Specific Magnetic Isolation of E6 HPV16 Modified Magnetizable Particles Coupled with PCR and Electrochemical Detection
Abstract
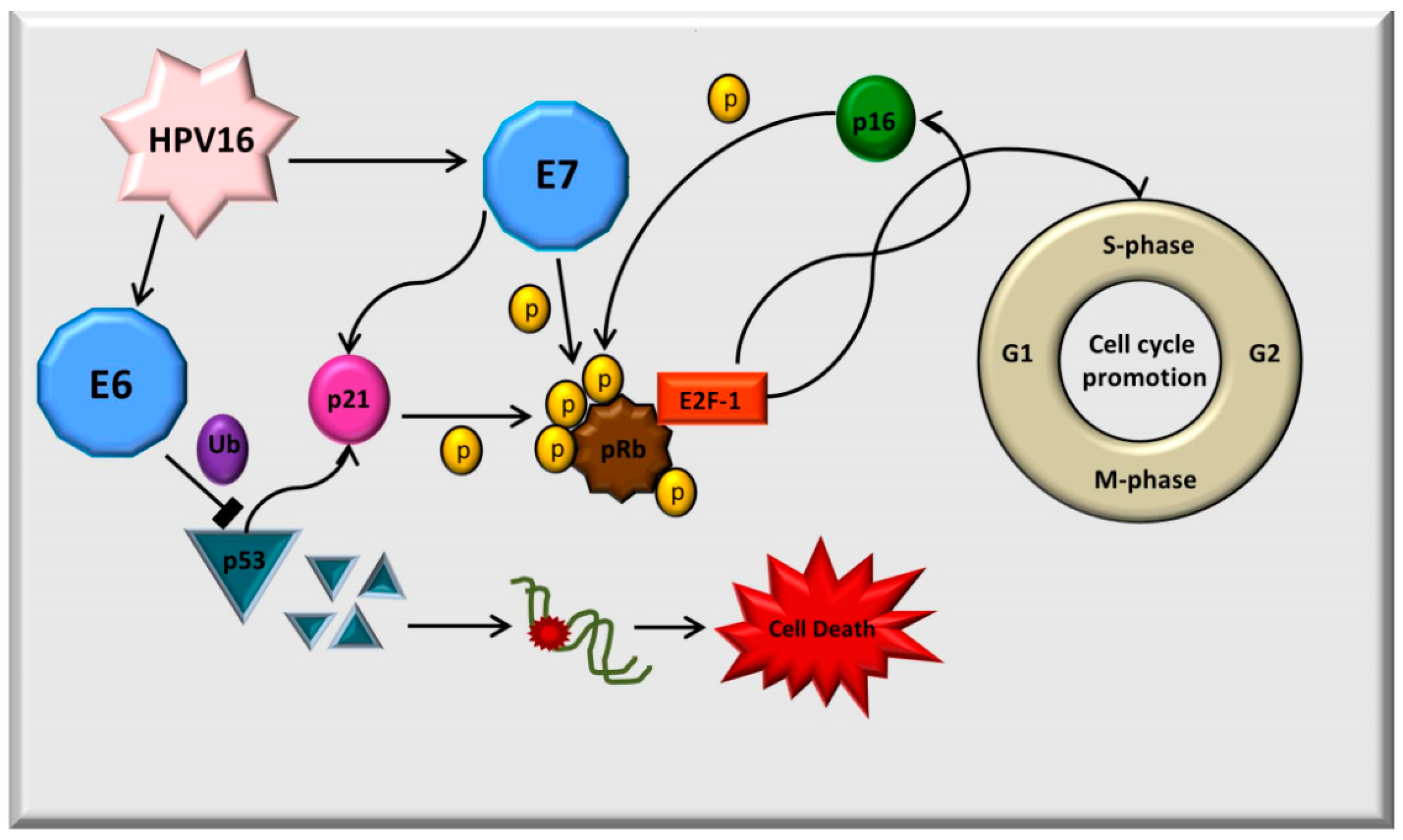
:1. Introduction
2. Results
2.1. Polymerase Chain Reaction (PCR) Optimization of E6-HPV16 (Human Papillomavirus 16) Oncogene
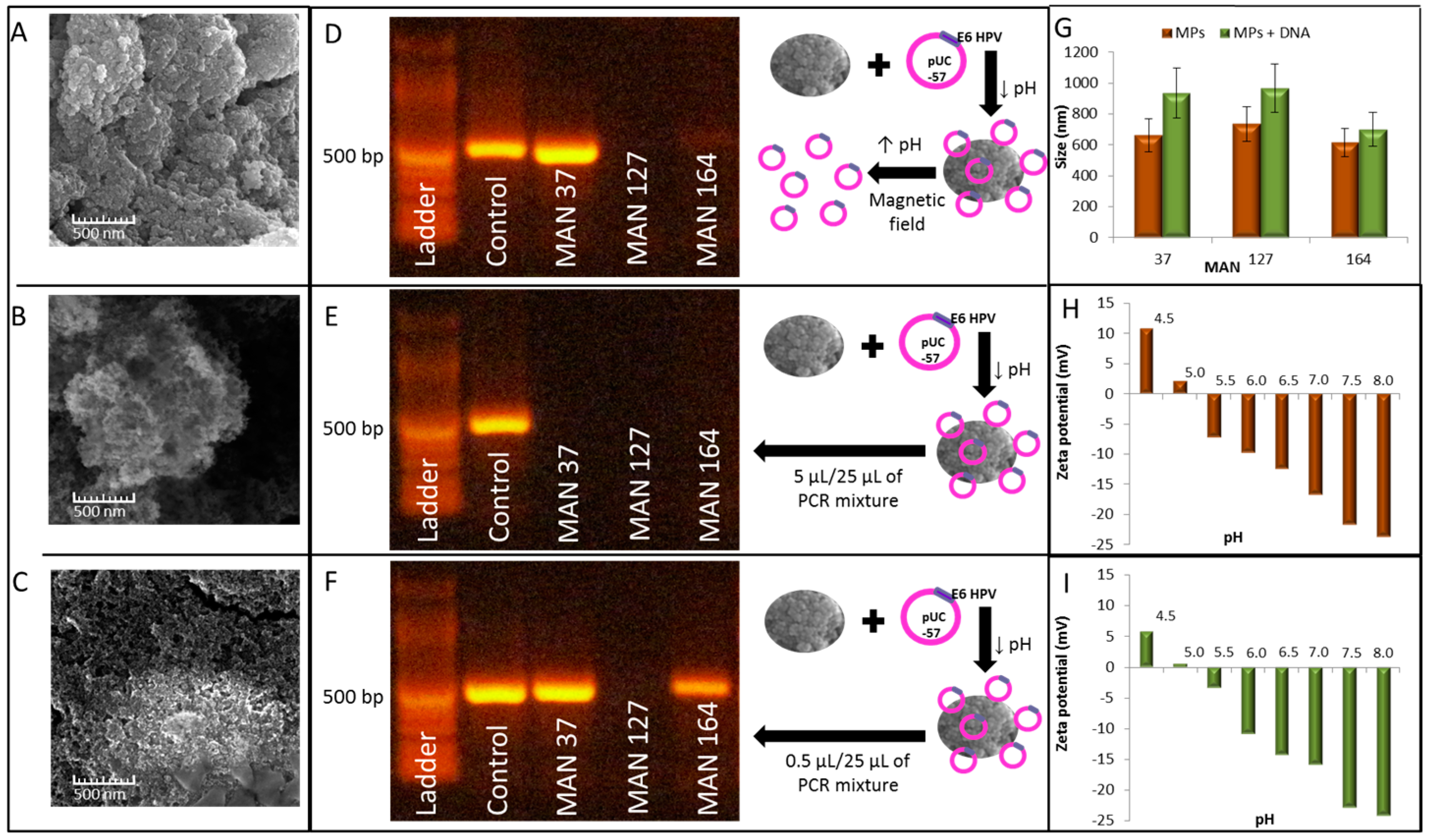
2.2. Isolation of E6-HPV16 Gene Using Commercial MPs with PCR Detection
2.3. Isolation of E6-HPV 16 Gene in a Plasmid Using Different “ Homemade” MPs with PCR Amplification and Detection
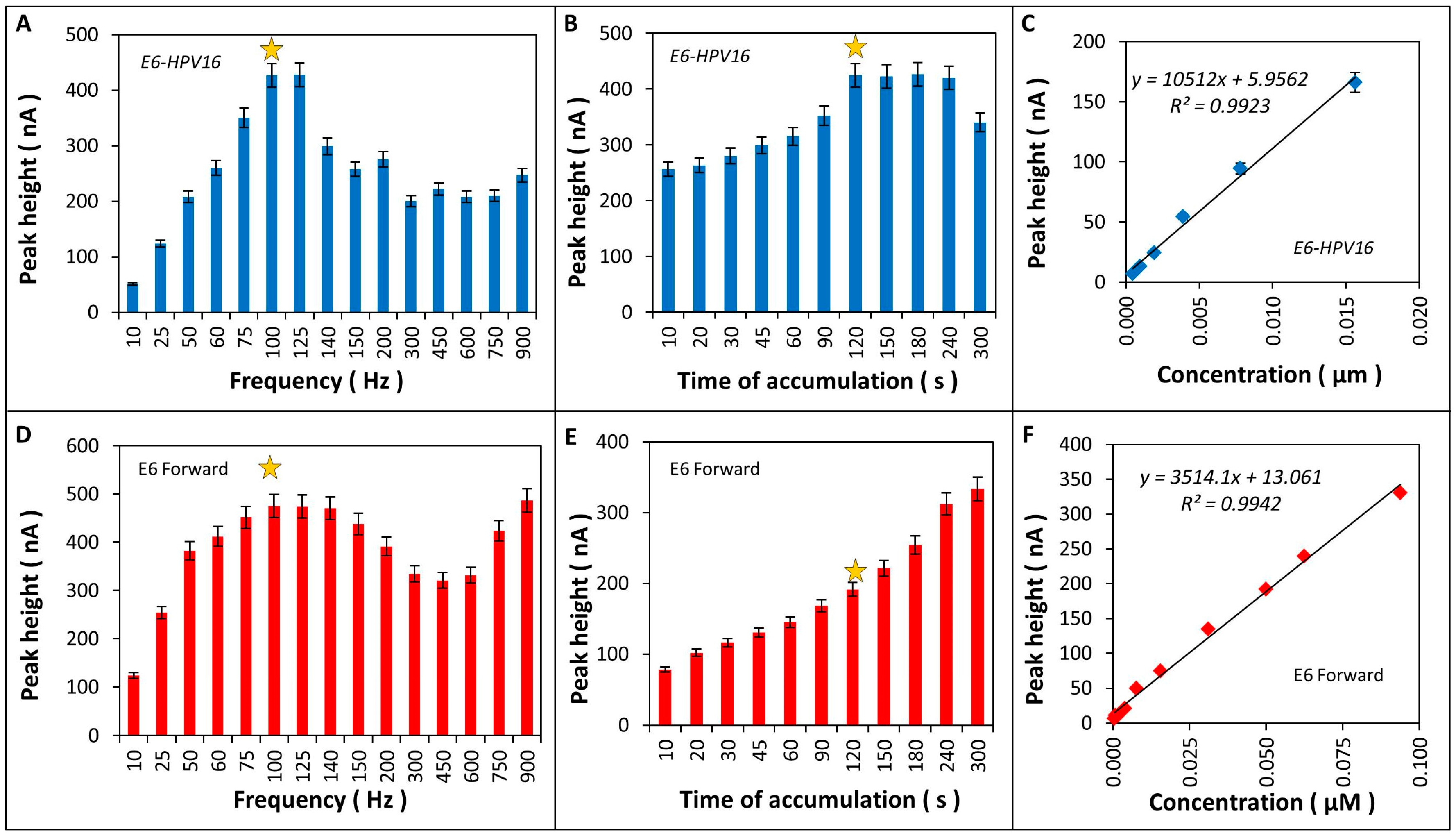
2.4. Optimization of Electrochemical Detection of of Cytosine-Adenine (CA) Peak of the Product of Amplification
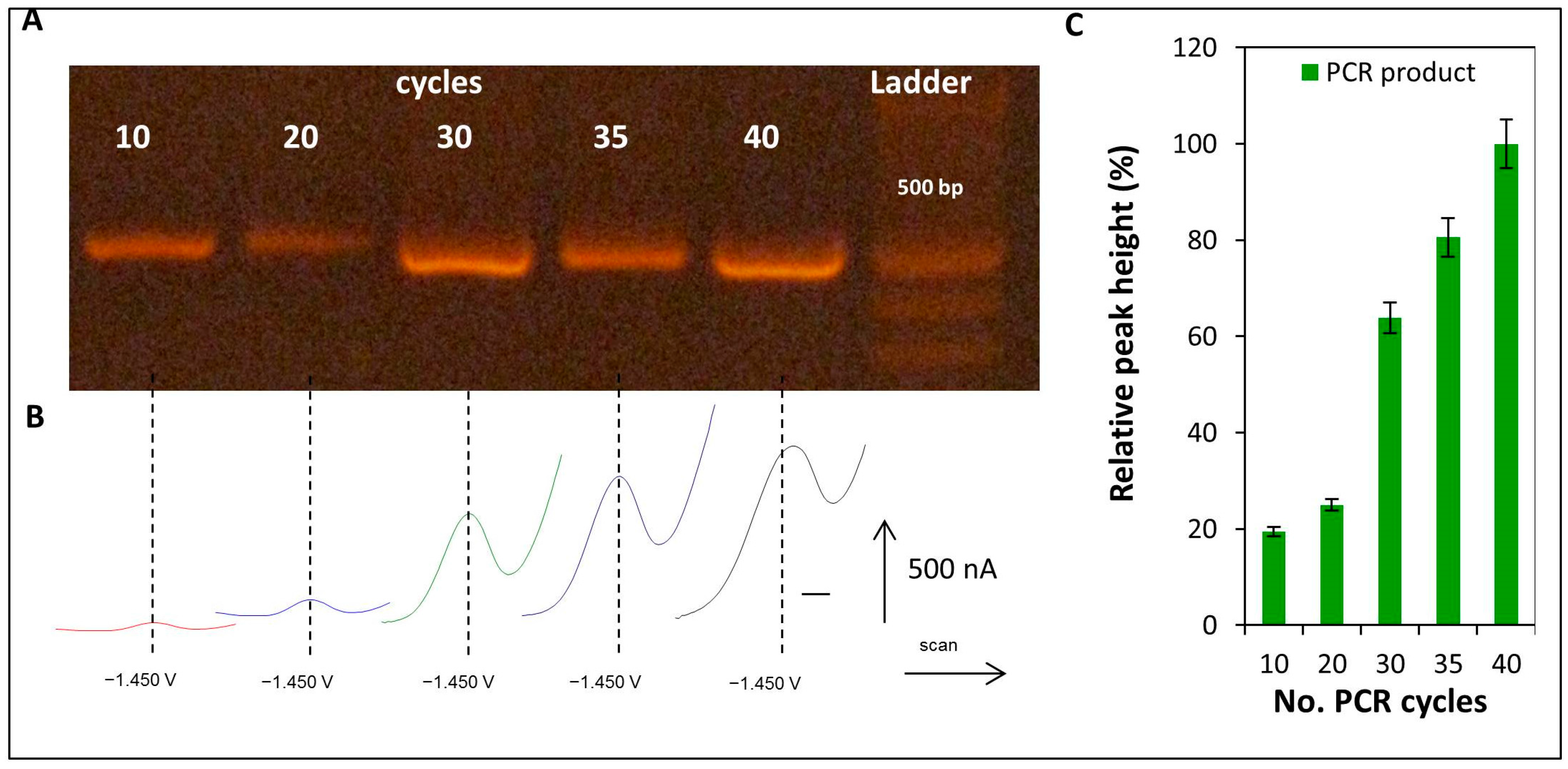
2.5. Detection of CA Peak in the Product of PCR Amplification of E6-HPV16 Gene
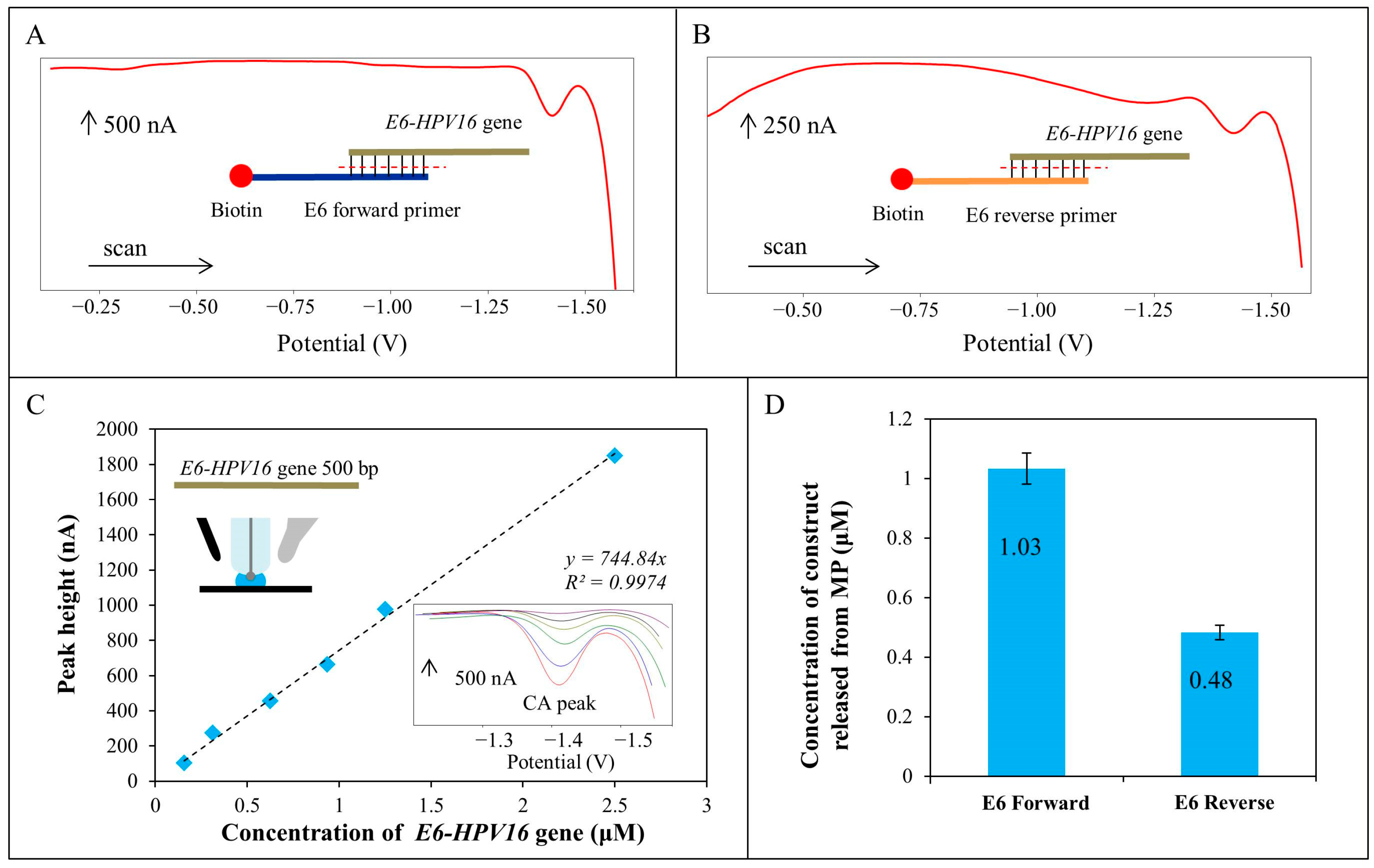
2.6. Electrochemical Characterization of E6-HPV16 Gene as a Product after Magnetic Isolation
3. Discussion
4. Materials and Methods
4.1. Chemicals
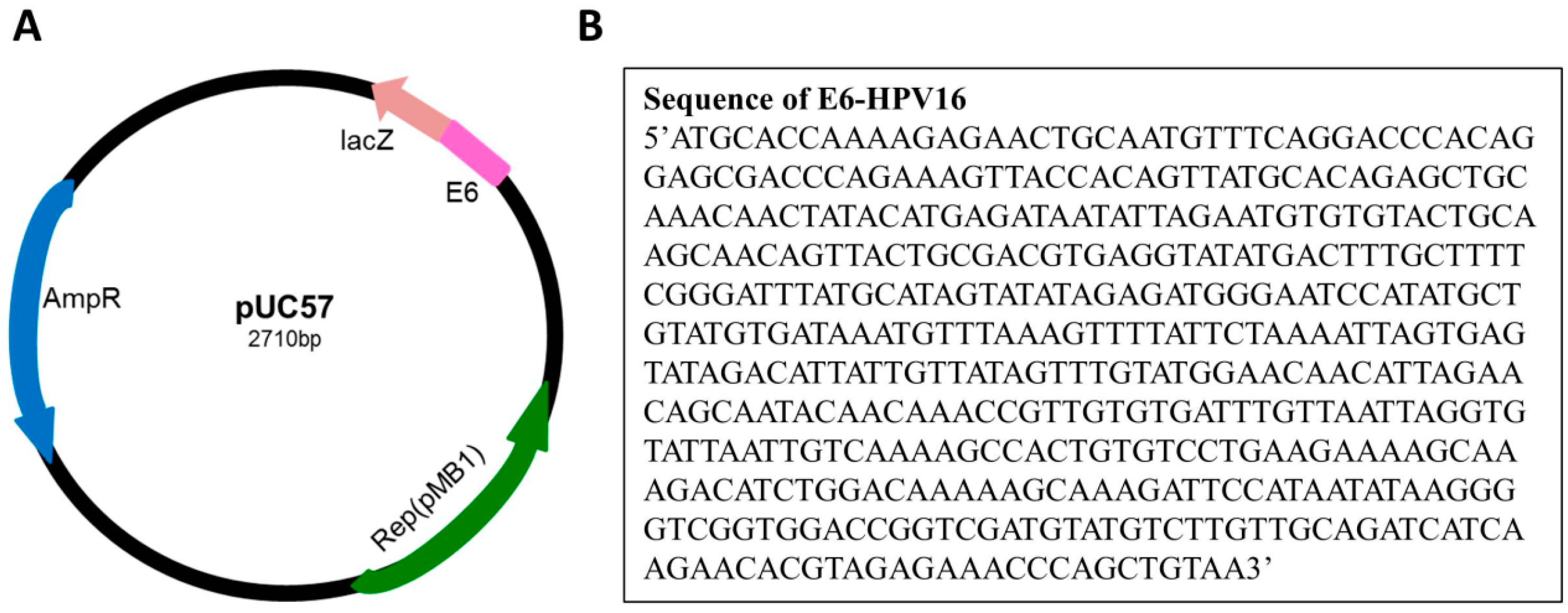
4.2. Cloning of the E6 Oncoprotein of Human Papillomavirus 16
4.3. Isolation and PCR Amplification of E6 Human Papillomavirus 16
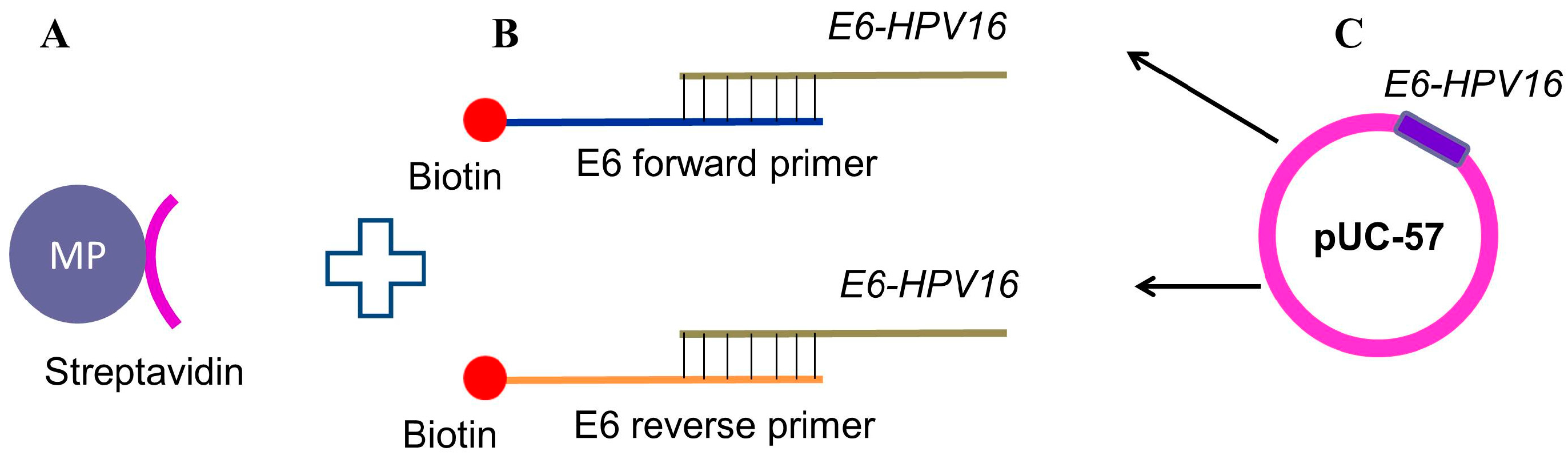
4.4. Biotinylation of Oligonucleotides
4.5. Magnetizable Particles and Their Preparation
4.5.1. MAN-37
4.5.2. MAN-127
4.5.3. MAN-164
4.6. Characterization of Particle Size
4.7. Scanning Electron Microscopy (SEM)
4.8. Binding of DNA with Magnetizable Particles
4.8.1. E6-HPV16 Gene Isolation Using Commercial Dynabeads
4.8.2. Isolation of E6-HPV16-pUC57 Plasmid Using Laboratory-Synthesized Magnetized Particles
4.9. Electrochemical Analysis
4.9.1. Optimization of Electrochemical Detection of CA Peak in Product of Amplification
