Role of Glucokinase in the Subcellular Localization of Glucokinase Regulatory Protein
Abstract
:1. Introduction
2. Results and Discussion
2.1. Sub-Cellular Localization of Glucokinase Regulatory Protein (GCKR) in Hepatic Cell Lines

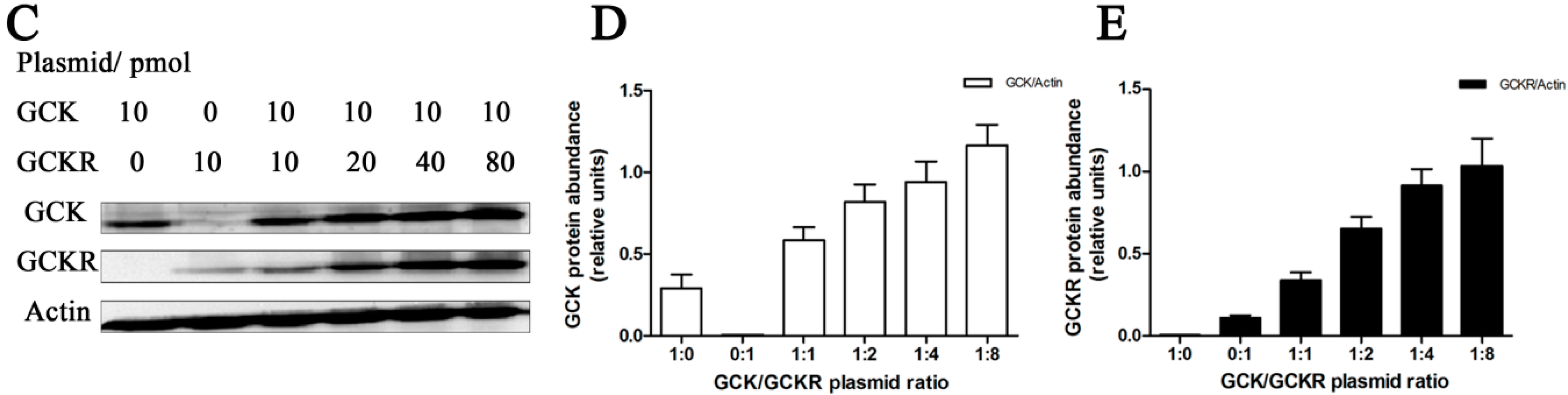
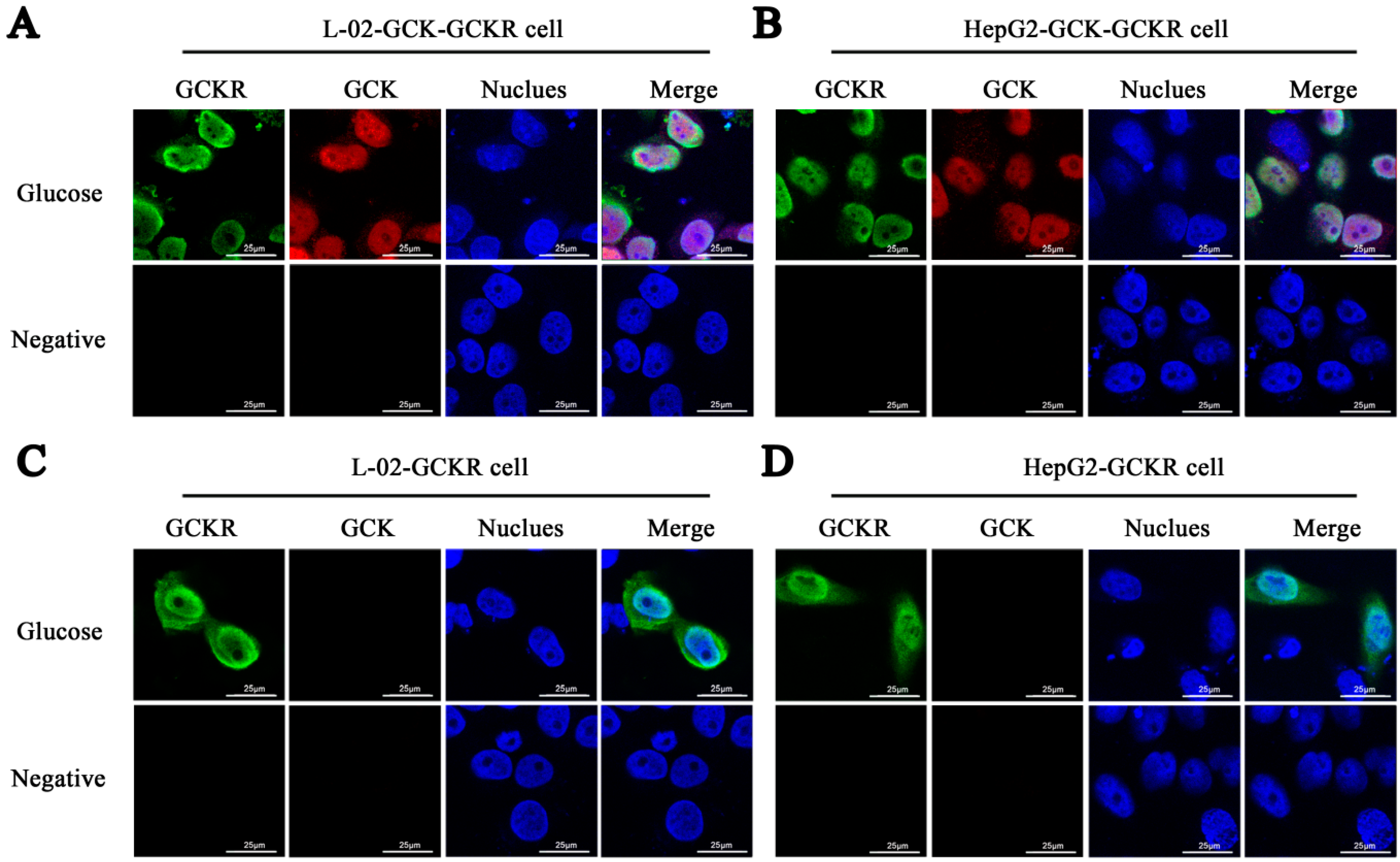
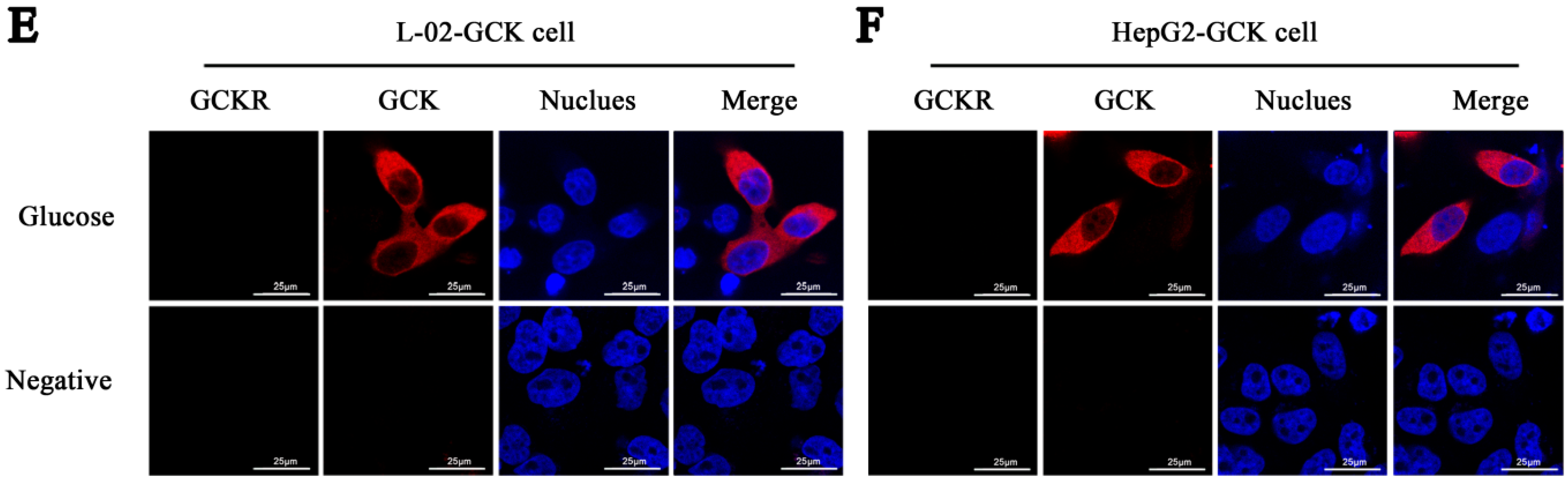
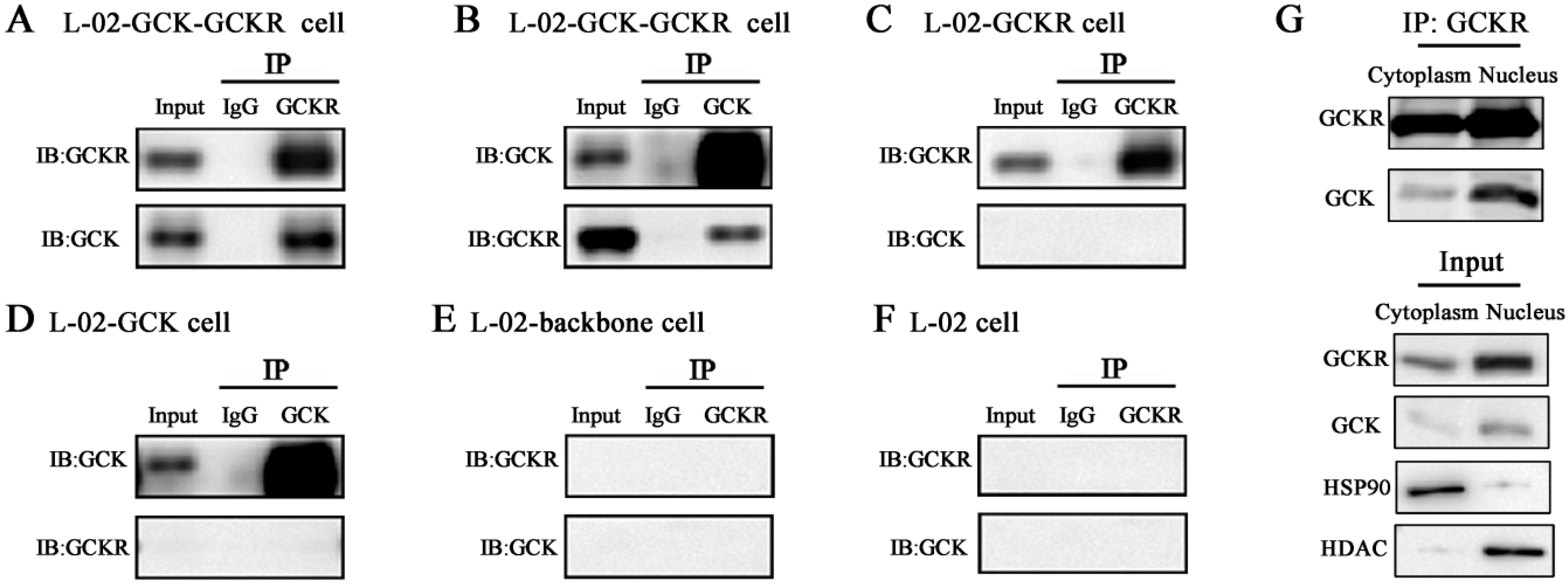
2.2. Direct Interaction of GCKR and Glucokinase (GCK) in the Nuclei of Co-Transfected Hepatic Cell Lines
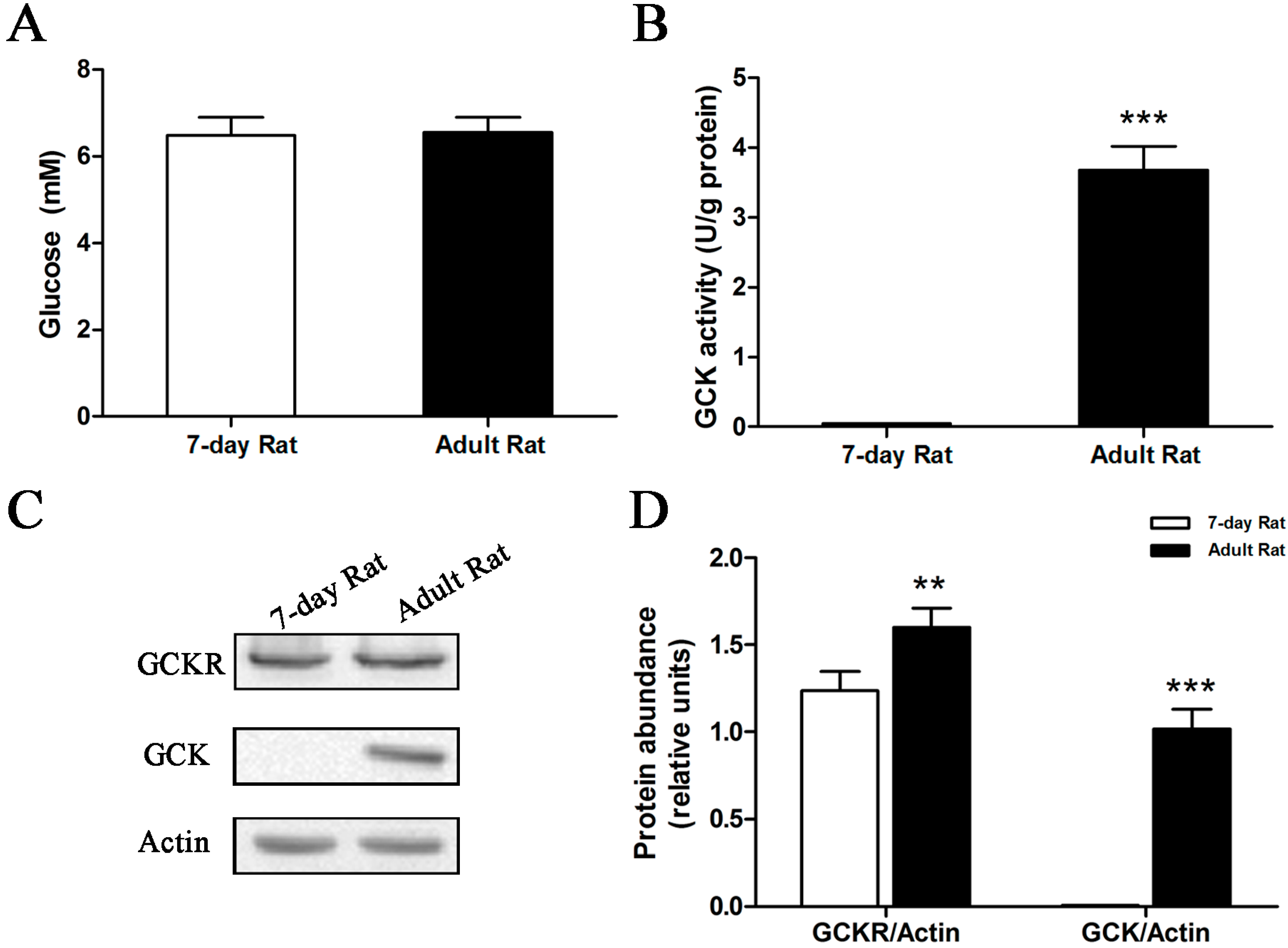
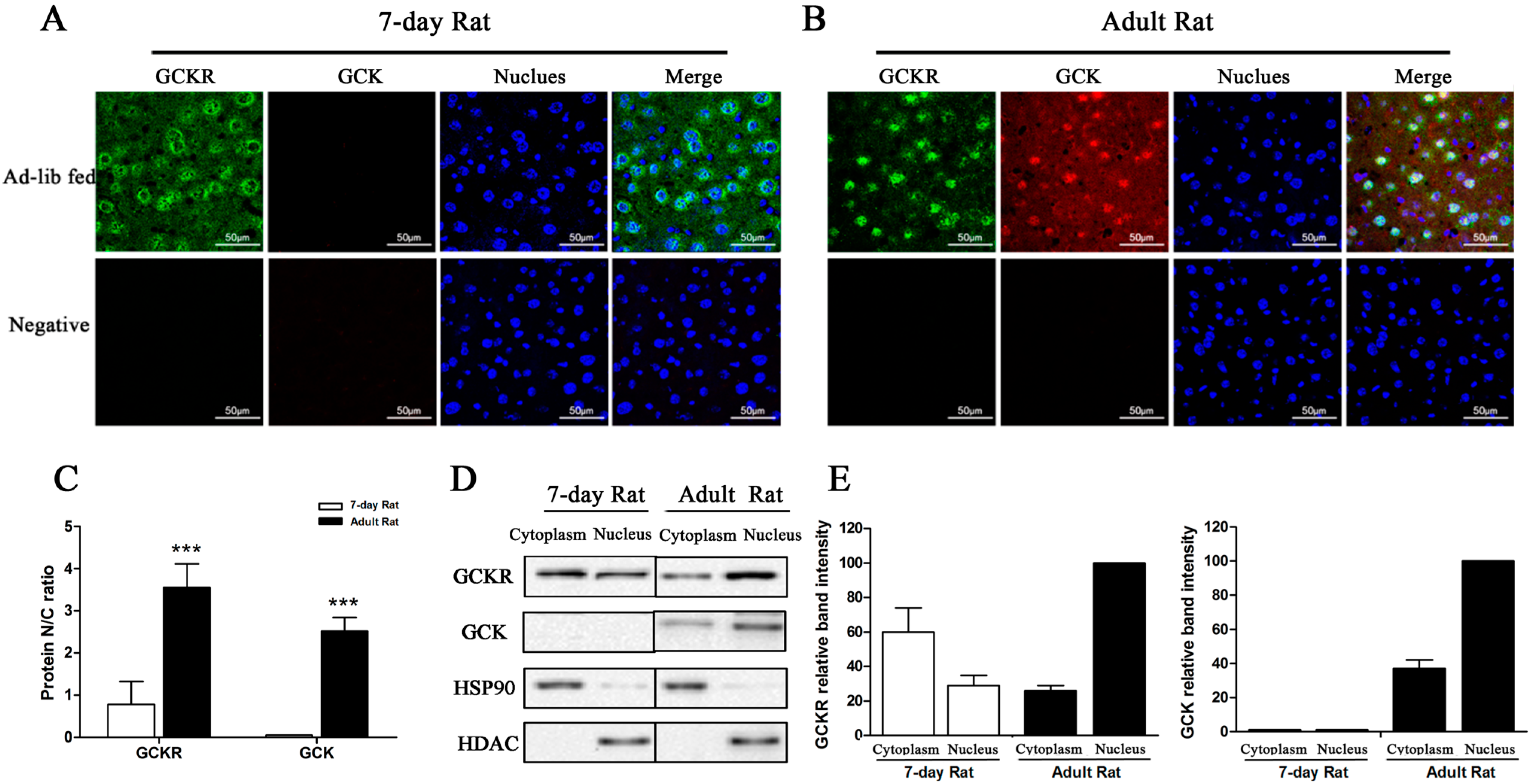
2.3. Sub-Cellular Localization of GCKR in Seven-Day Old Adult Rats
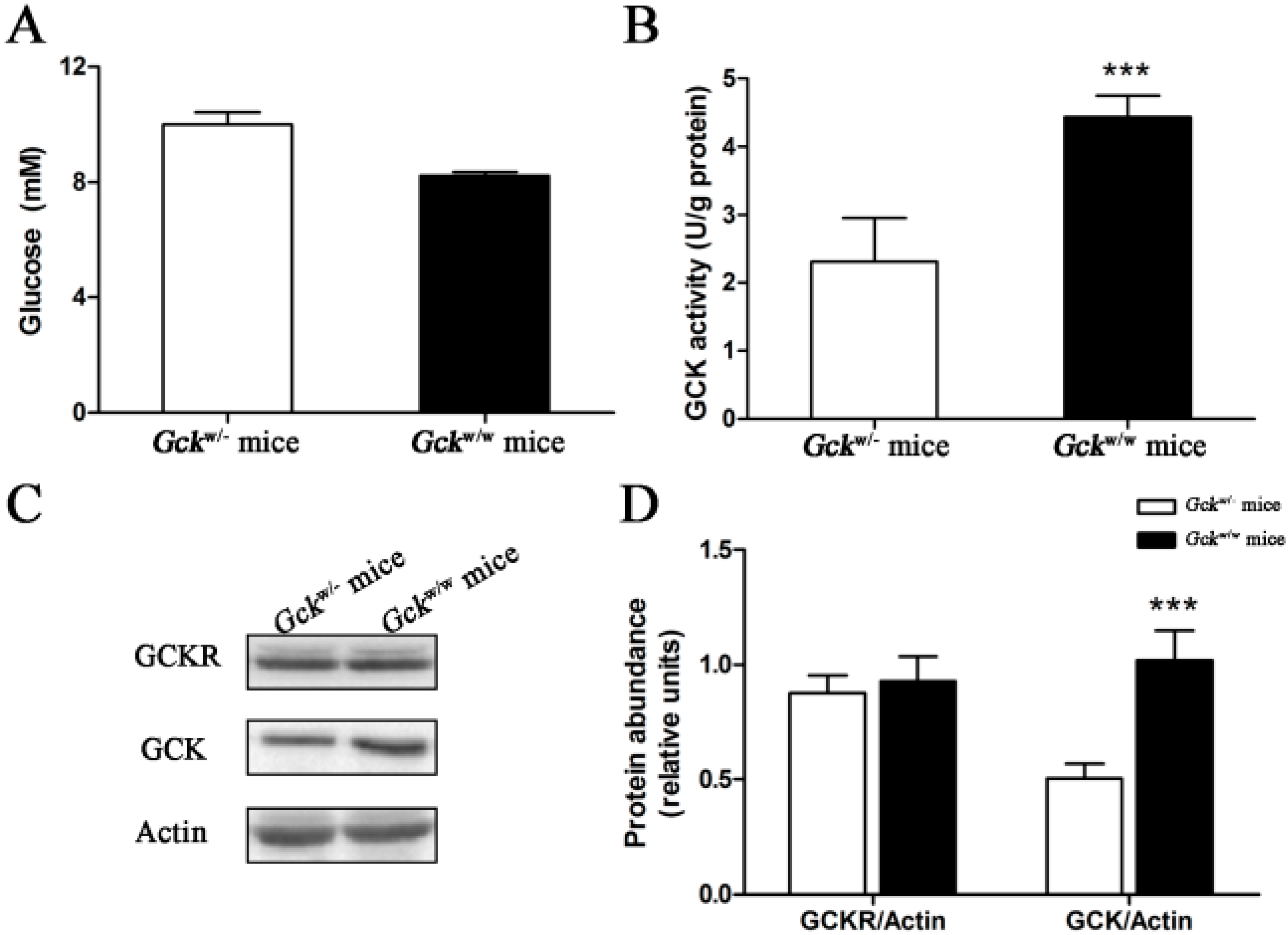
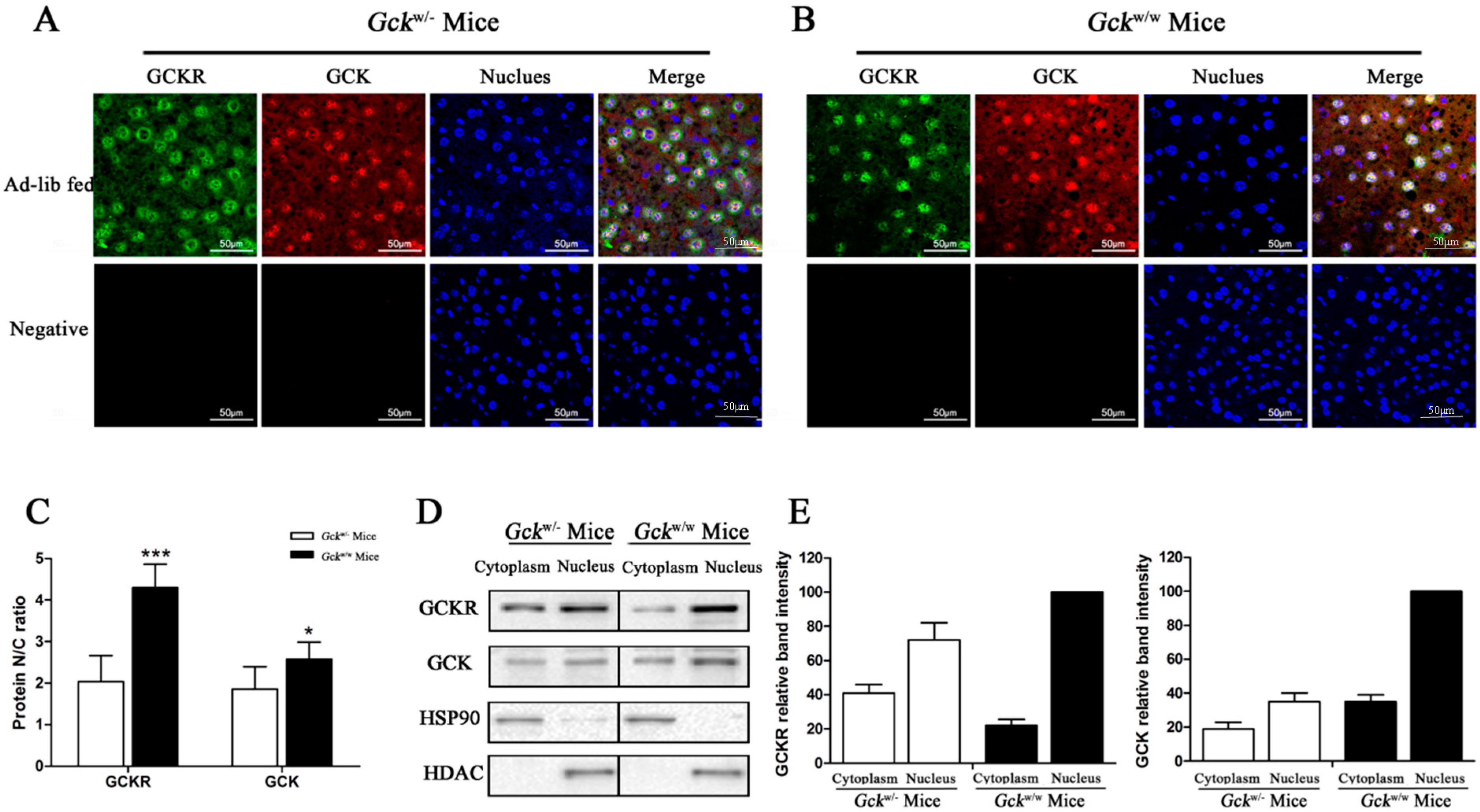
2.4. Sub-Cellular Localization of GCKR in Liver-Specific Gck Gene Knockout (Gckw/−) Mice
3. Discussion
4. Experimental Section
4.1. Cell Culture, Plasmids and Transfection
4.2. Animals
4.3. Blood Glucose and Glucokinase Activity
4.4. Immunofluorescence and Confocal Microscopy
4.5. Preparation of Total, Cytoplasmic, and Nuclear Protein Extracts
4.6. Co-Immunoprecipitation
4.7. Western Blotting
4.8. Isolation of Total RNA, Quantitative RT-PCR and RT-PCR
| Gene | Primer |
|---|---|
| Human Glucokinase (hGck) | F-GAATGACACGGTGGCCACGATG |
| R-CACTCGGTATTGACGCACATGCG | |
| Mouse Glucokinase (mGck) | F-GAATCTTCTGTTCCACGGAG |
| R-AGTGCTCAGGATGTTAAGGA | |
| Human Glucokinase regulatory protein (hGckr) | F-GGTGGAAGTGCCACCAAGATTCTGC |
| R-GTCTGCCAGCCAACCAGGTACACG | |
| Mouse Glucokinase regulatory protein (mGckr) | F-AGGCATTTCCGTGGGACTCTC |
| R-ACCGGATTGAAGCCAACCAG | |
| Human Actin | F-TTCGAGGCTTTCAACACACCAG |
