A Novel Sucrose-Regulatory MADS-Box Transcription Factor GmNMHC5 Promotes Root Development and Nodulation in Soybean (Glycine max [L.] Merr.)
Abstract
:1. Introduction
2. Results
2.1. Gene Cloning and Phylogenic Analysis of the GmNMHC5 Protein
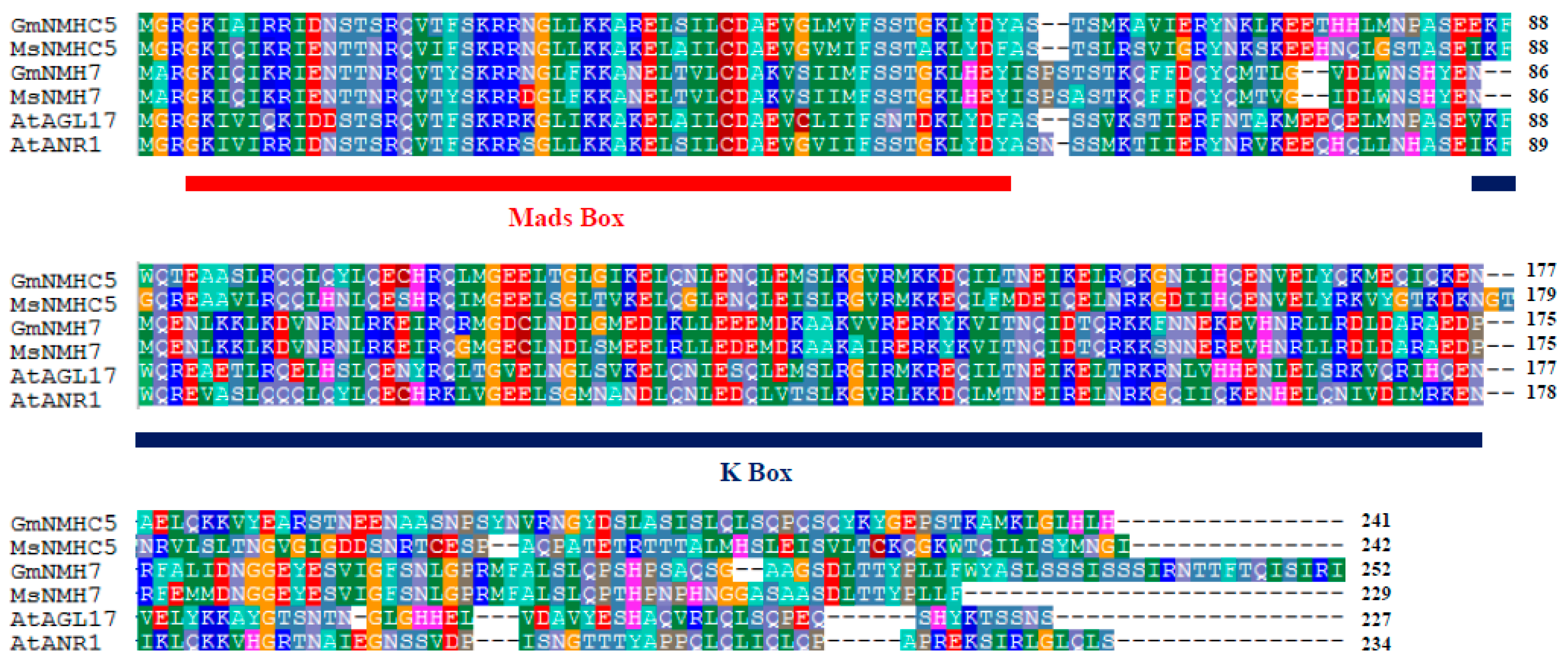
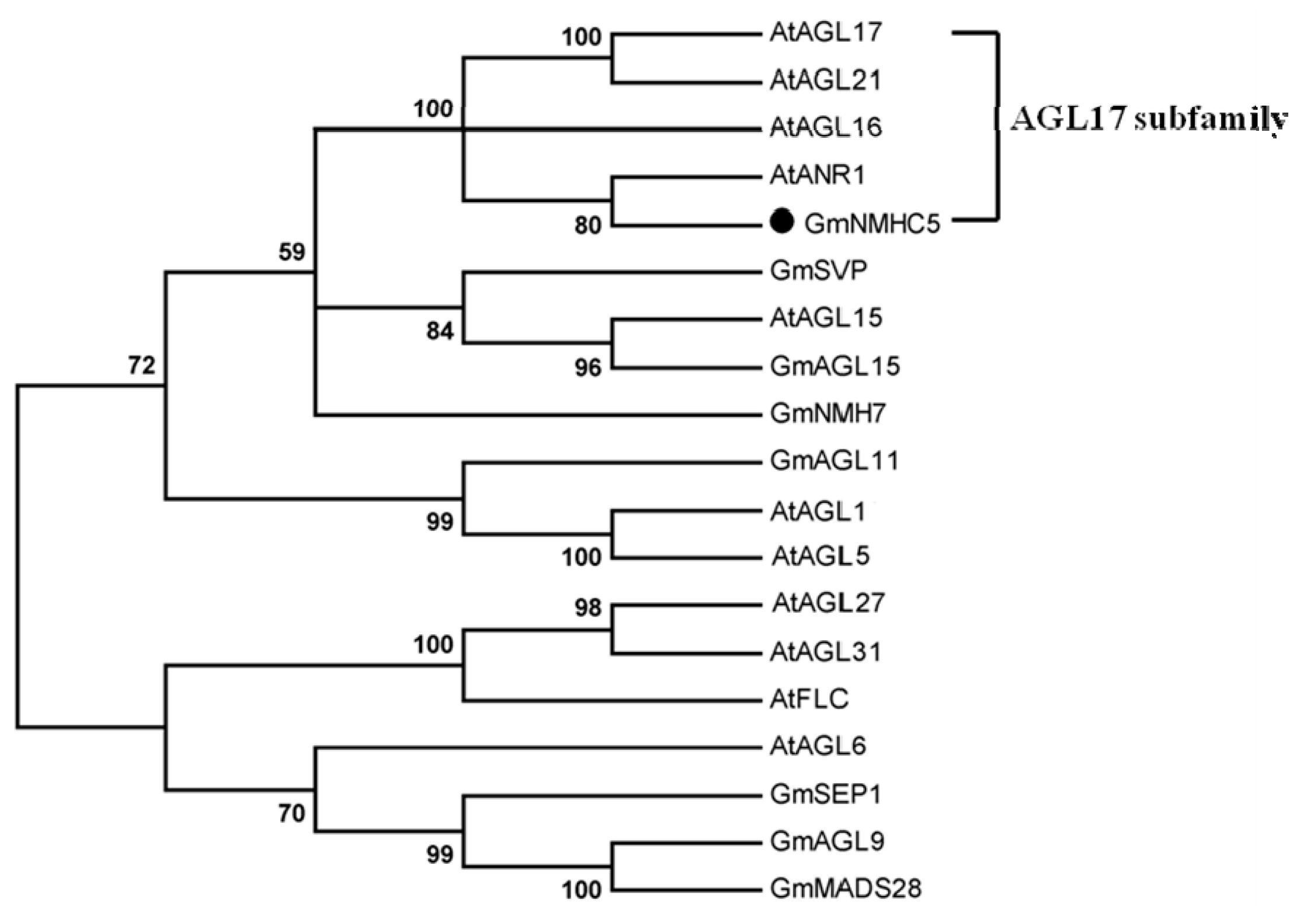
2.2. Subcellular Localization of GmNMHC5

2.3. Expression Analysis of GmNMHC5
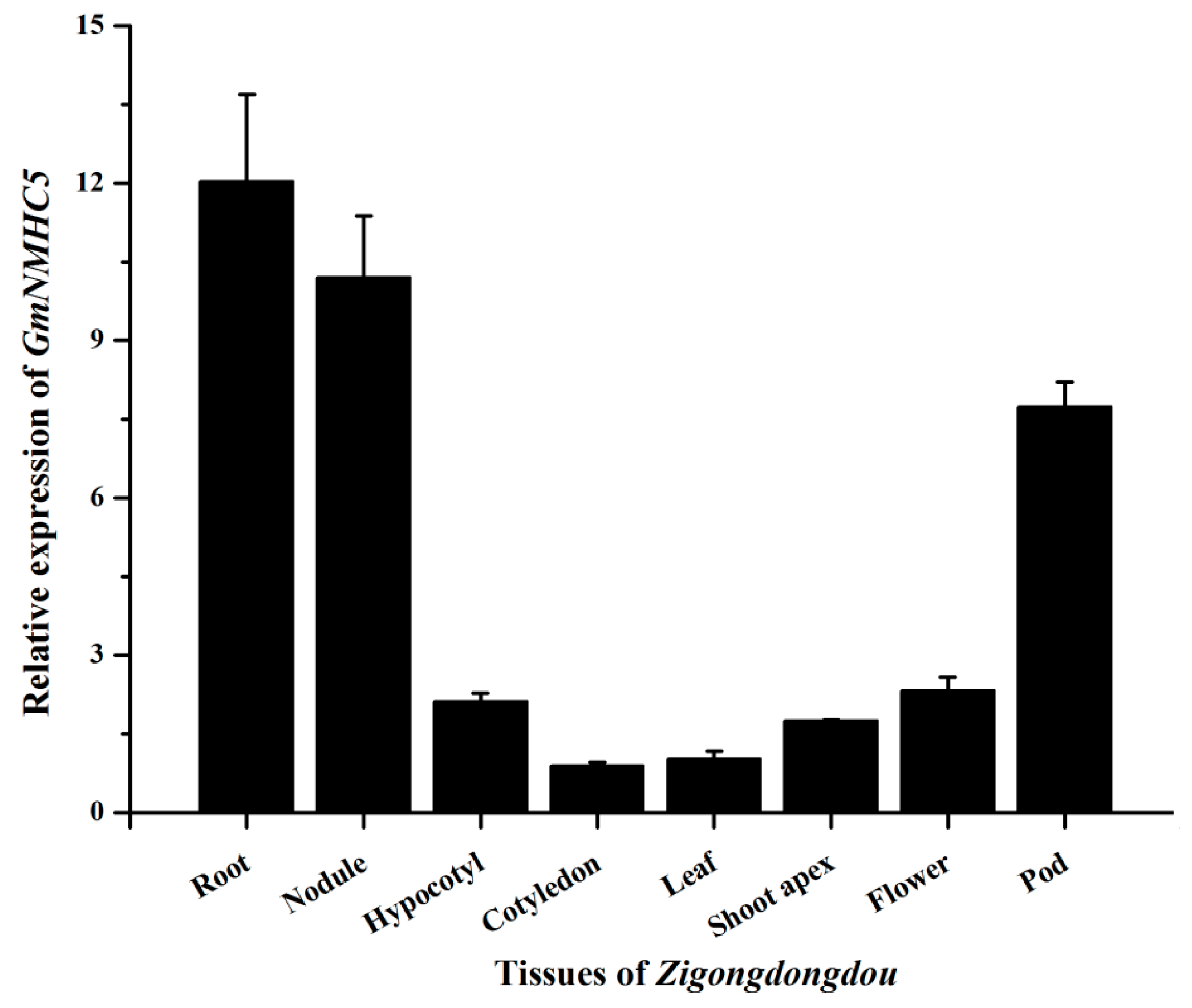
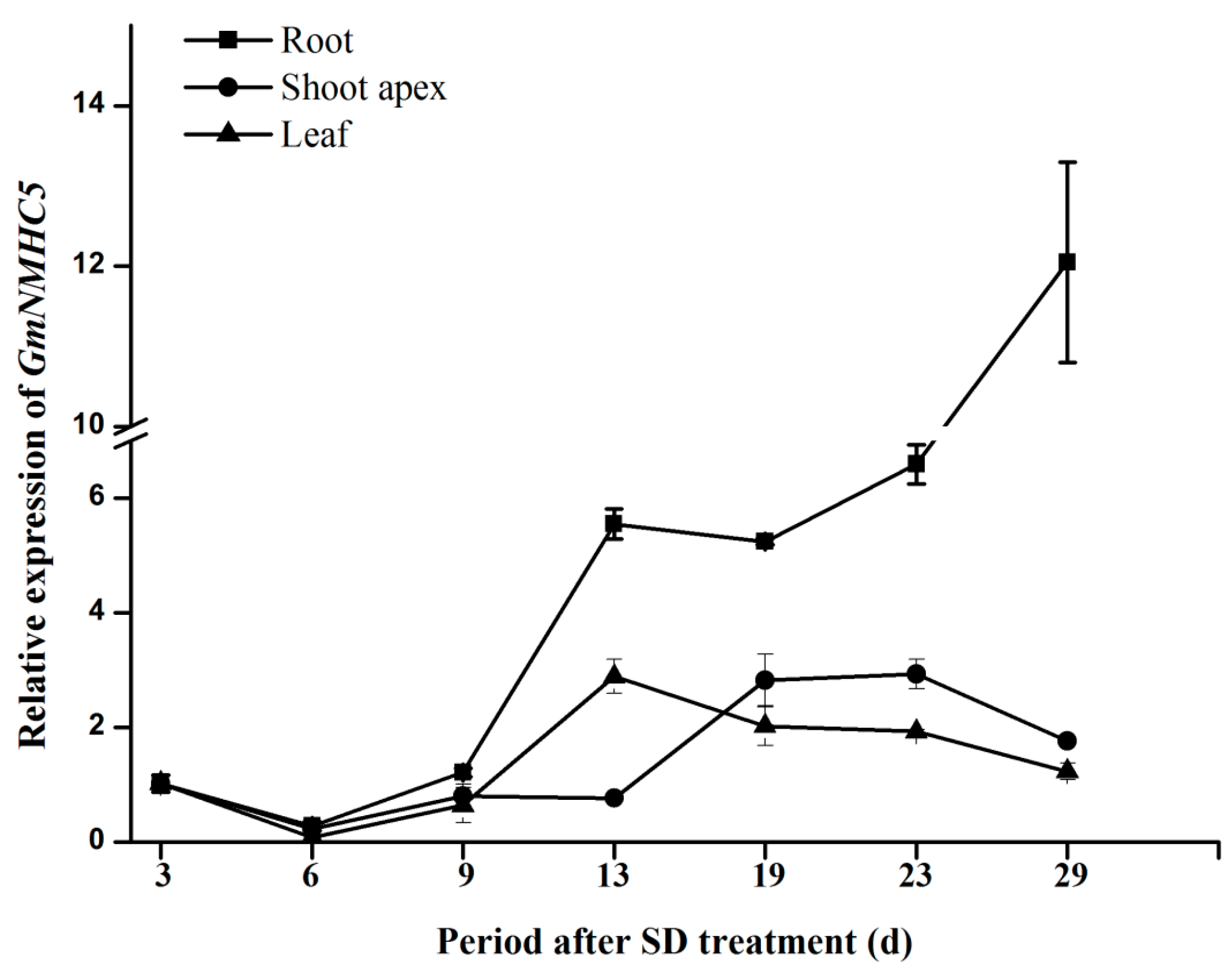
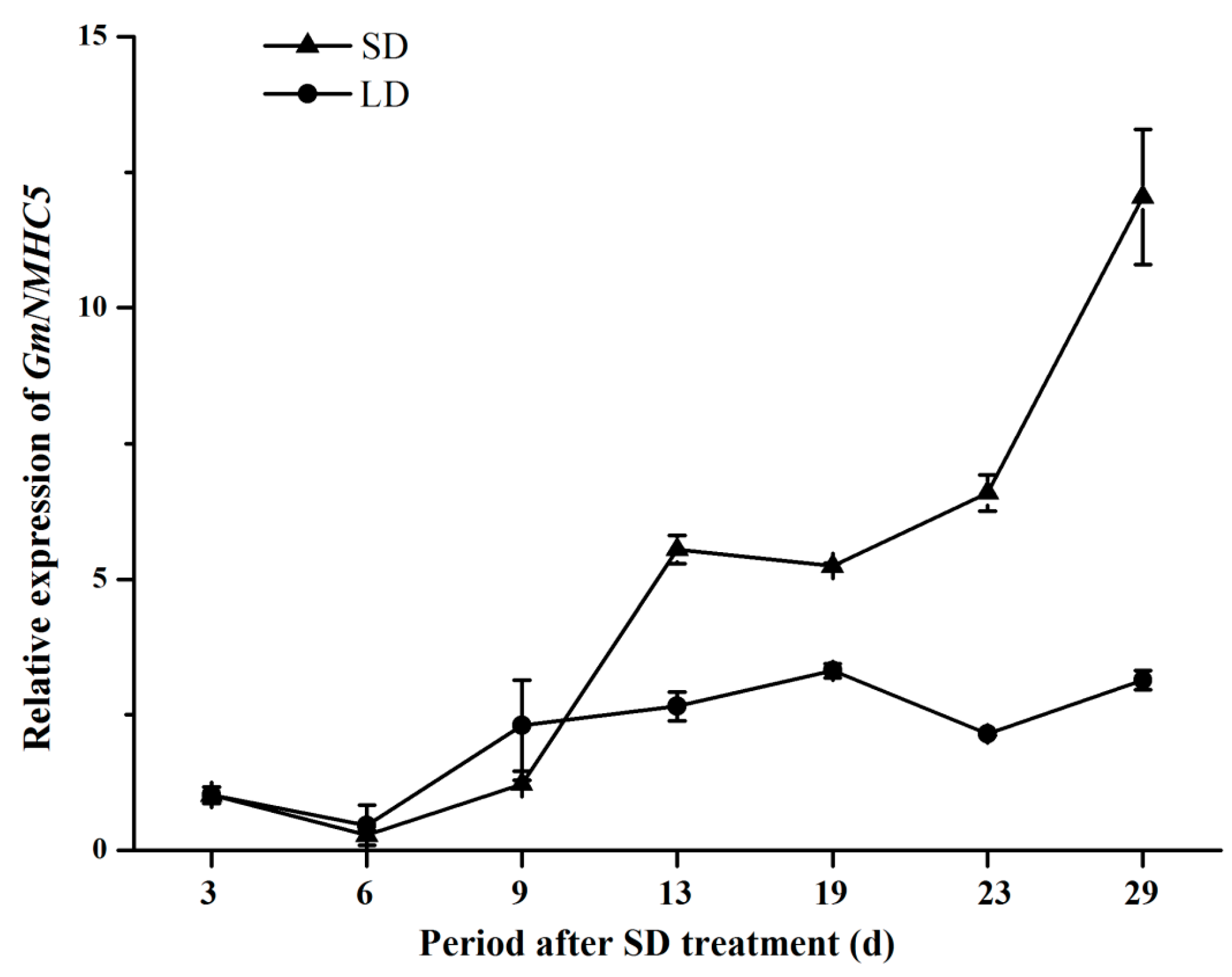
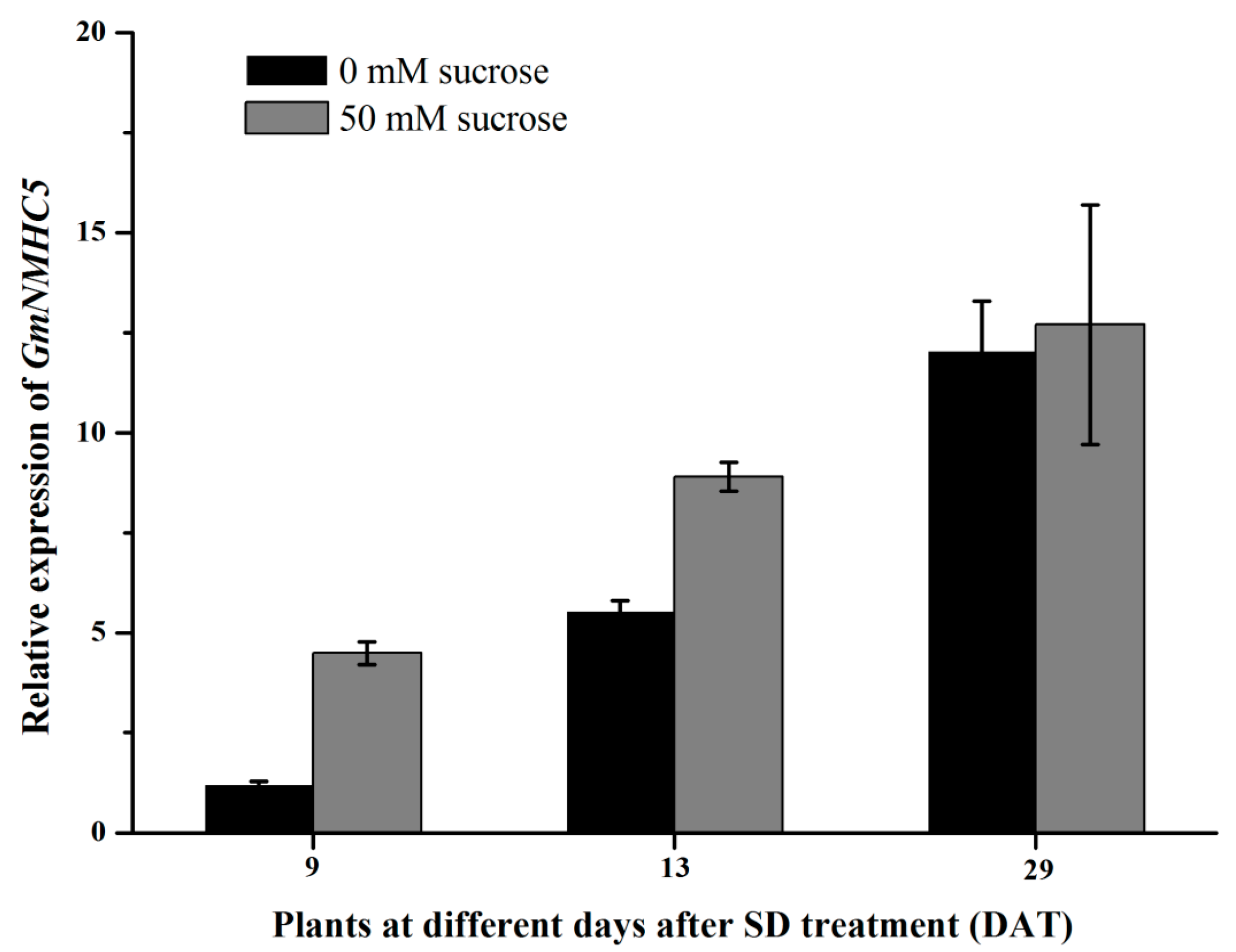
2.4. Sucrose Upregulates GmNMHC5
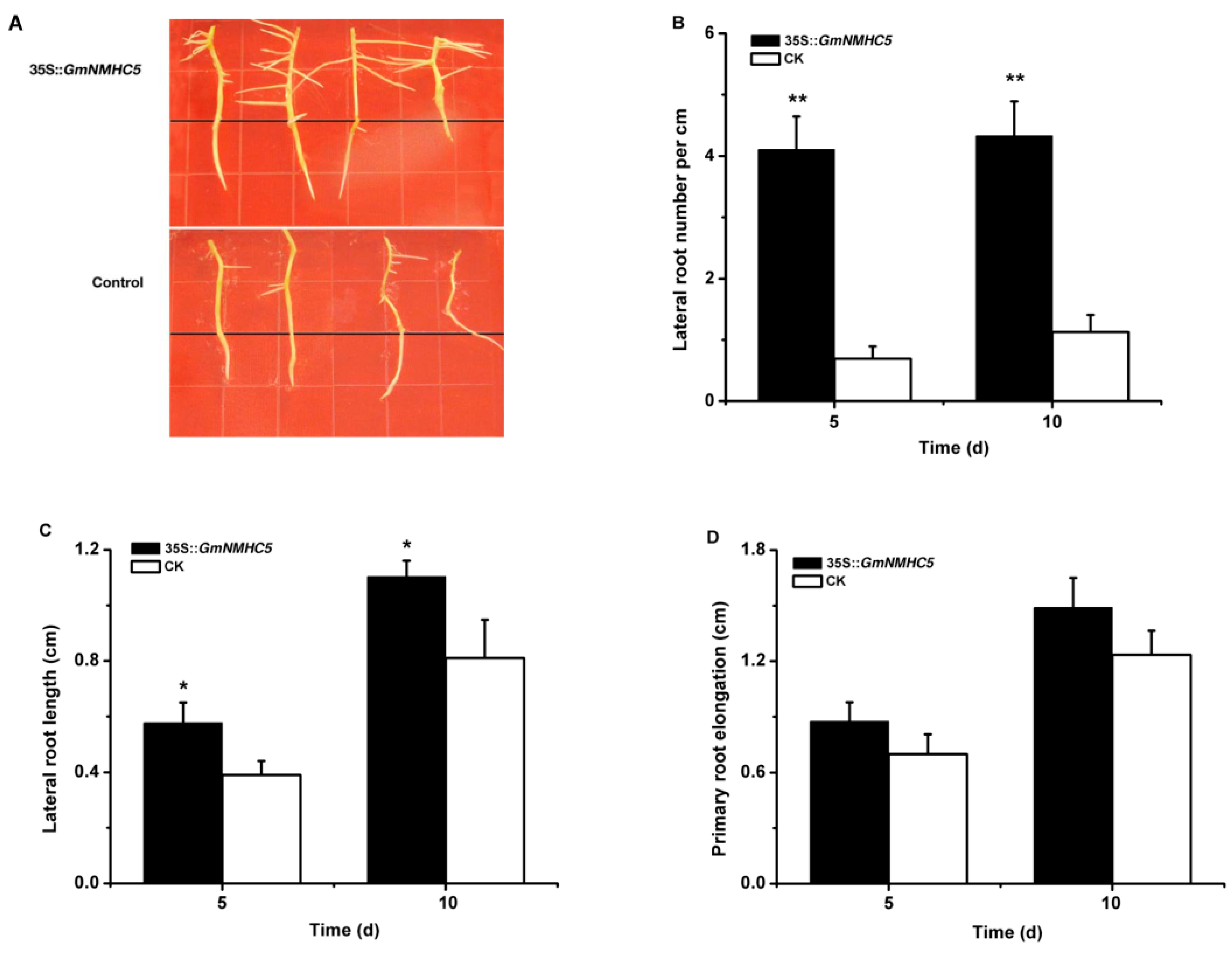
2.5. GmNMHC5 Promotes Lateral Root Development
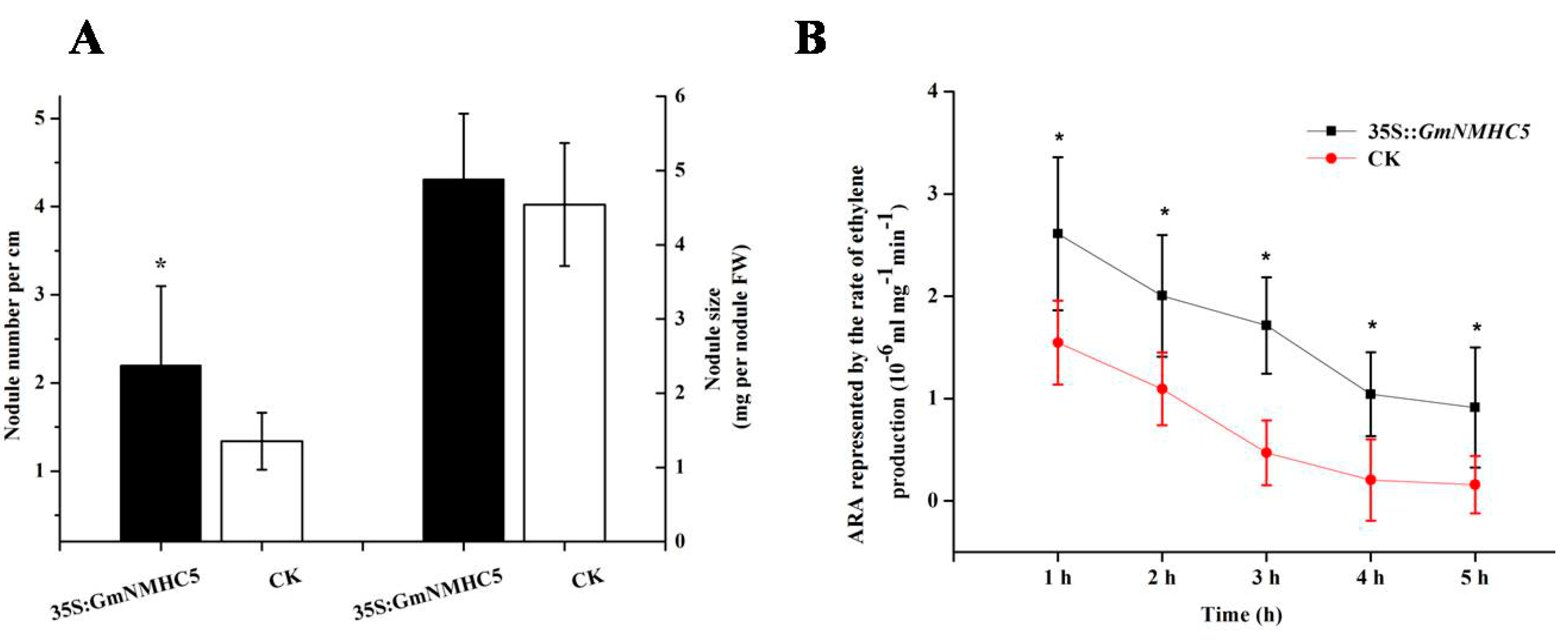
2.6. GmNMHC5 Promotes Nodulation

3. Discussion
3.1. Overexpression of GmNMHC5 Promotes Lateral Roots of Hairy Root Growth and Nodulation in Soybean
3.2. Expression of GmNMHC5 Is Induced in Floral Initiation and the Podding Period
4. Experimental Section
4.1. Plant Materials and Growth Conditions
4.2. Cloning of the GmNMHC5 Gene
4.3. Transient Expression of a GmNMHC5-GFP Fusion Protein in Onion Epidermal Cells
4.4. Bioinformatic Analysis of the GmNMHC5 Promoter
4.5. Construction of 35S::GmNMHC5 for Overexpression of the Gene
4.6. Expression Analysis of the GmNMHC5 Gene
4.7. Sucrose Treatment
4.8. Agrobacterium rhizogenes-Mediated Hairy Root Transformation
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Dong, Z. Soybean Yield Physiology; Agricultural Press: Beijing, China, 2000; pp. 192–195. [Google Scholar]
