Molecular Characterization of Three Gonadotropin Subunits and Their Expression Patterns during Ovarian Maturation in Cynoglossus semilaevis
Abstract
:1. Introduction
2. Results and Discussion
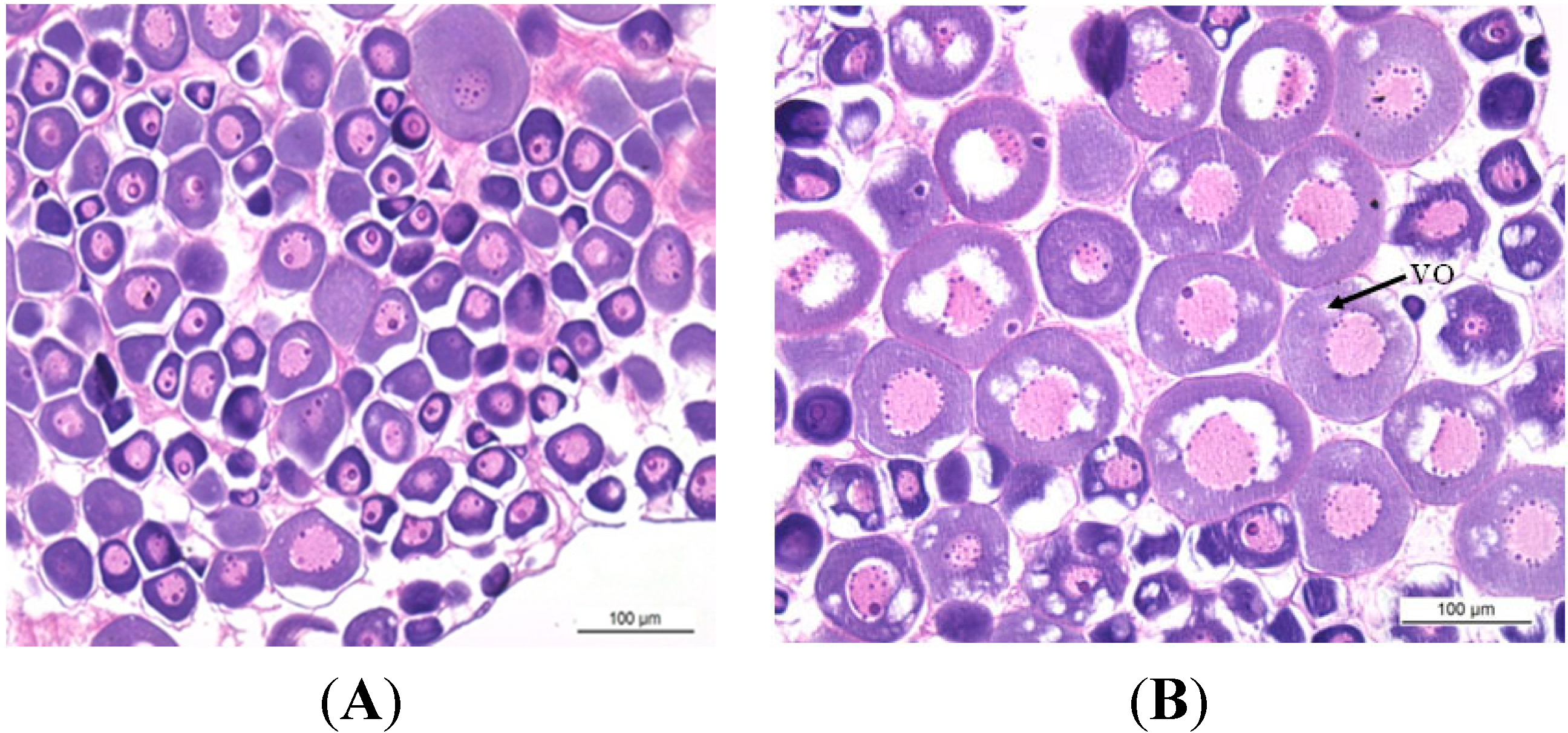