| R-GGCATGAGGCAGGGCATAAC | |
| Mouse Actin | F-CATCCGTAAAGACCTCTATGCCAAC |
| R-ATGGAGCCACCGATCCACA |
| Gene | Primer |
|---|---|
| Glucokinase (Gck) | F-ATGTCACAAGGAGCCAGGCCCAGA |
| R-ACTTCTGAGCCTTCTGGGGTGGA | |
| Glucokinase vector | F-GCGTGTACGGTGGGAGGTCTATAT |
| R-ACTTCTGAGCCTTCTGGGGTGGA | |
| Glucokinase regulatory protein (Gckr) | F-GCAAGTGGGAGTTGTCTGGGTAC |
| R-CAATCCCGTGCAAGGCACTATCT | |
| Glucokinase regulatory protein vector | F-AGGCGTGTACGGTGGGAGGTCTAT |
| R-GGCTCCTTCAGCACTTCCTGAACTT |
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Vandercammen, A.; van Schaftingen, E. Species and tissue distribution of the regulatory protein of glucokinase. Biochem. J. 1993, 294, 551–556. [Google Scholar]
- Van Schaftingen, E. A protein from rat liver confers to glucokinase the property of being antagonistically regulated by fructose 6-phosphate and fructose 1-phosphate. Eur. J. Biochem. 1989, 179, 179–184. [Google Scholar]
- Vandercammen, A.; van Schaftingen, E. The mechanism by which rat liver glucokinase is inhibited by the regulatory protein. Eur. J. Biochem. 1990, 191, 483–489. [Google Scholar] [CrossRef]
- Farrelly, D.; Brown, K.S.; Tieman, A.; Ren, J.; Lira, S.A.; Hagan, D.; Gregg, R.; Mookhtiar, K.A.; Hariharan, N. Mice mutant for glucokinase regulatory protein exhibit decreased liver glucokinase: A sequestration mechanism in metabolic regulation. Proc. Natl. Acad. Sci. USA 1999, 96, 14511–14516. [Google Scholar] [CrossRef]
- Grimsby, J.; Coffey, J.W.; Dvorozniak, M.T.; Magram, J.; Li, G.; Matschinsky, F.M.; Shiota, C.; Kaur, S.; Magnuson, M.A.; Grippo, J.F. Characterization of glucokinase regulatory protein-deficient mice. J. Boil. Chem. 2000, 275, 7826–7831. [Google Scholar] [CrossRef]