- Lee, H.; Suh, S.S.; Park, E.; Cho, E.; Ahn, J.H.; Kim, S.G.; Lee, J.S.; Kwon, Y.M.; Lee, I. The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis. Genes Dev. 2000, 14, 2366–2376. [Google Scholar] [CrossRef] [PubMed]
- Yoo, S.K.; Lee, J.S.; Ahn, J.H. Overexpression of AGAMOUS-LIKE 28 (AGL28) promotes flowering by upregulating expression of floral promoters within the autonomous pathway. Biochem. Biophys. Res. Commun. 2006, 348, 929–936. [Google Scholar] [CrossRef] [PubMed]
- Seo, E.; Lee, H.; Jeon, J.; Park, H.; Kim, J.; Noh, Y.S.; Lee, I. Crosstalk between cold response and flowering in Arabidopsis is mediated through the flowering-time gene SOC1 and its upstream negative regulator FLC. Plant Cell Online 2009, 21, 3185–3197. [Google Scholar] [CrossRef] [PubMed]
- Deng, W.; Ying, H.; Helliwell, C.A.; Taylor, J.M.; Peacock, W.J.; Dennis, E.S. FLOWERING LOCUS C (FLC) regulates development pathways throughout the life cycle of Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 6680–6685. [Google Scholar] [CrossRef] [PubMed]
- Adamczyk, B.J.; Lehti-Shiu, M.D.; Fernandez, D.E. The MADS domain factors AGL15 and AGL18 act redundantly as repressors of the floral transition in Arabidopsis. Plant J. 2007, 50, 1007–1019. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Yoo, S.J.; Park, S.H.; Hwang, I.; Lee, J.S.; Ahn, J.H. Role of SVP in the control of flowering time by ambient temperature in Arabidopsis. Genes Dev. 2007, 21, 397–402. [Google Scholar] [CrossRef] [PubMed]
