Genetic Variants in the Apoptosis Gene BCL2L1 Improve Response to Interferon-Based Treatment of Hepatitis C Virus Genotype 3 Infection
Abstract
:1. Introduction
2. Results
2.1. Host Genotypes
2.2. Treatment Responses
2.2.1. Study Population and Association of BCL2L1 with SVR and HCV Viral Load
| Characteristic | Value, n (%) |
|---|---|
| Male | 132 (66) |
| HCV genotype | |
| 1 | 100 (50) |
| 3 | 101 (50) |
| Age at treatment initiation, years (median, IQR) | 48 (43, 53) |
| HCV viral load at treatment initiation, log (IU/mL) (median, IQR) | 6.1 (5.4, 6.7) |
| IL28B genotype, rs12979860 (n = 198 *) | |
| CC | 75 (38) |
| CT | 106 (53) |
| TT | 17 (9) |
| Completion of treatment | |
| As scheduled | 118 (59) |
| With dose reduction | 36 (18) |
| Terminated before scheduled | 47 (23) |
| Fibrosis (n = 102 **) | |
| None/light | 43 (39) |
| Moderate/advanced | 30 (32) |
| Cirrhosis | 29 (29) |
| Ribavirin dose, mg/day (n = 199 ***) | |
| ≤800 | 78 (39) |
| 1000 | 57 (29) |
| ≥1200 | 64 (32) |
| Pegylated interferon α | |
| 2a | 133 (66) |
| 2b | 68 (34) |
| Characteristic | HCV Genotype 1, n = 100 | HCV Genotype 3, n = 101 | ||||
|---|---|---|---|---|---|---|
| SVR n = 46 (46%) | Non-Response, n = 54 (54%) | p-Value | SVR, n = 71 (70%) | Non-Response, n = 30 (30%) | p-Value | |
| Sex | 0.7 | 1 | ||||
| Female | 16 (50) | 16 (50) | 26 (70) | 11 (30) | ||
| Male | 30 (44) | 38 (56) | 45 (70) | 19 (30) | ||
| Age at treatment initiation | 0.2 | 0.03 | ||||
| <40 years | 14 (58) | 10 (42) | 36 (82) | 8 (18) | ||
| ≥40 years | 32 (42) | 44 (58) | 35 (61) | 22 (39) | ||
| HCV viral load at treatment initiation * | 0.3 | 0.06 | ||||
| <5.8 log10 IU/mL | 16 (53) | 14 (47) | 38 (79) | 10 (21) | ||
| ≥5.8 log10 IU/mL | 30 (43) | 40 (57) | 33 (62) | 20 (38) | ||
| IL28B genotype | 0.01 | 0.2 | ||||
| CC | 19 (70) | 8 (30) | 32 (67) | 16 (33) | ||
| TC | 22 (37) | 37 (63) | 33 (70) | 14 (30) | ||
| TT | 3 (27) | 8 (73) | 6 (100) | 0 | ||
| BCL2L1 genotype | 0.01 | 0.2 | ||||
| TT | 17 (36) | 30 (64) | 35 (65) | 19 (35) | ||
| TC | 15 (41) | 22 (59) | 31 (74) | 11 (26) | ||
| CC | 10 (83) | 2 (17) | 5 (100) | 0 | ||
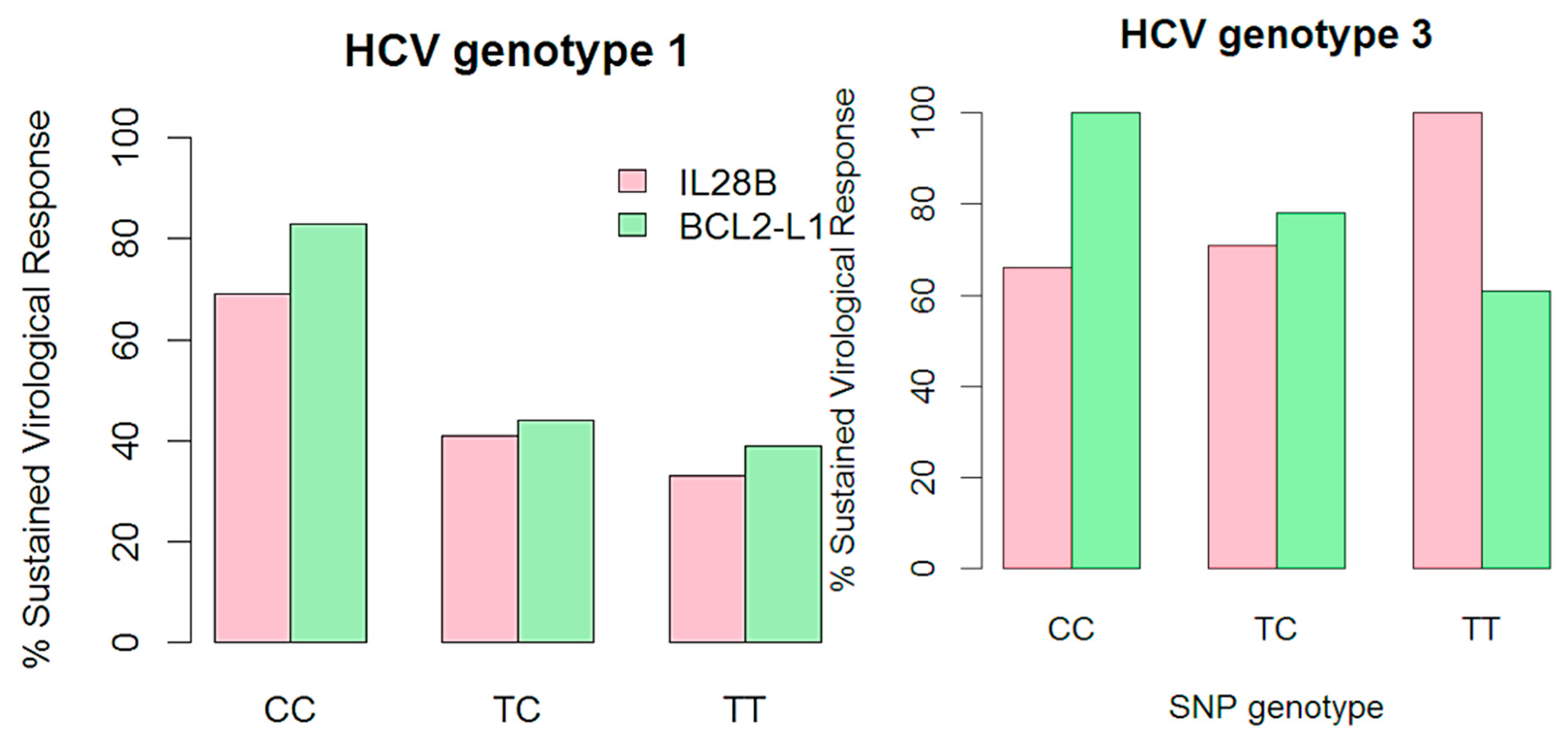