- Slosberg, E.D.; Desai, U.J.; Fanelli, B.; St Denny, I.; Connelly, S.; Kaleko, M.; Boettcher, B.R.; Caplan, S.L. Treatment of type 2 diabetes by adenoviral-mediated overexpression of the glucokinase regulatory protein. Diabetes 2001, 50, 1813–1820. [Google Scholar] [CrossRef]
- Bosco, D.; Meda, P.; Iynedjian, P.B. Glucokinase and glucokinase regulatory protein: Mutual dependence for nuclear localization. Biochem. J. 2000, 348, 215–222. [Google Scholar] [CrossRef] [PubMed]
- Shiota, C.; Coffey, J.; Grimsby, J.; Grippo, J.F.; Magnuson, M.A. Nuclear import of hepatic glucokinase depends upon glucokinase regulatory protein, whereas export is due to a nuclear export signal sequence in glucokinase. J. Biol. Chem. 1999, 274, 37125–37130. [Google Scholar] [CrossRef] [PubMed]
- Vandercammen, A.; Detheux, M.; van Schaftingen, E. Binding of sorbitol 6-phosphate and of fructose 1-phosphate to the regulatory protein of liver glucokinase. Biochem. J. 1992, 286, 253–256. [Google Scholar] [PubMed]
- Walker, D.G.; Holland, G. The development of hepatic glucokinase in the neonatal rat. Biochem. J. 1965, 97, 845–854. [Google Scholar] [PubMed]
- Guo, T.; Mao, Y.; Li, H.; Wang, X.; Xu, W.; Song, R.; Jia, J.; Lei, Z.; Irwin, D.M.; Niu, G.; et al. Characterization of the gene expression profile of heterozygous liver-specific glucokinase knockout mice at a young age. Biomed. Pharmacother. 2012, 66, 587–596. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.L.; Tan, X.H.; Xiao, M.F.; Li, H.; Mao, Y.Q.; Yang, X.; Tan, H.R. Establishment of liver specific glucokinase gene knockout mice: A new animal model for screening anti-diabetic drugs. Acta Pharmacol. Sin. 2004, 25, 1659–1665. [Google Scholar] [PubMed]