4.9.2. Detection of CA Peak in the Product of PCR Amplification of E6-HPV16 Gene
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| AdTS | Adsorptive transfer stripping |
| APTES | 3-aminopropyltriethoxysilane |
| CA | Cytosine-adenine |
| CV | Cyclic voltammetry |
| DPV | Differential pulse voltammetry |
| EV | Elimination voltammetry |
| HPV | Human papillomavirus |
| MPs | Magnetizable particles |
| PBS | phosphate buffered saline |
| PCR | Polymerase chain reaction |
| SEM | Scanning electron microscopy |
| SWV | Square wave voltammetry |
References
- Asiaf, A.; Ahmad, S.T.; Mohammad, S.O.; Zargar, M.A. Review of the current knowledge on the epidemiology, pathogenesis, and prevention of human papillomavirus infection. Eur. J. Cancer Prev. 2014, 23, 206–224. [Google Scholar] [CrossRef] [PubMed]
- Parfenov, M.; Pedamallu, C.S.; Gehlenborg, N.; Freeman, S.S.; Danilova, L.; Bristow, C.A.; Lee, S.; Hadjipanayis, A.G.; Ivanova, E.V.; Wilkerson, M.D.; et al. Characterization of HPV and host genome interactions in primary head and neck cancers. Proc. Natl. Acad. Sci. USA 2014, 111, 15544–15549. [Google Scholar] [CrossRef] [PubMed]
- Ruttkay-Nedecky, B.; Jimenez, A.M.J.; Nejdl, L.; Chudobova, D.; Gumulec, J.; Masarik, M.; Adam, V.; Kizek, R. Relevance of infection with human papillomavirus: The role of the p53 tumor suppressor protein and E6/E7 zinc finger proteins. Int. J. Oncol. 2013, 43, 1754–1762. [Google Scholar] [PubMed]
- Venuti, A.; Paolini, F. HPV detection methods in head and neck cancer. Head Neck Pathol. 2012, 6, S63–S74. [Google Scholar] [CrossRef] [PubMed]
- Syrjanen, S. Human papillomavirus (HPV) in head and neck cancer. J. Clin. Virol. 2005, 32, S59–S66. [Google Scholar] [CrossRef] [PubMed]