- Gan, Y.; Filleur, S.; Rahman, A.; Gotensparre, S.; Forde, B.G. Nutritional regulation of ANR1 and other root-expressed MADS-box genes in Arabidopsis thaliana. Planta 2005, 222, 730–742. [Google Scholar] [CrossRef] [PubMed]
- Burgeff, C.; Liljegren, S.J.; Tapia-López, R.; Yanofsky, M.F.; Alvarez-Buylla, E.R. MADS-box gene expression in lateral primordia, meristems and differentiated tissues of Arabidopsis thaliana roots. Planta 2002, 214, 365–372. [Google Scholar] [CrossRef] [PubMed]
- Han, P.; García-Ponce, B.; Fonseca-Salazar, G.; Alvarez-Buylla, E.R.; Yu, H. AGAMOUS-LIKE 17, a novel flowering promoter, acts in a FT-independent photoperiod pathway. Plant J. 2008, 55, 253–265. [Google Scholar] [CrossRef] [PubMed]
- Tapia-López, R.; García-Ponce, B.; Dubrovsky, J.G.; Garay-Arroyo, A.; Pérez-Ruíz, R.V.; Kim, S.H.; Acevedo, F.; Pelaz, S.; Alvarez-Buylla, E.R. An AGAMOUS-related MADS-box gene, XAL1 (AGL12), regulates root meristem cell proliferation and flowering transition in Arabidopsis. Plant Physiol. 2008, 146, 1182–1192. [Google Scholar] [CrossRef] [PubMed]
- Kutter, C.; Schöb, H.; Stadler, M.; Meins, F.; Si-Ammour, A. MicroRNA-mediated regulation of stomatal development in Arabidopsis. Plant Cell 2007, 19, 2417–2429. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.Y.; Zhou, Y.; He, F.; Dong, X.; Liu, L.Y.; Coupland, G.; Turck, F.; de Meaux, J. miR824-regulated AGAMOUS-LIKE16 contributes to flowering time repression in Arabidopsis. Plant Cell Online 2014, 26, 2024–2037. [Google Scholar] [CrossRef] [PubMed]