- Wang, R.; Gao, H.; Xu, W.; Li, H.; Mao, Y.; Wang, Y.; Guo, T.; Wang, X.; Song, R.; Li, Z.; et al. Differential expression of genes and changes in glucose metabolism in the liver of liver-specific glucokinase gene knockout mice. Gene 2013, 516, 248–254. [Google Scholar] [CrossRef]
- Jetton, T.L.; Shiota, M.; Knobel, S.M.; Piston, D.W.; Cherrington, A.D.; Magnuson, M.A. Substrate-induced nuclear export and peripheral compartmentalization of hepatic glucokinase correlates with glycogen deposition. Int. J. Exp. Diabetes Res. 2001, 2, 173–186. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Novell, J.M.; Castel, S.; Bellido, D.; Ferrer, J.C.; Vilaro, S.; Guinovart, J.J. Intracellular distribution of hepatic glucokinase and glucokinase regulatory protein during the fasted to refed transition in rats. FEBS Lett. 1999, 459, 211–214. [Google Scholar] [CrossRef] [PubMed]
- Kaminski, M.T.; Schultz, J.; Waterstradt, R.; Tiedge, M.; Lenzen, S.; Baltrusch, S. Glucose-induced dissociation of glucokinase from its regulatory protein in the nucleus of hepatocytes prior to nuclear export. Biochim. Biophys. Acta 2014, 1843, 554–564. [Google Scholar] [CrossRef] [PubMed]
- Rees, M.G.; Ng, D.; Ruppert, S.; Turner, C.; Beer, N.L.; Swift, A.J.; Morken, M.A.; Below, J.E.; Blech, I.; Mullikin, J.C.; et al. Correlation of rare coding variants in the gene encoding human glucokinase regulatory protein with phenotypic, cellular, and kinetic outcomes. J. Clin. Investig. 2012, 122, 205–217. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Guo, T.; Zhao, S.; Li, Z.; Mao, Y.; Li, H.; Wang, X.; Wang, R.; Xu, W.; Song, R.; et al. Expression of the human glucokinase gene: Important roles of the 5' flanking and intron 1 sequences. PLoS ONE 2012, 7, e45824. [Google Scholar] [CrossRef] [PubMed]
- Knowles, B.B.; Howe, C.C.; Aden, D.P. Human hepatocellular carcinoma cell lines secrete the major plasma proteins and hepatitis B surface antigen. Science 1980, 209, 497–499. [Google Scholar] [CrossRef] [PubMed]
- Beer, N.L.; Tribble, N.D.; McCulloch, L.J.; Roos, C.; Johnson, P.R.; Orho-Melander, M.; Gloyn, A.L. The P446L variant in GCKR associated with fasting plasma glucose and triglyceride levels exerts its effect through increased glucokinase activity in liver. Hum. Mol. Genet. 2009, 18, 4081–4088. [Google Scholar] [CrossRef] [PubMed]
- De la Iglesia, N.; Veiga-da-Cunha, M.; van Schaftingen, E.; Guinovart, J.J.; Ferrer, J.C. Glucokinase regulatory protein is essential for the proper subcellular localisation of liver glucokinase. FEBS Lett. 1999, 456, 332–338. [Google Scholar] [CrossRef] [PubMed]
- Lefrancois-Martinez, A.M.; Diaz-Guerra, M.J.; Vallet, V.; Kahn, A.; Antoine, B. Glucose-dependent regulation of the l-pyruvate kinase gene in a hepatoma cell line is independent of insulin and cyclic AMP. FASEB J. 1994, 8, 89–96. [Google Scholar]
- Ishihara, H.; Asano, T.; Tsukuda, K.; Katagiri, H.; Inukai, K.; Anai, M.; Kikuchi, M.; Yazaki, Y.; Miyazaki, J.; Oka, Y. Overexpression of hexokinase I but not GLUT1 glucose transporter alters concentration dependence of glucose-stimulated insulin secretion in pancreatic β-cell line MIN6. J. Biol. Chem. 1994, 269, 3081–3087. [Google Scholar] [PubMed]