- Avci, G.A. Genomic organization and proteins of human papillomavirus. Mikrobiyol. Bul. 2012, 46, 507–515. [Google Scholar]
- Yuan, C.H.; Filippova, M.; Duerksen-Hughes, P. Modulation of Apoptotic Pathways by human papillomaviruses (HPV): Mechanisms and implications for therapy. Viruses 2012, 4, 3831–3850. [Google Scholar] [CrossRef] [PubMed]
- Nejdl, L.; Skalickova, S.; Kudr, J.; Ruttkay-Nedecky, B.; Nguyen, H.V.; Rodrigo, M.A.M.; Kopel, P.; Konecna, M.; Adam, V.; Kizek, R. Interaction of E6 gene from human papilloma virus 16 (HPV-16) with CdS quantum dots. Chromatographia 2014, 77, 1433–1439. [Google Scholar] [CrossRef]
- Polanska, H.; Raudenska, M.; Gumulec, J.; Sztalmachova, M.; Adam, V.; Kizek, R.; Masarik, M. Clinical significance of head and neck squamous cell cancer biomarkers. Oral Oncol. 2014, 50, 168–177. [Google Scholar] [CrossRef] [PubMed]
- Moreas, H.; Tsiambas, E.; Lazaris, A.C.; Nonni, A.; Karameris, A.; Metaxas, G.E.; Armatas, H.E.; Patsouris, E. Impact of HPV detection in colorectal adenocarcinoma: HPV protein and chromogenic in situ hybridization analysis based on tissue microarrays. J. Buon 2014, 19, 91–96. [Google Scholar] [PubMed]
- Ramlee, M.K.; Yan, T.D.; Cheung, A.M.S.; Chuah, C.T.H.; Li, S. High-throughput genotyping of CRISPR/Cas9-mediated mutants using fluorescent PCR-capillary gel electrophoresis. Sci. Rep. 2015, 5, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.L.; Lin, X.X.; Lin, L.Y.; Yi, L.L.; Li, H.F.; Lin, J.M. A comparative study of three different nucleic acid amplification techniques combined with microchip electrophoresis for HPV16 E6/E7 mRNA detection. Analyst 2015, 140, 6736–6741. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.F.; Cheng, R.; Shi, Z.L.; Jin, Y. A PCR-free fluorescence strategy for detecting telomerase activity via double amplification strategy. Biosens. Bioelectron. 2016, 75, 101–107. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.Y.; Lu, X.H.; Su, F.X.; Wang, L.M.; Liu, C.H.; Duan, X.R.; Li, Z.P. Real-time fluorescence ligase chain reaction for sensitive detection of single nucleotide polymorphism based on fluorescence resonance energy transfer. Biosens. Bioelectron. 2015, 74, 705–710. [Google Scholar] [CrossRef] [PubMed]
- Miyaguchi, H.; Yamamuro, T.; Ohta, H.; Nakahara, H.; Suzuki, S. Genotyping of toxic pufferfish based on specific PCR-RFLP products as determined by liquid chromatography/quadrupole-orbitrap hybrid mass spectrometry. J. Agric. Food Chem. 2015, 63, 9363–9371. [Google Scholar] [CrossRef] [PubMed]
- Yin, R.; Sun, Y.J.; Yu, S.; Wang, Y.; Zhang, M.P.; Xu, Y.W.; Xue, J.; Xu, N. A validated strip-based lateral flow assay for the confirmation of sheep-specific PCR products for the authentication of meat. Food Control 2016, 60, 146–150. [Google Scholar] [CrossRef]
- Loh, Q.T.; Omar, N.; Glokler, J.; Lim, T.S. IQPA: Isothermal nucleic acid amplification-based immunoassay using DNAzyme as the reporter system. Anal. Biochem. 2014, 463, 67–69. [Google Scholar] [CrossRef] [PubMed]
- Napper, A.D. Perspectives in ASSAY and drug development technologies. Assay Drug Dev. Technol. 2015, 13, 241–241. [Google Scholar] [CrossRef] [PubMed]
- Terenzi, A.; Ducani, C.; Blanco, V.; Zerzankova, L.; Westendorf, A.F.; Peinador, C.; Quintela, J.M.; Bednarski, P.J.; Barone, G.; Hannon, M.J. DNA Binding studies and cytotoxicity of a dinuclear PtII diazapyrenium-based metallo-supramolecular rectangular box. Chem. Eur. J. 2012, 18, 10983–10990. [Google Scholar] [CrossRef] [PubMed]