- Pinyopich, A.; Ditta, G.S.; Savidge, B.; Liljegren, S.J.; Baumann, E.; Wisman, E.; Yanofsky, M.F. Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature 2003, 424, 85–88. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Risueno, M.A.; van Norman, J.M.; Moreno, A.; Zhang, J.; Ahnert, S.E.; Benfey, P.N. Oscillating gene expression determines competence for periodic Arabidopsis root branching. Science 2010, 329, 1306–1311. [Google Scholar] [CrossRef] [PubMed]
- Koo, S.C.; Bracko, O.; Park, M.S.; Schwab, R.; Chun, H.J.; Park, K.M.; Seo, J.S.; Grbic, V.; Balasubramanian, S.; Schmid, M.; et al. Control of lateral organ development and flowering time by the Arabidopsis thaliana MADS-box Gene AGAMOUS-LIKE6. Plant J. 2010, 62, 807–816. [Google Scholar] [CrossRef] [PubMed]
- Yoo, S.K.; Wu, X.; Lee, J.S.; Ahn, J.H. AGAMOUS-LIKE 6 is a floral promoter that negatively regulates the FLC/MAF clade genes and positively regulates FT in Arabidopsis. Plant J. 2011, 65, 62–76. [Google Scholar] [CrossRef] [PubMed]
- Heard, J.; Caspi, M.; Dunn, K. Evolutionary diversity of symbiotically induced nodule MADS box genes: Characterization of nmhC5, a member of a novel subfamily. Mol. Plant Microbe Interact. 1997, 10, 665–676. [Google Scholar] [CrossRef] [PubMed]
- Heard, J.; Dunn, K. Symbiotic induction of a MADS-box gene during development of alfalfa root nodules. Proc. Natl. Acad. Sci. USA 1995, 92, 5273–5277. [Google Scholar] [CrossRef] [PubMed]