- Ferber, S.; BeltrandelRio, H.; Johnson, J.H.; Noel, R.J.; Cassidy, L.E.; Clark, S.; Becker, T.C.; Hughes, S.D.; Newgard, C.B. GLUT-2 gene transfer into insulinoma cells confers both low and high affinity glucose-stimulated insulin release. Relationship to glucokinase activity. J. Biol. Chem. 1994, 269, 11523–11529. [Google Scholar] [PubMed]
- Rees, M.G.; Wincovitch, S.; Schultz, J.; Waterstradt, R.; Beer, N.L.; Baltrusch, S.; Collins, F.S.; Gloyn, A.L. Cellular characterisation of the Gckr P446L variant associated with type 2 diabetes risk. Diabetologia 2012, 55, 114–122. [Google Scholar] [CrossRef] [PubMed]
- Toyoda, Y.; Ito, Y.; Yoshie, S.; Miwa, I. Shuttling of glucokinase between the nucleus and the cytoplasm in primary cultures of rat hepatocytes: Possible involvement in the regulation of the glucose metabolism. Arch. Histol. Cytol. 1997, 60, 307–316. [Google Scholar] [CrossRef] [PubMed]
- Marfori, M.; Mynott, A.; Ellis, J.J.; Mehdi, A.M.; Saunders, N.F.; Curmi, P.M.; Forwood, J.K.; Boden, M.; Kobe, B. Molecular basis for specificity of nuclear import and prediction of nuclear localization. Biochim. Biophys. Acta 2011, 1813, 1562–1577. [Google Scholar] [CrossRef] [PubMed]
- La Cour, T.; Kiemer, L.; Molgaard, A.; Gupta, R.; Skriver, K.; Brunak, S. Analysis and prediction of leucine-rich nuclear export signals. Protein Eng. Des. Sel. 2004, 17, 527–536. [Google Scholar]
- Dingwall, C.; Robbins, J.; Dilworth, S.M.; Roberts, B.; Richardson, W.D. The nucleoplasmin nuclear location sequence is larger and more complex than that of SV-40 large T antigen. J. Cell Biol. 1988, 107, 841–849. [Google Scholar] [CrossRef] [PubMed]
