RNase 7 in Cutaneous Defense
Abstract
:1. Introduction
2. Antimicrobial Activity of RNase 7
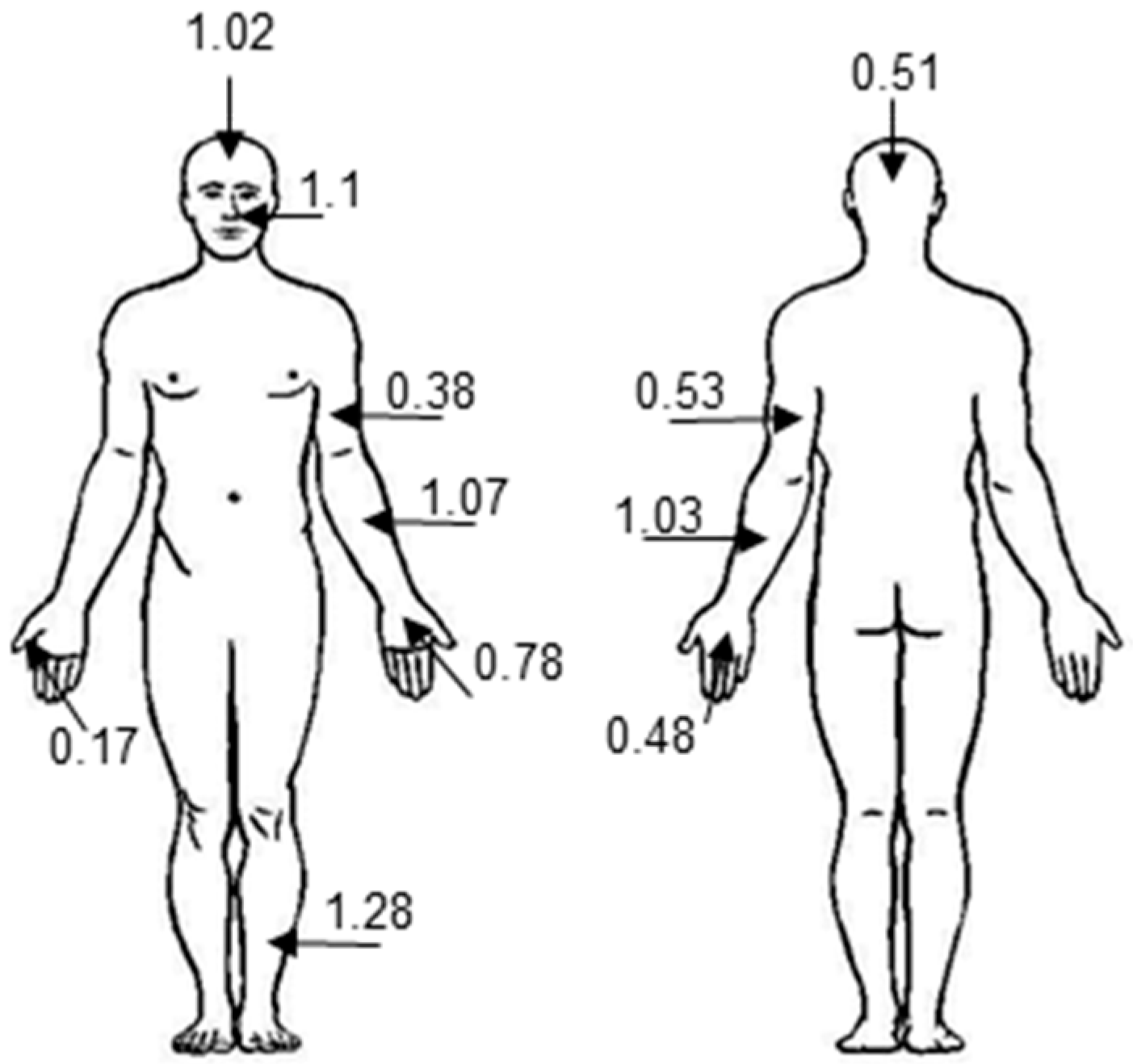
2.1. Microbial Targets
2.2. Mechanism
2.3. Bacterial Strategies to Subvert the Antimicrobial Activity of RNase 7
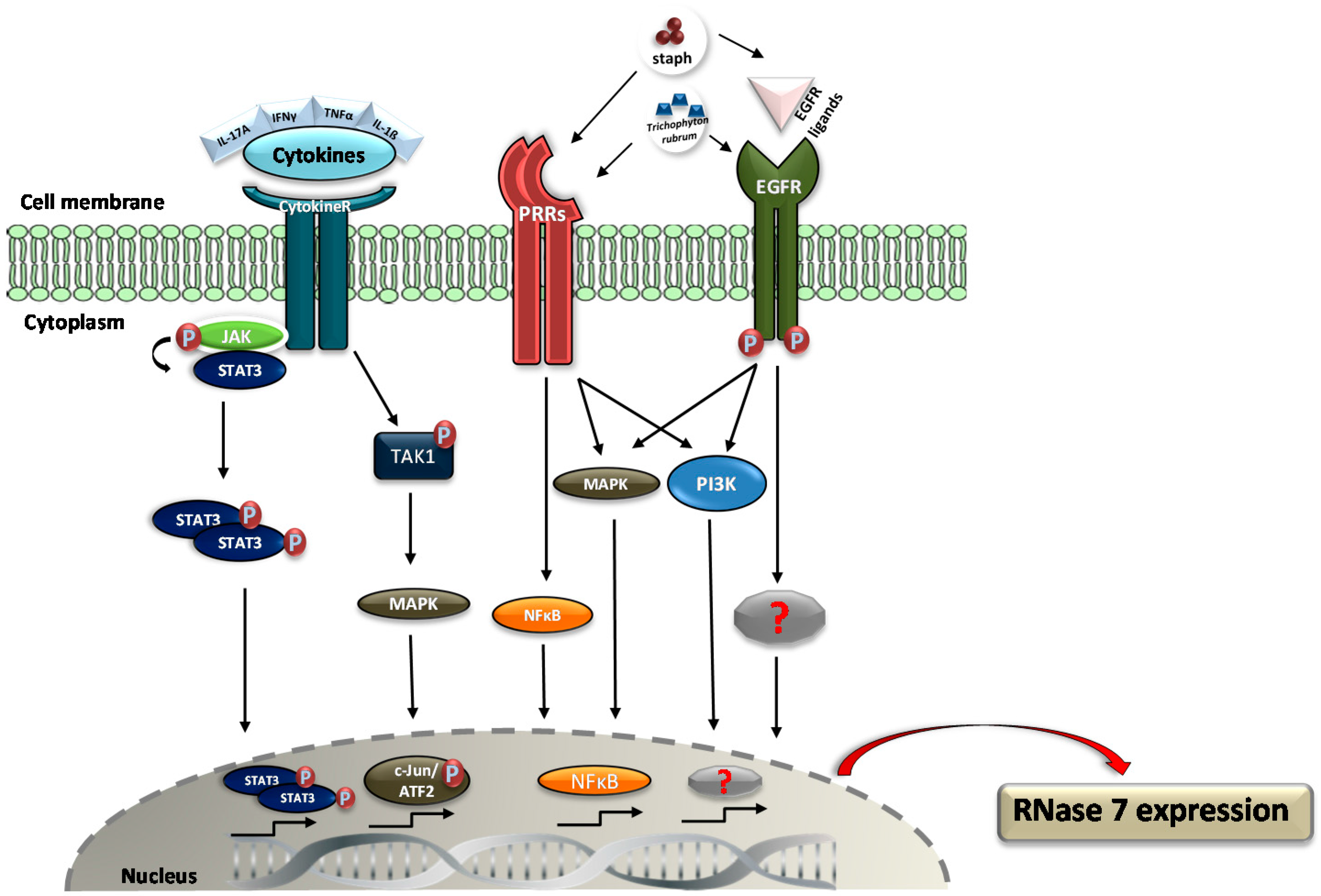
3. Regulation of RNase 7 Expression
4. Role of RNase 7 in Skin Diseases
4.1. Atopic Dermatitis
4.2. Psoriasis
4.3. Acne Vulgaris
4.4. Wounds
4.5. Viral Infections
4.6. Skin Cancer
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| AD | Atopic dermatitis |
| AMPs | Antimicrobial peptides and proteins |
| EGFR | Epidermal growth factor |
| ECP | Eosinophil cationic protein |
| EDN | Eosinophil-derived neurotoxin |
| IL | Interleukin |
| IFN | Interferon |
| hBD | Human beta-defensin |
| TLR | Toll-like receptor |
| SCV | Small-colony variant |
References
- Harder, J.; Schröder, J.-M. RNase 7, a novel innate immune defense antimicrobial protein of healthy human skin. J. Biol. Chem. 2002, 277, 46779–46784. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Dyer, K.D.; Rosenberg, H.F. Human RNase 7: A new cationic ribonuclease of the RNase A superfamily. Nucleic Acids Res. 2003, 31, 602–607. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, H.F. RNase A ribonucleases and host defense: An evolving story. J. Leukoc. Biol. 2008, 83, 1079–1087. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.; Beintema, J.J.; Zhang, J. The ribonuclease A superfamily of mammals and birds: Identifying new members and tracing evolutionary histories. Genomics 2005, 85, 208–220. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, H.F.; Domachowske, J.B. Eosinophils, eosinophil ribonucleases, and their role in host defense against respiratory virus pathogens. J. Leukoc. Biol. 2001, 70, 691–698. [Google Scholar] [PubMed]
- Rosenberg, H.F. Eosinophil-derived neurotoxin (EDN/RNase 2) and the mouse eosinophil-associated RNases (mEars): Expanding roles in promoting host defense. Int. J. Mol. Sci. 2015, 16, 15442–15455. [Google Scholar] [CrossRef] [PubMed]
- Simanski, M.; Köten, B.; Schröder, J.M.; Gläser, R.; Harder, J. Antimicrobial RNases in cutaneous defense. J. Innate Immun. 2012, 4, 241–247. [Google Scholar] [CrossRef] [PubMed]