- Zucchero, J.C.; Caspi, M.; Dunn, K. ngl9: A third MADS box gene expressed in alfalfa root nodules. Mol. Plant Microbe Interact. 2001, 14, 1463–1467. [Google Scholar] [CrossRef] [PubMed]
- Páez-Valencia, J.; Sánchez-Gómez, C.; Valencia-Mayoral, P.; Contreras-Ramos, A.; Hernández-Lucas, I.; Orozco-Segovia, A.; Gamboa-deBuen, A. Localization of the MADS domain transcriptional factor NMH7 during seed, seedling and nodule development of Medicago sativa. Plant Sci. 2008, 175, 596–603. [Google Scholar] [CrossRef]
- Páez-Valencia, J.; Valencia-Mayoral, P.; Sánchez-Gómez, C.; Contreras-Ramos, A.; Hernández-Lucas, I.; Martínez-Barajas, E.; Gamboa-deBuen, A. Identification of fructose-1, 6-bisphosphate aldolase cytosolic class I as an NMH7 MADS domain associated protein. Biochem. Biophys. Res. Commun. 2008, 376, 700–705. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.X.; Ma, Q.B.; Yam, K.M.; Cheung, M.Y.; Xu, Y.Y.; Han, T.F.; Lam, H.M.; Chong, K. In situ expression of the GmNMH7 gene is photoperiod-dependent in a unique soybean (Glycine max [L.] Merr.) flowering reversion system. Planta 2006, 223, 725–735. [Google Scholar] [CrossRef] [PubMed]
- Becker, A.; Theißen, G. The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Mol. Phylogenet. Evol. 2003, 29, 464–489. [Google Scholar] [CrossRef]
- Riechmann, J.L.; Meyerowitz, E.M. MADS domain proteins in plant development. Biol. Chem. 1997, 378, 1079–1102. [Google Scholar] [PubMed]