- Brown, K.S.; Kalinowski, S.S.; Megill, J.R.; Durham, S.K.; Mookhtiar, K.A. Glucokinase regulatory protein may interact with glucokinase in the hepatocyte nucleus. Diabetes 1997, 46, 179–186. [Google Scholar] [CrossRef] [PubMed]
- Mukhtar, M.; Stubbs, M.; Agius, L. Evidence for glucose and sorbitol-induced nuclear export of glucokinase regulatory protein in hepatocytes. FEBS Lett. 1999, 462, 453–458. [Google Scholar] [CrossRef] [PubMed]
- Toyoda, Y.; Miwa, I.; Satake, S.; Anai, M.; Oka, Y. Nuclear location of the regulatory protein of glucokinase in rat liver and translocation of the regulator to the cytoplasm in response to high glucose. Biochem. Biophys. Res. Commun. 1995, 215, 467–473. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Li, H.; Wang, R.; Lei, Z.; Mao, Y.; Wang, X.; Zhang, Y.; Guo, T.; Song, R.; Zhang, X.; et al. Differential expression of genes associated with the progression of renal disease in the kidneys of liver-specific glucokinase gene knockout mice. Int. J. Mol. Sci. 2013, 14, 6467–6486. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, Y.; Torres, T.P.; Donahue, E.P.; Shiota, M. Glucose toxicity is responsible for the development of impaired regulation of endogenous glucose production and hepatic glucokinase in Zucker diabetic fatty rats. Diabetes 2006, 55, 2479–2490. [Google Scholar] [CrossRef] [PubMed]
- Ueta, K.; O’Brien, T.P.; McCoy, G.A.; Kim, K.; Healey, E.C.; Farmer, T.D.; Donahue, E.P.; Condren, A.B.; Printz, R.L.; Shiota, M. Glucotoxicity targets hepatic glucokinase in Zucker diabetic fatty rats, a model of type 2 diabetes associated with obesity. Am. J. Physiol. Endocrinol. Metab. 2014, 306, E1225–E1238. [Google Scholar] [CrossRef] [PubMed]