- Bartosik, M.; Hrstka, R.; Palecek, E.; Vojtesek, B. Adsorptive transfer stripping for quick electrochemical determination of microRNAs in total RNA samples. Electroanalysis 2014, 26, 2558–2562. [Google Scholar] [CrossRef]
- Fojta, M.; Jelen, F.; Havran, L.; Palecek, E. Electrochemical stripping techniques in analysis of nucleic acids and their constituents. Curr. Anal. Chem. 2008, 4, 250–262. [Google Scholar] [CrossRef]
- Palecek, E.; Wang, J. Electrochemistry of nucleic acids and proteins-towards electrochemical sensors for genomics and proteomics preface. In Electrochemistry of Nucleic Acids and Proteins: Towards Electrochemical Sensors for Genomics and Proteomics; Palecek, E., Scheller, F., Wang, J., Eds.; Elsevier Science Bv: Amsterdam, The Netherlands, 2005; Volume 1, pp. XV–XVII. [Google Scholar]
- Ribeiro, J.A.; Pereira, C.M.; Silva, F. Electrochemistry of the interaction between bioactive drugs daunorubicin and dopamine and DNA at a water/oil interface. Electrochim. Acta 2015, 180, 687–694. [Google Scholar] [CrossRef]
- Masarik, M.; Kizek, R.; Kramer, K.J.; Billova, S.; Brazdova, M.; Vacek, J.; Bailey, M.; Jelen, F.; Howard, J.A. Application of avidin-biotin technology and adsorptive transfer stripping square-wave voltammetry for detection of DNA hybridization and avidin in transgenic avidin maize. Anal. Chem. 2003, 75, 2663–2669. [Google Scholar] [CrossRef] [PubMed]
- Gammoh, N.; Grm, H.S.; Massimi, P.; Banks, L. Regulation of human papillomavirus type 16 E7 activity through direct protein interaction with the E2 transcriptional activator. J. Virol. 2006, 80, 1787–1797. [Google Scholar] [CrossRef] [PubMed]