- Gan, Y.; Bernreiter, A.; Filleur, S.; Abram, B.; Forde, B.G. Overexpressing the ANR1 MADS-box gene in transgenic plants provides new insights into its role in the nitrate regulation of root development. Plant Cell Physiol. 2012, 53, 1003–1016. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Forde, B.G. An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science 1998, 279, 407–409. [Google Scholar] [CrossRef] [PubMed]
- Li, X.M.; Wu, C.X.; Ma, Q.B.; Zhang, S.; Li, C.L.; Zhang, X.Y.; Han, T.F. Morphology and anatomy of the differentiation of flower buds and the process of flowering reversion in soybean cv. Zigongdongdou. Acta Agron. Sin. 2005, 31, 1437–1442. [Google Scholar]
- Sun, H.; Jia, Z.; Cao, D.; Jiang, B.; Wu, C.; Hou, W.; Liu, Y.; Fei, Z.; Zhao, D.; Han, T. GmFT2a, a soybean homolog of FLOWERING LOCUS T, is involved in flowering transition and maintenance. PLoS ONE 2011, 12, e29238. [Google Scholar] [CrossRef] [PubMed]
- D’haeseleer, K.; de Keyser, A.; Goormachtig, S.; Holsters, M. Transcription factor MtATB2: About nodulation, sucrose and senescence. Plant Cell Physiol. 2010, 51, 1416–1424. [Google Scholar] [CrossRef] [PubMed]
- D’Aloia, M.; Bonhomme, D.; Bouché, F.; Tamseddak, K.; Ormenese, S.; Torti, S.; Coupland, G.; Périlleux, C. Cytokinin promotes flowering of Arabidopsis via transcriptional activation of the FT paralogue TSF. Plant J. 2011, 65, 972–979. [Google Scholar] [CrossRef] [PubMed]
- Eveland, A.L.; Jackson, D.P. Sugars, signalling, and plant development. J. Exp. Bot. 2012, 67, 3367–3377. [Google Scholar] [CrossRef] [PubMed]