2.2.2. Logistic Regression Analysis of BCL2L1 Association to SVR
| Characteristic | HCV Genotype 1 (n = 91) | HCV Genotype 3 (n = 91) | ||
|---|---|---|---|---|
| Adjusted Odds Ratio (95% CI) | p-Value | Adjusted Odds Ratio (95% CI) | p-Value | |
| IL28B, rs12979860 | 0.01 | 0.6 | ||
| TT | 1 | 1 | ||
| TC | 0.97 (0.2, 4.9) | 0.4 (0, 3.1) | ||
| CC | 5.7 (1.0, 32.5) | 0.4 (0, 3.4) | ||
| IL28B, rs12979860 | 0.002 | 0.8 | ||
| TC + TT | 1 | 1 | ||
| CC | 5.9 (1.9, 17.3) | 1.1 (0.4, 3.4) | ||
| BCL2L1, rs1484994 | 0.049 | 0.06 | ||
| TT | 1 | 1 | ||
| TC | 1.5 (0.5, 4.3) | 0.4 | 2.8 (0.9, 9.7) | 0.08 |
| CC | 9.0 (1.6, 52.3) | 0.01 | 2.9 (0.5, infinity) | 0.4 |
| BCL2L1, rs1484994 | 0.1 | 0.02 | ||
| TT | 1 | 1 | ||
| TC + CC | 2.2 (0.8, 5.9) | 3.4 (1.2, 9.8) | ||
2.3. Ability of Gene- and Non-Gene Classifiers to Predict Treatment Response
3. Discussion
Identification of SNPs in the BCL2L1 Gene that Are Predictive of SVR to pegIFN/RBV Treatment of Genotype 3 Chronic Hepatitis C
4. Experimental Section
4.1. Study Subjects
4.2. SNP Selection and Genotyping
4.3. Statistical Analyses
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Mohd, H.K.; Groeger, J.; Flaxman, A.D.; Wiersma, S.T. Global epidemiology of hepatitis C virus infection: New estimates of age-specific antibody to HCV seroprevalence. Hepatology 2013, 57, 1333–1342. [Google Scholar] [CrossRef] [PubMed]
- Fischer, R.; Baumert, T.; Blum, H.E. Hepatitis C virus infection and apoptosis. World J. Gastroenterol. 2007, 13, 4865–4872. [Google Scholar] [PubMed]
- Canbay, A.; Friedman, S.; Gores, G.J. Apoptosis: The nexus of liver injury and fibrosis. Hepatology 2004, 39, 273–278. [Google Scholar] [CrossRef] [PubMed]
- TaqMan® SNP Genotyping Assays. Available online: http://www.ncbi.nlm.nih.gov/projects/SNP (accessed on 6 December 2014).
- Chattopadhyay, K.; Ramagopal, U.A.; Mukhopadhaya, A.; Malashkevich, V.N.; Dilorenzo, T.P.; Brenowitz, M.; Nathenson, S.G.; Almo, S.C. Assembly and structural properties of glucocorticoid-induced TNF receptor ligand: Implications for function. Proc. Natl. Acad. Sci. USA 2007, 104, 19452–19457. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Li, Z.; Mahesh, S.P.; Pantanelli, S.; Hwang, F.S.; Siu, W.O.; Nussenblatt, R.B. Glucocorticoid-induced tumor necrosis factor receptor negatively regulates activation of human primary natural killer (NK) cells by blocking proliferative signals and increasing NK cell apoptosis. J. Biol. Chem. 2008, 283, 8202–8210. [Google Scholar] [CrossRef] [PubMed]
- Volkmann, X.; Cornberg, M.; Wedemeyer, H.; Lehner, F.; Manns, M.P.; Schulze-Osthoff, K.; Bantel, H. Caspase activation is required for antiviral treatment response in chronic hepatitis C virus infection. Hepatology 2006, 43, 1311–1316. [Google Scholar] [CrossRef] [PubMed]
- Schiavon, L.L.; Narciso-Schiavon, J.L.; Carvalho-Filho, R.J.; Sampaio, J.P.; El Batah, P.N.; Silva, G.A.; Carvente, C.T.; Silva, A.E.; Ferraz, M.L. Evidence of a significant role for Fas-mediated apoptosis in HCV clearance during pegylated interferon plus ribavirin combination therapy. Antivir. Ther. 2011, 16, 291–298. [Google Scholar] [CrossRef] [PubMed]
- Anatol, P.; Danuta, P.; Janusz, D.; Bozena, P. Expression of bcl-2 protein in chronic hepatitis C: Effect of interferon alpha 2b with ribavirin therapy. World J. Gastroenterol. 2005, 11, 2949–2952. [Google Scholar] [CrossRef] [PubMed]
- Shaker, O.G.; Eskander, E.F.; Yahya, S.M.; Mohamed, M.S.; Abd-Rabou, A.A. Genetic variation in BCL-2 and response to interferon in hepatitis C virus type 4 patients. Clin. Chim. Acta 2011, 412, 593–598. [Google Scholar] [CrossRef] [PubMed]
- Lawitz, E.; Mangia, A.; Wyles, D.; Rodriguez-Torres, M.; Hassanein, T.; Gordon, S.C.; Schultz, M.; Davis, M.N.; Kayali, Z.; Reddy, K.R.; et al. Sofosbuvir for previously untreated chronic hepatitis C infection. N. Engl. J. Med. 2013, 368, 1878–1887. [Google Scholar]
- Lawitz, E.; Poordad, F.; Brainard, D. Sofosbuvir in combination with pegIFN and ribavirin for 12 weeks provides high SVR rates in HCV-infected genotype 2 or 3 treatment experienced patients with and without compensated cirrhosis: Results from the LONESTAR-2 study. In Proceedings of the 64th Annual Meeting of the American Association for the Study of Liver Diseases, Washington, DC, USA, 1–5 November 2013.