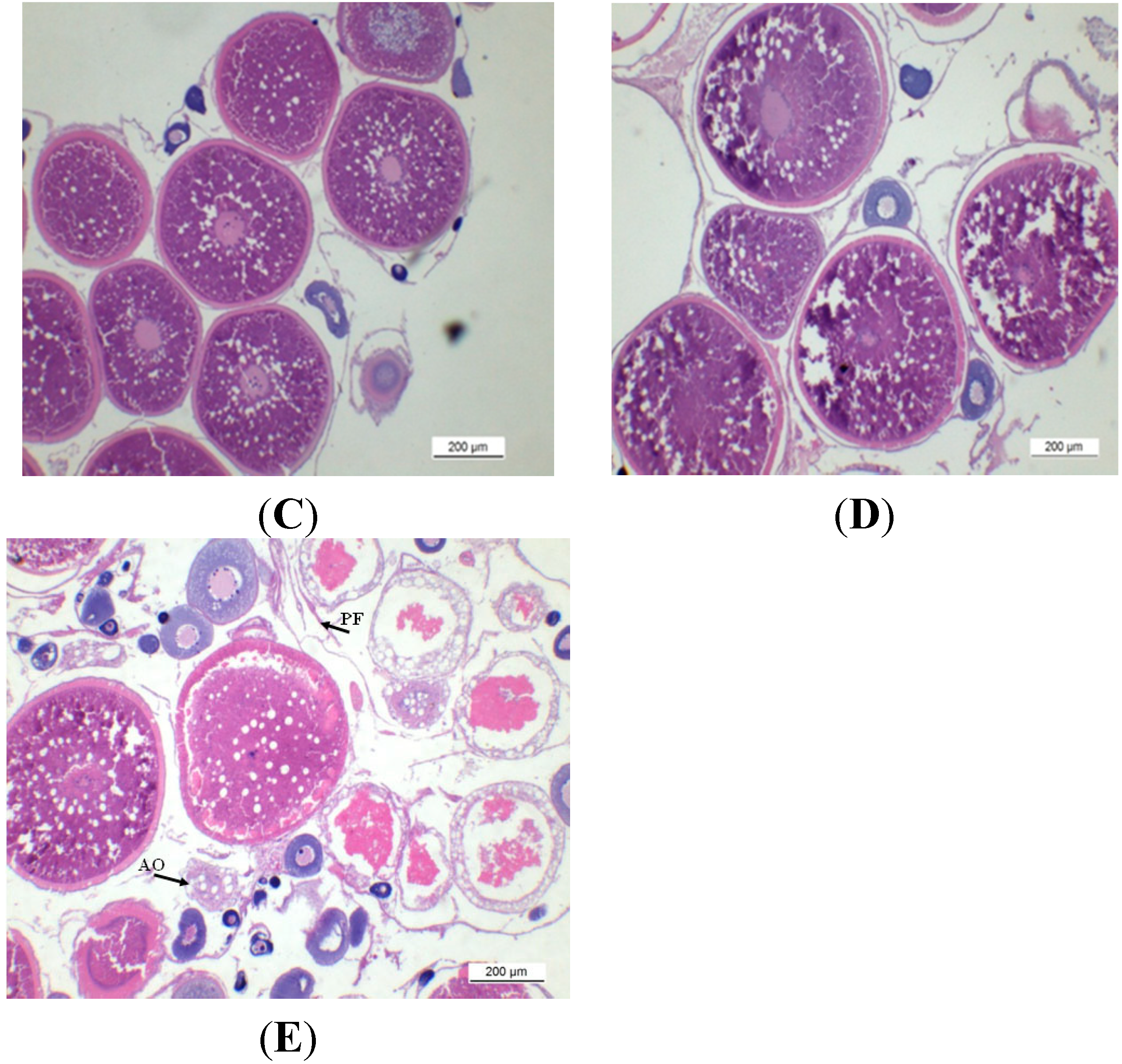
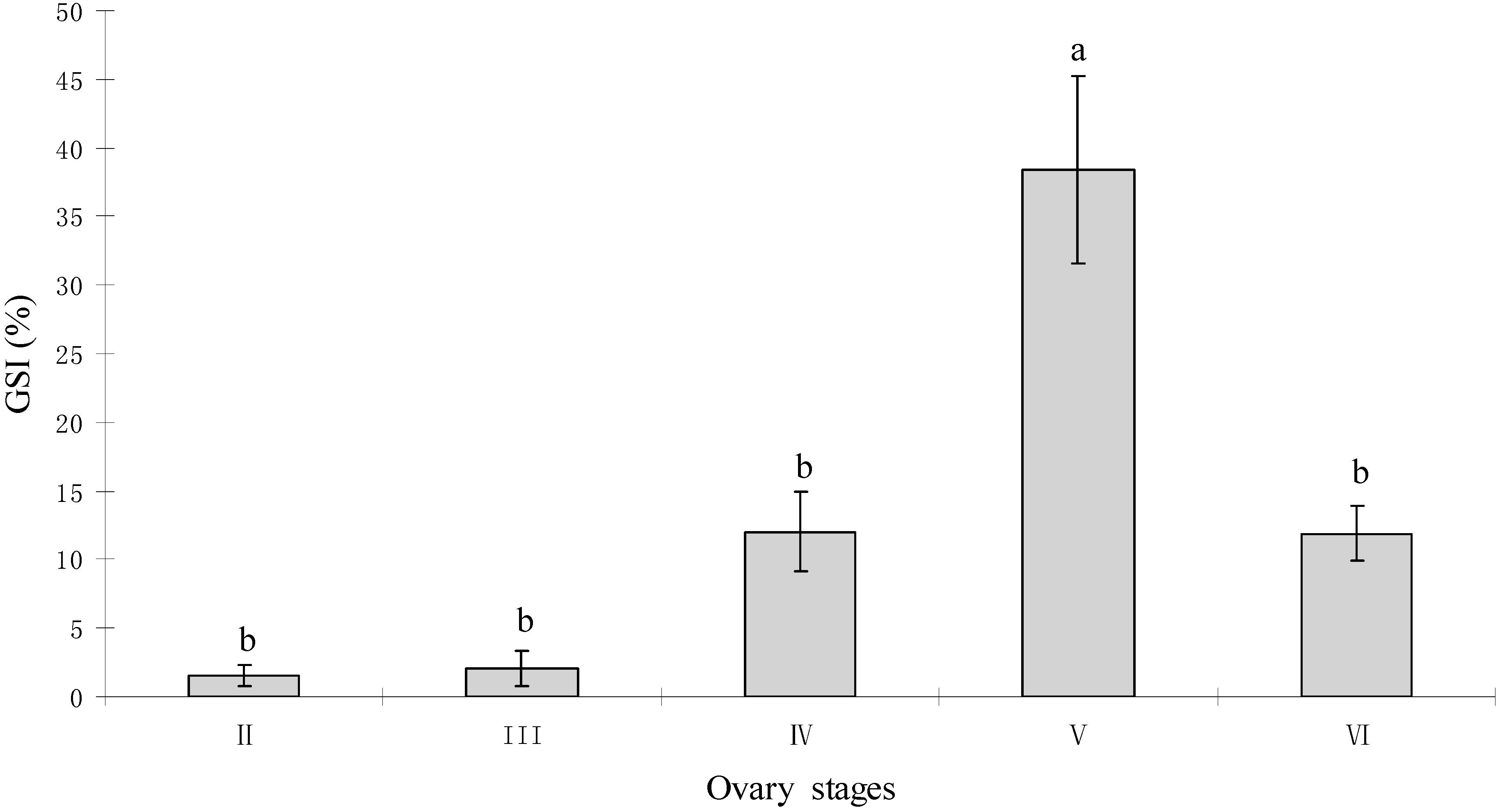
2.1. Gonadal Stages and Changes in Gonadosomatic Index (GSI)