- Gibson, S.I. Control of plant development and gene expression by sugar signaling. Curr. Opin. Plant Biol. 2005, 8, 93–102. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.F.; Xiong, A.S.; Peng, R.H.; Liu, J.G.; Gao, F.; Chen, J.M.; Yao, Q.H. OsAREB1, an ABRE-binding protein responding to ABA and glucose, has multiple functions in Arabidopsis. BMB Rep. 2010, 43, 34–39. [Google Scholar] [CrossRef] [PubMed]
- Krizek, B.A. Auxin regulation of Arabidopsis flower development involves members of the AINTEGUMENTA-LIKE/PLETHORA (AIL/PLT) family. J. Exp. Bot. 2011, 62, 3311–3319. [Google Scholar] [CrossRef] [PubMed]
- Oliva, M.; Farcot, E.; Vernoux, T. Plant hormone signaling during development: Insights from computational models. Curr. Opin. Plant Biol. 2013, 16, 19–24. [Google Scholar] [CrossRef] [PubMed]
- Han, T.; Wang, J. Studies on the post-flowering photoperiodic responses in soybean. Acta Bot. Sin. 1994, 37, 863–869. [Google Scholar]
- Sudha, P.M.; Low, S.; Kwang, J.; Gong, Z. Multiple tissue transformation in adult zebrafish by gene gun bombardment and muscular injection of naked DNA. Mar. Biotechnol. 2001, 3, 119–125. [Google Scholar] [CrossRef] [PubMed]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Cao, D.; Hou, W.S.; Liu, W.; Yao, W.W.; Wu, C.X.; Liu, X.B.; Han, T.F. Overexpression of TaNHX2 enhances salt tolerance of ‘composite’ and whole transgenic soybean plants. Plant Cell Tissue Organ Cult. 2011, 107, 541–552. [Google Scholar] [CrossRef]