- Rudolph, B.; Podschun, R.; Sahly, H.; Schubert, S.; Schroder, J.M.; Harder, J. Identification of RNase 8 as a novel human antimicrobial protein. Antimicrob. Agents Chemother. 2006, 50, 3194–3196. [Google Scholar] [CrossRef] [PubMed]
- Hooper, L.V.; Stappenbeck, T.S.; Hong, C.V.; Gordon, J.I. Angiogenins: A new class of microbicidal proteins involved in innate immunity. Nat. Immunol. 2003, 4, 269–273. [Google Scholar] [CrossRef] [PubMed]
- Malik, A.; Batra, J.K. Antimicrobial activity of human eosinophil granule proteins: Involvement in host defence against pathogens. Crit. Rev. Microbiol. 2012, 38, 168–181. [Google Scholar] [CrossRef] [PubMed]
- Spencer, J.D.; Schwaderer, A.L.; Dirosario, J.D.; Mchugh, K.M.; Mcgillivary, G.; Justice, S.S.; Carpenter, A.R.; Baker, P.B.; Harder, J.; Hains, D.S. Ribonuclease 7 is a potent antimicrobial peptide within the human urinary tract. Kidney Int. 2011, 80, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Schwaderer, A.L.; Kline, J.; Spencer, J.D.; Kline, D.; Hains, D.S. Contribution of structural domains to the activity of ribonuclease 7 against uropathogenic bacteria. Antimicrob. Agents Chemother. 2013, 57, 766–774. [Google Scholar] [CrossRef] [PubMed]
- Köten, B.; Simanski, M.; Gläser, R.; Podschun, R.; Schröder, J.M.; Harder, J. RNase 7 contributes to the cutaneous defense against Enterococcus faecium. PLoS ONE 2009, 4, e6424. [Google Scholar] [CrossRef] [PubMed]
- Simanski, M.; Dressel, S.; Gläser, R.; Harder, J. RNase 7 Protects Healthy Skin from Staphylococcus aureus Colonization. J. Investig. Dermatol. 2010, 130, 2836–2838. [Google Scholar] [CrossRef] [PubMed]
- Fritz, P.; Beck-Jendroschek, V.; Brasch, J. Inhibition of dermatophytes by the antimicrobial peptides human β-defensin-2, ribonuclease 7 and psoriasin. Med. Mycol. 2012, 50, 579–584. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.C.; Lin, Y.M.; Chang, T.W.; Wu, S.J.; Lee, Y.S.; Chang, M.D.T.; Chen, C.; Wu, S.H.; Liao, Y. The flexible and clustered lysine residues of human ribonuclease 7 are critical for membrane permeability and antimicrobial activity. J. Biol. Chem. 2007, 282, 4626–4633. [Google Scholar] [CrossRef] [PubMed]
- Zanger, P.; Holzer, J.; Schleucher, R.; Steffen, H.; Schittek, B.; Gabrysch, S. Constitutive expression of the antimicrobial peptide RNase 7 is associated with Staphylococcus aureus infection of the skin. J. Infect. Dis. 2009, 200, 1907–1915. [Google Scholar] [CrossRef] [PubMed]