- Zeuzem, S.; Dusheiko, G.M.; Salupere, R. Sofosbuvir + ribavirin for 12 or 24 weeks for patients with HCV genotype 2 or 3: The VALENCE trial. In Proceedings of the 64th Annual Meeting of the American Association for the Study of Liver Diseases, Washington, DC, USA, 1–5 November 2013.
- Prokunina-Olsson, L.; Muchmore, B.; Tang, W.; Pfeiffer, R.M.; Park, H.; Dickensheets, H.; Hergott, D.; Porter-Gill, P.; Mumy, A.; Kohaar, I.; et al. A variant upstream of IFNL3 (IL28B) creating a new interferon gene IFNL4 is associated with impaired clearance of hepatitis C virus. Nat. Genet. 2013, 45, 164–171. [Google Scholar]
- Guo, C.J.; Pan, Q.; Jiang, B.; Chen, G.Y.; Li, D.G. Effects of upregulated expression of microRNA-16 on biological properties of culture-activated hepatic stellate cells. Apoptosis 2009, 14, 1331–1340. [Google Scholar] [CrossRef] [PubMed]
- Bota, S.; Sporea, I.; Sirli, R.; Neghina, A.M.; Popescu, A.; Strain, M. Role of interleukin-28B polymorphism as a predictor of sustained virological response in patients with chronic hepatitis C treated with triple therapy: A systematic review and meta-analysis. Clin. Drug Investig. 2013, 33, 325–331. [Google Scholar] [CrossRef] [PubMed]
- Holmes, J.A.; Desmond, P.V.; Thompson, A.J. Does IL28B genotyping still have a role in the era of direct-acting antiviral therapy for chronic hepatitis C infection? J. Viral Hepat. 2012, 19, 677–684. [Google Scholar] [CrossRef] [PubMed]
- Muir, A.J. IL28B in the era of direct-acting antivirals for hepatitis C. J. Clin. Gastroenterol. 2013, 47, 222–227. [Google Scholar] [CrossRef] [PubMed]
- Thompson, A.J.; McHutchison, J.G. Will IL28B polymorphism remain relevant in the era of direct-acting antiviral agents for hepatitis C virus? Hepatology 2012, 56, 373–381. [Google Scholar] [CrossRef] [PubMed]
- Ge, D.; Fellay, J.; Thompson, A.J.; Simon, J.S.; Shianna, K.V.; Urban, T.J.; Heinzen, E.L.; Qiu, P.; Bertelsen, A.H.; Muir, A.J.; et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature 2009, 461, 399–401. [Google Scholar]
- Rauch, A.; Kutalik, Z.; Descombes, P.; Cai, T.; Di, I.J.; Mueller, T.; Bochud, M.; Battegay, M.; Bernasconi, E.; Borovicka, J.; et al. Genetic variation in IL28B is associated with chronic hepatitis C and treatment failure: A genome-wide association study. Gastroenterology 2010, 138, 1338–1345. [Google Scholar]
- Suppiah, V.; Moldovan, M.; Ahlenstiel, G.; Berg, T.; Weltman, M.; Abate, M.L.; Bassendine, M.; Spengler, U.; Dore, G.J.; Powell, E.; et al. IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat. Genet. 2009, 41, 1100–1104. [Google Scholar]
- Tanaka, Y.; Nishida, N.; Sugiyama, M.; Kurosaki, M.; Matsuura, K.; Sakamoto, N.; Nakagawa, M.; Korenaga, M.; Hino, K.; Hige, S.; et al. Genome-wide association of IL28B with response to pegylated interferon-α and ribavirin therapy for chronic hepatitis C. Nat. Genet. 2009, 41, 1105–1109. [Google Scholar]
