassmann AA, Rizzi C, Schuch RA, Jorge S, Oliveira TL, McBride AJA, Hartwig DD. Reverse Vaccinology: An Approach for Identifying Leptospiral Vaccine Candidates. International Journal of Molecular Sciences. 2017; 18(1):158. https://doi.org/10.3390/ijms18010158
Chicago/Turabian StyleDellagostin, Odir A., André A. Grassmann, Caroline Rizzi, Rodrigo A. Schuch, Sérgio Jorge, Thais L. Oliveira, Alan J. A. McBride, and Daiane D. Hartwig. 2017. "Reverse Vaccinology: An Approach for Identifying Leptospiral Vaccine Candidates" International Journal of Molecular Sciences 18, no. 1: 158. https://doi.org/10.3390/ijms18010158
APA StyleDellagostin, O. A., Grassmann, A. A., Rizzi, C., Schuch, R. A., Jorge, S., Oliveira, T. L., McBride, A. J. A., & Hartwig, D. D. (2017). Reverse Vaccinology: An Approach for Identifying Leptospiral Vaccine Candidates. International Journal of Molecular Sciences, 18(1), 158. https://doi.org/10.3390/ijms18010158