- Pulido, D.; Torrent, M.; Andreu, D.; Nogues, M.V.; Boix, E. Two human host defense ribonucleases against mycobacteria, the eosinophil cationic protein (RNase 3) and RNase 7. Antimicrob. Agents Chemother. 2013, 57, 3797–3805. [Google Scholar] [CrossRef] [PubMed]
- Torrent, M.; Pulido, D.; Valle, J.; Nogués, M.V.; Andreu, D.; Boix, E. Ribonucleases as a host-defence family: Evidence of evolutionarily conserved antimicrobial activity at the N-terminus. Biochem. J. 2013, 456, 99–108. [Google Scholar] [CrossRef] [PubMed]
- Torrent, M.; Sánchez, D.; Buzón, V.; Nogués, M.V.; Cladera, J.; Boix, E. Comparison of the membrane interaction mechanism of two antimicrobial RNases: RNase 3/ECP and RNase 7. Biochim. Biophys. Acta 2009, 1788, 1116–1125. [Google Scholar] [CrossRef] [PubMed]
- Torrent, M.; Badia, M.; Moussaoui, M.; Sanchez, D.; Nogués, M.V.; Boix, E. Comparison of human RNase 3 and RNase 7 bactericidal action at the Gram-negative and Gram-positive bacterial cell wall. FEBS J. 2010, 277, 1713–1725. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.M.; Wu, S.J.; Chang, T.W.; Wang, C.F.; Suen, C.S.; Hwang, M.J.; Chang, M.D.T.; Chen, Y.T.; Liao, Y. Outer membrane protein I of Pseudomonas aeruginosa is a target of cationic antimicrobial peptide/protein. J. Biol. Chem. 2010, 285, 8985–8994. [Google Scholar] [CrossRef] [PubMed]
- Iyer, S.; Holloway, D.E.; Kumar, K.; Shapiro, R.; Acharya, K.R. Molecular recognition of human eosinophil-derived neurotoxin (RNase 2) by placental ribonuclease inhibitor. J. Mol. Biol. 2005, 347, 637–655. [Google Scholar] [CrossRef] [PubMed]
- Johnson, R.J.; McCoy, J.G.; Bingman, C.A.; Phillips, G.N., Jr.; Raines, R.T. Inhibition of human pancreatic ribonuclease by the human ribonuclease inhibitor protein. J. Mol. Biol. 2007, 368, 434–449. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, R.; Vallee, B.L. Human placental ribonuclease inhibitor abolishes both angiogenic and ribonucleolytic activities of angiogenin. Proc. Natl. Acad. Sci. USA 1987, 84, 2238–2241. [Google Scholar] [CrossRef] [PubMed]
- Abtin, A.; Eckhart, L.; Mildner, M.; Ghannadan, M.; Harder, J.; Schröder, J.-M.; Tschachler, E. Degradation by stratum corneum proteases prevents endogenous RNase inhibitor from blocking antimicrobial activities of RNase 5 and RNase 7. J. Investig. Dermatol. 2009, 129, 2193–2201. [Google Scholar] [CrossRef] [PubMed]