- Fåhraeus, G. The infection of clover root hairs by nodule bacteria studied by a simple glass slide technique. J. Gen. Microbiol. 1957, 16, 374–381. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, W.; Han, X.; Zhan, G.; Zhao, Z.; Feng, Y.; Wu, C. A Novel Sucrose-Regulatory MADS-Box Transcription Factor GmNMHC5 Promotes Root Development and Nodulation in Soybean (Glycine max [L.] Merr.). Int. J. Mol. Sci. 2015, 16, 20657-20673. https://doi.org/10.3390/ijms160920657
Liu W, Han X, Zhan G, Zhao Z, Feng Y, Wu C. A Novel Sucrose-Regulatory MADS-Box Transcription Factor GmNMHC5 Promotes Root Development and Nodulation in Soybean (Glycine max [L.] Merr.). International Journal of Molecular Sciences. 2015; 16(9):20657-20673. https://doi.org/10.3390/ijms160920657
Chicago/Turabian StyleLiu, Wei, Xiangdong Han, Ge Zhan, Zhenfang Zhao, Yongjun Feng, and Cunxiang Wu. 2015. "A Novel Sucrose-Regulatory MADS-Box Transcription Factor GmNMHC5 Promotes Root Development and Nodulation in Soybean (Glycine max [L.] Merr.)" International Journal of Molecular Sciences 16, no. 9: 20657-20673. https://doi.org/10.3390/ijms160920657
APA StyleLiu, W., Han, X., Zhan, G., Zhao, Z., Feng, Y., & Wu, C. (2015). A Novel Sucrose-Regulatory MADS-Box Transcription Factor GmNMHC5 Promotes Root Development and Nodulation in Soybean (Glycine max [L.] Merr.). International Journal of Molecular Sciences, 16(9), 20657-20673. https://doi.org/10.3390/ijms160920657