- Van Etten, R.A.; Jackson, P.K.; Baltimore, D.; Sanders, M.C.; Matsudaira, P.T.; Janmey, P.A. The COOH terminus of the c-Abl tyrosine kinase contains distinct F- and G-actin binding domains with bundling activity. J. Cell Biol. 1994, 124, 325–340. [Google Scholar]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jin, L.; Guo, T.; Li, Z.; Lei, Z.; Li, H.; Mao, Y.; Wang, X.; Zhou, N.; Zhang, Y.; Hu, R.; et al. Role of Glucokinase in the Subcellular Localization of Glucokinase Regulatory Protein. Int. J. Mol. Sci. 2015, 16, 7377-7393. https://doi.org/10.3390/ijms16047377
Jin L, Guo T, Li Z, Lei Z, Li H, Mao Y, Wang X, Zhou N, Zhang Y, Hu R, et al. Role of Glucokinase in the Subcellular Localization of Glucokinase Regulatory Protein. International Journal of Molecular Sciences. 2015; 16(4):7377-7393. https://doi.org/10.3390/ijms16047377
Chicago/Turabian StyleJin, Ling, Tingting Guo, Zhixin Li, Zhen Lei, Hui Li, Yiqing Mao, Xi Wang, Na Zhou, Yizhuang Zhang, Ruobi Hu, and et al. 2015. "Role of Glucokinase in the Subcellular Localization of Glucokinase Regulatory Protein" International Journal of Molecular Sciences 16, no. 4: 7377-7393. https://doi.org/10.3390/ijms16047377
APA StyleJin, L., Guo, T., Li, Z., Lei, Z., Li, H., Mao, Y., Wang, X., Zhou, N., Zhang, Y., Hu, R., Zhang, X., Niu, G., Irwin, D. M., & Tan, H. (2015). Role of Glucokinase in the Subcellular Localization of Glucokinase Regulatory Protein. International Journal of Molecular Sciences, 16(4), 7377-7393. https://doi.org/10.3390/ijms16047377