- Spencer, J.D.; Schwaderer, A.L.; Eichler, T.; Wang, H.; Kline, J.; Justice, S.S.; Cohen, D.M.; Hains, D.S. An endogenous ribonuclease inhibitor regulates the antimicrobial activity of ribonuclease 7 in the human urinary tract. Kidney Int. 2014, 85, 1179–1191. [Google Scholar] [CrossRef] [PubMed]
- Simanski, M.; Gläser, R.; Köten, B.; Meyer-Hoffert, U.; Wanner, S.; Weidenmaier, C.; Peschel, A.; Harder, J. Staphylococcus aureus subverts cutaneous defense by d-alanylation of teichoic acids. Exp. Dermatol. 2013, 22, 294–296. [Google Scholar] [CrossRef] [PubMed]
- Proctor, R.A.; von Eiff, C.; Kahl, B.C.; Becker, K.; McNamara, P.; Herrmann, M.; Peters, G. Small colony variants: A pathogenic form of bacteria that facilitates persistent and recurrent infections. Nat. Rev. Microbiol. 2006, 4, 295–305. [Google Scholar] [CrossRef] [PubMed]
- Vaudaux, P.; Kelley, W.L.; Lew, D.P. Staphylococcus aureus small colony variants: Difficult to diagnose and difficult to treat. Clin. Infect. Dis. 2006, 43, 968–970. [Google Scholar] [CrossRef] [PubMed]
- Gläser, R.; Becker, K.; von Eiff, C.; Meyer-Hoffert, U.; Harder, J. Decreased susceptibility of Staphylococcus aureus small-colony variants toward human antimicrobial peptides. J. Investig. Dermatol. 2014, 134, 2347–2350. [Google Scholar] [CrossRef] [PubMed]
- Sadowska, B.; Bonar, A.; Eiff, C.; Proctor, R.A.; Chmiela, M.; Rudnicka, W.; Rozalska, B. Characteristics of Staphylococcus aureus, isolated from airways of cystic fibrosis patients, and their small colony variants. FEMS Immunol. Med. Microbiol. 2002, 32, 191–197. [Google Scholar] [CrossRef] [PubMed]
- Spencer, J.D.; Schwaderer, A.L.; Wang, H.; Bartz, J.; Kline, J.; Eichler, T.; DeSouza, K.R.; Sims-Lucas, S.; Baker, P.; Hains, D.S. Ribonuclease 7, an antimicrobial peptide upregulated during infection, contributes to microbial defense of the human urinary tract. Kidney Int. 2013, 83, 615–625. [Google Scholar] [CrossRef] [PubMed]
- Schuster, C.; Gläser, R.; Fiala, C.; Eppel, W.; Harder, J.; Schröder, J.-M.; Elbe-Bürger, A. Prenatal human skin expresses the antimicrobial peptide RNase 7. Arch. Dermatol. Res. 2013, 305, 545–549. [Google Scholar] [CrossRef] [PubMed]