- Hansen, N.; Obel, N.; Christensen, P.B.; Krarup, H.; Laursen, A.L.; Clausen, M.R.; Lunding, S.; Moller, A.; Schlichting, P.; Kromann-Andersen, H.; et al. Predictors of antiviral treatment initiation in hepatitis C virus-infected patients: A Danish cohort study. J. Viral Hepat. 2009, 16, 659–665. [Google Scholar]
- Cordell, H.J.; Clayton, D.G. Genetic association studies. Lancet 2005, 366, 1121–1131. [Google Scholar] [CrossRef] [PubMed]
- Fried, M.W.; Shiffman, M.L.; Reddy, K.R.; Smith, C.; Marinos, G.; Goncales, F.L.; Haussinger, D.; Diago, M.; Carosi, G.; Dhumeaux, D.; et al. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N. Engl. J. Med. 2002, 347, 975–982. [Google Scholar]
- Hansen, N.; Obel, N.; Christensen, P.B.; Kjaer, M.; Laursen, A.L.; Krarup, H.B.; Moller, A.; Schlichting, P.; Bukh, J.; Weis, N. Effectiveness of treatment with pegylated interferon and ribavirin in an unselected population of patients with chronic hepatitis C: A Danish nationwide cohort study. BMC Infect. Dis. 2011, 11. [Google Scholar] [CrossRef] [PubMed]
- Manns, M.P.; McHutchison, J.G.; Gordon, S.C.; Rustgi, V.K.; Shiffman, M.; Reindollar, R.; Goodman, Z.D.; Koury, K.; Ling, M.; Albrecht, J.K. Peginterferon α-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: A randomised trial. Lancet 2001, 358, 958–965. [Google Scholar] [CrossRef] [PubMed]
- Nijman, I.J.; Kuipers, S.; Verheul, M.; Guryev, V.; Cuppen, E. A genome-wide SNP panel for mapping and association studies in the rat. BMC Genomics 2008, 9. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Clausen, L.N.; Weis, N.; Ladelund, S.; Madsen, L.; Lunding, S.; Tarp, B.; Christensen, P.B.; Krarup, H.B.; Møller, A.; Gerstoft, J.; et al. Genetic Variants in the Apoptosis Gene BCL2L1 Improve Response to Interferon-Based Treatment of Hepatitis C Virus Genotype 3 Infection. Int. J. Mol. Sci. 2015, 16, 3213-3225. https://doi.org/10.3390/ijms16023213
Clausen LN, Weis N, Ladelund S, Madsen L, Lunding S, Tarp B, Christensen PB, Krarup HB, Møller A, Gerstoft J, et al. Genetic Variants in the Apoptosis Gene BCL2L1 Improve Response to Interferon-Based Treatment of Hepatitis C Virus Genotype 3 Infection. International Journal of Molecular Sciences. 2015; 16(2):3213-3225. https://doi.org/10.3390/ijms16023213
Chicago/Turabian StyleClausen, Louise Nygaard, Nina Weis, Steen Ladelund, Lone Madsen, Suzanne Lunding, Britta Tarp, Peer Brehm Christensen, Henrik Bygum Krarup, Axel Møller, Jan Gerstoft, and et al. 2015. "Genetic Variants in the Apoptosis Gene BCL2L1 Improve Response to Interferon-Based Treatment of Hepatitis C Virus Genotype 3 Infection" International Journal of Molecular Sciences 16, no. 2: 3213-3225. https://doi.org/10.3390/ijms16023213
APA StyleClausen, L. N., Weis, N., Ladelund, S., Madsen, L., Lunding, S., Tarp, B., Christensen, P. B., Krarup, H. B., Møller, A., Gerstoft, J., Clausen, M. R., Benfield, T., & The DANHEP group. (2015). Genetic Variants in the Apoptosis Gene BCL2L1 Improve Response to Interferon-Based Treatment of Hepatitis C Virus Genotype 3 Infection. International Journal of Molecular Sciences, 16(2), 3213-3225. https://doi.org/10.3390/ijms16023213