- Haugg, A.M.; Rennspiess, D.; zur Hausen, A.; Speel, E.J.M.; Cathomas, G.; Becker, J.C.; Schrama, D. Fluorescence in situ hybridization and qPCR to detect Merkel cell polyomavirus physical status and load in Merkel cell carcinomas. Int. J. Cancer 2014, 135, 2804–2815. [Google Scholar] [CrossRef] [PubMed]
- Heidegger, I.; Pichler, R.; Muller, B.; Klocker, H.; Oswald, D.; Haid, B.; Zelger, B.; Horninger, W.; Oswald, J. Is real-time PCR the correct method to evaluate the incidence of human papillomavirus in prepuces of asymptomatic boys and men? World J. Urol. 2014, 32, 1199–1204. [Google Scholar] [CrossRef] [PubMed]
- Krejcova, L.; Nguyen, H.V.; Hynek, D.; Guran, R.; Adam, V.; Kizek, R. Paramagnetic particles and PNA probe for automated separation and electrochemical detection of influenza. Chromatographia 2014, 77, 1425–1432. [Google Scholar] [CrossRef]
- Krejcova, L.; Hynek, D.; Guran, R.; Michalek, P.; Moulick, A.; Kopel, P.; Tmejova, K.; Hoai, N.V.; Adam, V.; Hubalek, J.; et al. Beads based electrochemical assay for detection of hemagglutinin labeled by two different types of cadmium quantum dots. Int. J. Electrochem. Sci. 2014, 9, 3349–3363. [Google Scholar]
- Krejcova, L.; Nejdl, L.; Rodrigo, M.A.M.; Zurek, M.; Matousek, M.; Hynek, D.; Zitka, O.; Kopel, P.; Adam, V.; Kizek, R. 3D printed chip for electrochemical detection of influenza virus labeled with CdS quantum dots. Biosens. Bioelectron. 2014, 54, 421–427. [Google Scholar] [CrossRef] [PubMed]
- Abreu, A.L.P.; Souza, R.P.; Gimenes, F.; Consolaro, M.E.L. A review of methods for detect human Papillomavirus infection. Virol. J. 2012, 9. [Google Scholar] [CrossRef] [PubMed]