- Laudien, M.; Dressel, S.; Harder, J.; Gläser, R. Differential expression pattern of antimicrobial peptides in nasal mucosa and secretion. Rhinology 2011, 49, 107–111. [Google Scholar] [PubMed]
- Harder, J.; Dressel, S.; Wittersheim, M.; Cordes, J.; Meyer-Hoffert, U.; Mrowietz, U.; Fölster-Holst, R.; Proksch, E.; Schröder, J.-M.; Schwarz, T.; et al. Enhanced expression and secretion of antimicrobial peptides in atopic dermatitis and after superficial skin injury. J. Investig. Dermatol. 2010, 130, 1355–1364. [Google Scholar] [CrossRef] [PubMed]
- Burgey, C.; Kern, W.V.; Römer, W.; Rieg, S. Differential induction of innate defense antimicrobial peptides in primary nasal epithelial cells upon stimulation with inflammatory cytokines, Th17 cytokines or bacterial conditioned medium from Staphylococcus aureus isolates. Microb. Pathog. 2016, 90, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, I.; Yeung, A.; Abedin, A.; Hopkinson, A.; Dua, H.S. Signalling pathways involved in ribonuclease-7 expression. Cell. Mol. Life Sci. 2011, 68, 1941–1952. [Google Scholar] [CrossRef] [PubMed]
- Simanski, M.; Rademacher, F.; Schröder, L.; Schumacher, H.M.; Gläser, R.; Harder, J. IL-17A and IFN-g Synergistically Induce RNase 7 Expression via STAT3 in Primary Keratinocytes. PLoS ONE 2013, 8, e59531. [Google Scholar] [CrossRef] [PubMed]
- Minegishi, Y.; Saito, M.; Nagasawa, M.; Takada, H.; Hara, T.; Tsuchiya, S.; Agematsu, K.; Yamada, M.; Kawamura, N.; Ariga, T.; et al. Molecular explanation for the contradiction between systemic Th17 defect and localized bacterial infection in hyper-IgE syndrome. J. Exp. Med. 2009, 206, 1291–1301. [Google Scholar] [CrossRef] [PubMed]
- Wanke, I.; Steffen, H.; Christ, C.; Krismer, B.; Götz, F.; Peschel, A.; Schaller, M.; Schittek, B. Skin commensals amplify the innate immune response to pathogens by activation of distinct signaling pathways. J. Investig. Dermatol. 2011, 131, 382–390. [Google Scholar] [CrossRef] [PubMed]
- Firat, Y.H.; Simanski, M.; Rademacher, F.; Schröder, L.; Brasch, J.; Harder, J. Infection of keratinocytes with trichophytum rubrum induces epidermal growth factor-dependent RNase 7 and human β-defensin-3 expression. PLoS ONE 2014, 9, e93941. [Google Scholar] [CrossRef] [PubMed]
- Brasch, J.; Mörig, A.; Neumann, B.; Proksch, E. Expression of antimicrobial peptides and toll-like receptors is increased in tinea and pityriasis versicolor. Mycoses 2014, 57, 147–152. [Google Scholar] [CrossRef] [PubMed]
- Sato, K.; Yang, X.; Yudate, T.; Chung, J.-S.; Wu, J.; Luby-Phelps, K.; Kimberly, R.P.; Underhill, D.; Cruz, P.D.; Ariizumi, K. Dectin-2 is a pattern recognition receptor for fungi that couples with the Fc receptor γ chain to induce innate immune responses. J. Biol. Chem. 2006, 281, 38854–38866. [Google Scholar] [CrossRef] [PubMed]
- Lichtenberger, B.M.; Gerber, P.A.; Holcmann, M.; Buhren, B.A.; Amberg, N.; Smolle, V.; Schrumpf, H.; Boelke, E.; Ansari, P.; Mackenzie, C.; et al. Epidermal EGFR controls cutaneous host defense and prevents inflammation. Sci. Transl. Med. 2013, 5. [Google Scholar] [CrossRef] [PubMed]
- Eilers, R.E.; Gandhi, M.; Patel, J.D.; Mulcahy, M.F.; Agulnik, M.; Hensing, T.; Lacouture, M.E. Dermatologic infections in cancer patients treated with epidermal growth factor receptor inhibitor therapy. J. Natl. Cancer Inst. 2010, 102, 47–53. [Google Scholar] [CrossRef] [PubMed]
- Clausen, M.-L.; Slotved, H.-C.; Krogfelt, K.A.; Andersen, P.S.; Agner, T. In vivo expression of antimicrobial peptides in atopic dermatitis. Exp. Dermatol. 2016, 25, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Kopfnagel, V.; Harder, J.; Werfel, T. Expression of antimicrobial peptides in atopic dermatitis and possible immunoregulatory functions. Curr. Opin. Allergy Clin. Immunol. 2013, 13, 531–536. [Google Scholar] [CrossRef] [PubMed]
- Gambichler, T.; Skrygan, M.; Tomi, N.S.; Othlinghaus, N.; Brockmeyer, N.H.; Altmeyer, P.; Kreuter, A. Differential mRNA expression of antimicrobial peptides and proteins in atopic dermatitis as compared to psoriasis vulgaris and healthy skin. Int. Arch. Allergy Immunol. 2008, 147, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Jensen, J.-M.; Ahrens, K.; Meingassner, J.; Scherer, A.; Bräutigam, M.; Stütz, A.; Schwarz, T.; Fölster-Holst, R.; Harder, J.; Gläser, R.; et al. Differential suppression of epidermal antimicrobial protein expression in atopic dermatitis and in EFAD mice by pimecrolimus compared to corticosteroids. Exp. Dermatol. 2011, 20, 783–788. [Google Scholar] [CrossRef] [PubMed]
- Kisich, K.O.; Carspecken, C.W.; Fiéve, S.; Boguniewicz, M.; Leung, D.Y.M. Defective killing of Staphylococcus aureus in atopic dermatitis is associated with reduced mobilization of human β-defensin-3. J. Allergy Clin. Immunol. 2008, 122, 62–68. [Google Scholar] [CrossRef] [PubMed]