- Jampasa, S.; Wonsawat, W.; Rodthongkum, N.; Siangproh, W.; Yanatatsaneejit, P.; Vilaivan, T.; Chailapakul, O. Electrochemical detection of human papillomavirus DNA type 16 using a pyrrolidinyl peptide nucleic acid probe immobilized on screen-printed carbon electrodes. Biosens. Bioelectron. 2014, 54, 428–434. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Li, L.; Jin, H.; Yang, T.; Bao, W.; Huang, S.; Wang, J. Electrochemical detection of hepatitis B and papilloma virus DNAs using SWCNT array coated with gold nanoparticles. Biosens. Bioelectron. 2013, 41, 205–210. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Bai, W.; Dong, C.; Guo, R.; Liu, Z. An ultrasensitive electrochemical DNA biosensor based on graphene/Au nanorod/polythionine for human papillomavirus DNA detection. Biosens. Bioelectron. 2015, 68, 442–446. [Google Scholar] [CrossRef] [PubMed]
- Bartolome, J.P.; Echegoyen, L.; Fragoso, A. Reactive carbon nano-onion modified glassy carbon surfaces as DNA sensors for human papillomavirus oncogene detection with enhanced sensitivity. Anal. Chem. 2015, 87, 6744–6751. [Google Scholar] [CrossRef] [PubMed]
- Campos-Ferreira, D.S.; Souza, E.V.M.; Nascimento, G.A.; Zanforlin, D.M.L.; Arruda, M.S.; Beltrão, M.F.S.; Melo, A.L.; Bruneska, D.; Lima-Filho, J.L. Electrochemical DNA biosensor for the detection of human papillomavirus E6 gene inserted in recombinant plasmid. Arab. J. Chem. 2014, 1–8. [Google Scholar] [CrossRef]
- Sabzi, R.E.; Sehatnia, B.; Pournaghi-Azar, M.H.; Hejazi, M.S. Electrochemical detection of human papilloma virus (HPV) target DNA using MB on pencil graphite electrode. J. Iran Chem. Soc. 2008, 5, 476–483. [Google Scholar] [CrossRef]