- Harder, J.; Schröder, J.-M. Psoriatic scales: A promising source for the isolation of human skin-derived antimicrobial proteins. J. Leukoc. Biol. 2005, 77, 476–486. [Google Scholar] [CrossRef] [PubMed]
- Christophers, E.; Henseler, T. Contrasting disease patterns in psoriasis and atopic dermatitis. Arch. Dermatol. Res. 1987, 279, S48–S51. [Google Scholar] [CrossRef] [PubMed]
- Lai, Y.; Gallo, R.L. AMPed up immunity: How antimicrobial peptides have multiple roles in immune defense. Trends Immunol. 2009, 30, 131–141. [Google Scholar] [CrossRef] [PubMed]
- Hollox, E.J.; Huffmeier, U.; Zeeuwen, P.L.J.M.; Palla, R.; Lascorz, J.; Rodijk-Olthuis, D.; van de Kerkhof, P.C.M.; Traupe, H.; de Jongh, G.; den Heijer, M.; et al. Psoriasis is associated with increased β-defensin genomic copy number. Nat. Genet. 2008, 40, 23–25. [Google Scholar] [CrossRef] [PubMed]
- Lande, R.; Chamilos, G.; Ganguly, D.; Demaria, O.; Frasca, L.; Durr, S.; Conrad, C.; Schröder, J.; Gilliet, M. Cationic antimicrobial peptides in psoriatic skin cooperate to break innate tolerance to self-DNA. Eur. J. Immunol. 2015, 45, 203–213. [Google Scholar] [CrossRef] [PubMed]
- Bracke, S.; Carretero, M.; Guerrero-Aspizua, S.; Desmet, E.; Illera, N.; Navarro, M.; Lambert, J.; del Rio, M. Targeted silencing of DEFB4 in a bioengineered skin-humanized mouse model for psoriasis: Development of siRNA SECosome-based novel therapies. Exp. Dermatol. 2014, 23, 199–201. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Chen, Q.; Rosenberg, H.F.; Rybak, S.M.; Newton, D.L.; Wang, Z.Y.; Fu, Q.; Tchernev, V.T.; Wang, M.; Schweitzer, B.; et al. Human ribonuclease A superfamily members, eosinophil-derived neurotoxin and pancreatic ribonuclease, induce dendritic cell maturation and activation. J. Immunol. 2004, 173, 6134–6142. [Google Scholar] [CrossRef] [PubMed]
- Harder, J.; Tsuruta, D.; Murakami, M.; Kurokawa, I. What is the role of antimicrobial peptides (AMP) in acne vulgaris? Exp. Dermatol. 2013, 22, 386–391. [Google Scholar] [CrossRef] [PubMed]
- Reithmayer, K.; Meyer, K.C.; Kleditzsch, P.; Tiede, S.; Uppalapati, S.K.; Gläser, R.; Harder, J.; Schröder, J.-M.; Paus, R. Human hair follicle epithelium has an antimicrobial defence system that includes the inducible antimicrobial peptide psoriasin (S100A7) and RNase 7. Br. J. Dermatol. 2009, 161, 78–89. [Google Scholar] [CrossRef] [PubMed]