- Bo, Y.; Yang, H.; Hu, Y.; Yao, T.; Huang, S. A novel electrochemical DNA biosensor based on graphene and polyaniline nanowires. Electrochim. Acta 2011, 56, 2676–2681. [Google Scholar] [CrossRef]
- Zari, N.; Amine, A.; Ennaji, M.M. Label-free DNA biosensor for electrochemical detection of short DNA sequences related to human papilloma virus. Anal. Lett. 2009, 42, 519–535. [Google Scholar] [CrossRef]


© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jimenez Jimenez, A.M.; Ruttkay-Nedecky, B.; Dostalova, S.; Krejcova, L.; Michalek, P.; Richtera, L.; Adam, V. Specific Magnetic Isolation of E6 HPV16 Modified Magnetizable Particles Coupled with PCR and Electrochemical Detection. Int. J. Mol. Sci. 2016, 17, 585. https://doi.org/10.3390/ijms17050585
Jimenez Jimenez AM, Ruttkay-Nedecky B, Dostalova S, Krejcova L, Michalek P, Richtera L, Adam V. Specific Magnetic Isolation of E6 HPV16 Modified Magnetizable Particles Coupled with PCR and Electrochemical Detection. International Journal of Molecular Sciences. 2016; 17(5):585. https://doi.org/10.3390/ijms17050585
Chicago/Turabian StyleJimenez Jimenez, Ana Maria, Branislav Ruttkay-Nedecky, Simona Dostalova, Ludmila Krejcova, Petr Michalek, Lukas Richtera, and Vojtech Adam. 2016. "Specific Magnetic Isolation of E6 HPV16 Modified Magnetizable Particles Coupled with PCR and Electrochemical Detection" International Journal of Molecular Sciences 17, no. 5: 585. https://doi.org/10.3390/ijms17050585
APA StyleJimenez Jimenez, A. M., Ruttkay-Nedecky, B., Dostalova, S., Krejcova, L., Michalek, P., Richtera, L., & Adam, V. (2016). Specific Magnetic Isolation of E6 HPV16 Modified Magnetizable Particles Coupled with PCR and Electrochemical Detection. International Journal of Molecular Sciences, 17(5), 585. https://doi.org/10.3390/ijms17050585