- Zanger, P.; Nurjadi, D.; Vath, B.; Kremsner, P.G. Persistent nasal carriage of Staphylococcus aureus is associated with deficient induction of human β-defensin 3 after sterile wounding of healthy skin in vivo. Infect. Immun. 2011, 79, 2658–2662. [Google Scholar] [CrossRef] [PubMed]
- Zanger, P.; Holzer, J.; Schleucher, R.; Scherbaum, H.; Schittek, B.; Gabrysch, S. Severity of Staphylococcus aureus infection of the skin is associated with inducibility of human β-defensin 3 but not human β-defensin 2. Infect. Immun. 2010, 78, 3112–3117. [Google Scholar] [CrossRef] [PubMed]
- Dressel, S.; Harder, J.; Cordes, J.; Wittersheim, M.; Meyer-Hoffert, U.; Sunderkötter, C.; Gläser, R. Differential expression of antimicrobial peptides in margins of chronic wounds. Exp. Dermatol. 2010, 19, 628–632. [Google Scholar] [CrossRef] [PubMed]
- Surasombatpattana, P.; Hamel, R.; Patramool, S.; Luplertlop, N.; Thomas, F.; Desprès, P.; Briant, L.; Yssel, H.; Missé, D. Dengue virus replication in infected human keratinocytes leads to activation of antiviral innate immune responses. Infect. Genet. Evol. 2011, 11, 1664–1673. [Google Scholar] [CrossRef] [PubMed]
- Scola, N.; Gambichler, T.; Saklaoui, H.; Bechara, F.G.; Georgas, D.; Stücker, M.; Gläser, R.; Kreuter, A. The expression of antimicrobial peptides is significantly altered in cutaneous squamous cell carcinoma and precursor lesions. Br. J. Dermatol. 2012, 167, 591–597. [Google Scholar] [CrossRef] [PubMed]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rademacher, F.; Simanski, M.; Harder, J. RNase 7 in Cutaneous Defense. Int. J. Mol. Sci. 2016, 17, 560. https://doi.org/10.3390/ijms17040560
Rademacher F, Simanski M, Harder J. RNase 7 in Cutaneous Defense. International Journal of Molecular Sciences. 2016; 17(4):560. https://doi.org/10.3390/ijms17040560
Chicago/Turabian StyleRademacher, Franziska, Maren Simanski, and Jürgen Harder. 2016. "RNase 7 in Cutaneous Defense" International Journal of Molecular Sciences 17, no. 4: 560. https://doi.org/10.3390/ijms17040560
APA StyleRademacher, F., Simanski, M., & Harder, J. (2016). RNase 7 in Cutaneous Defense. International Journal of Molecular Sciences, 17(4), 560. https://doi.org/10.3390/ijms17040560