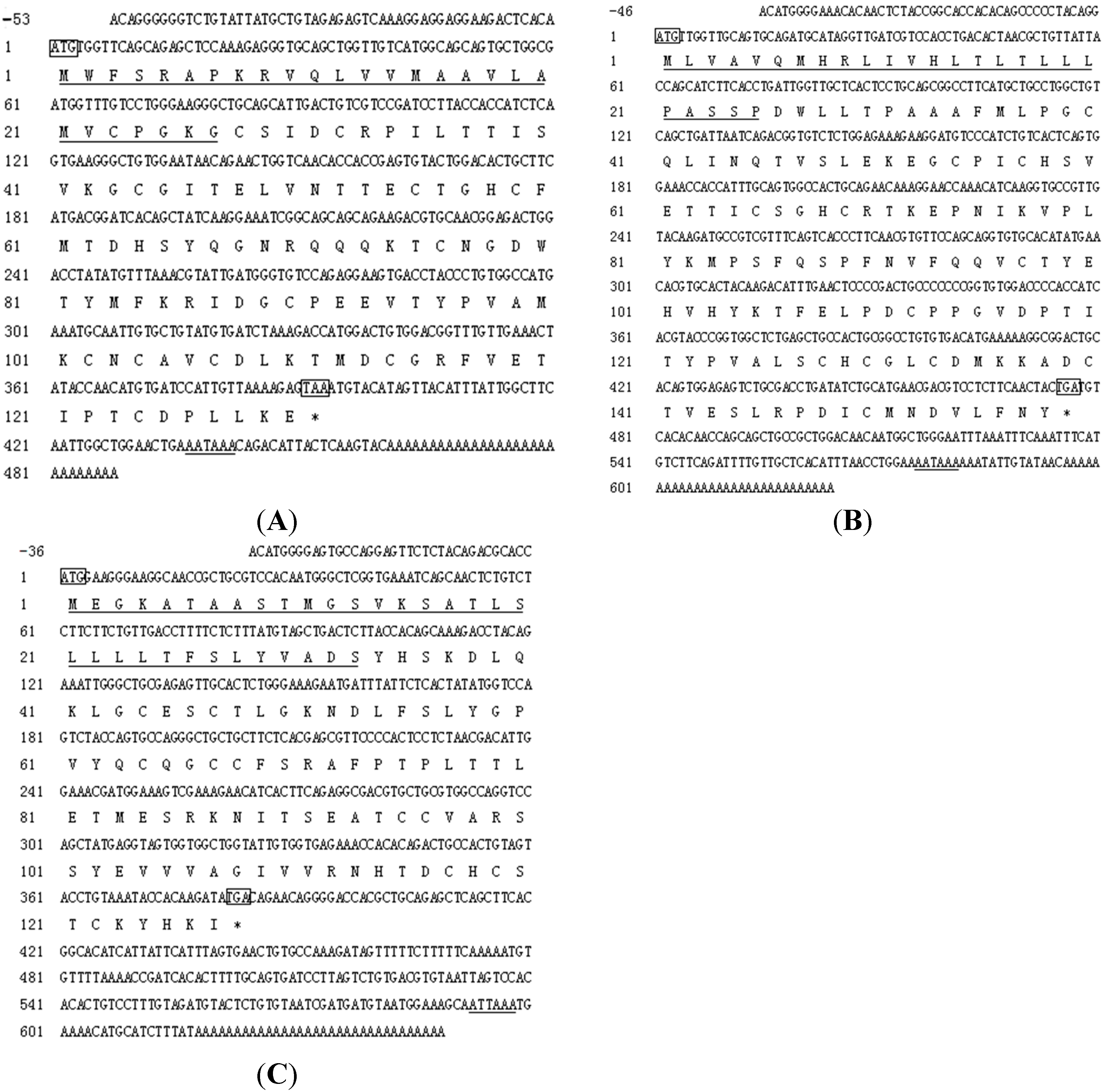
2.2. cDNA Cloning of Tongue Sole Gonadotropin Hormone (GTH) Subunits
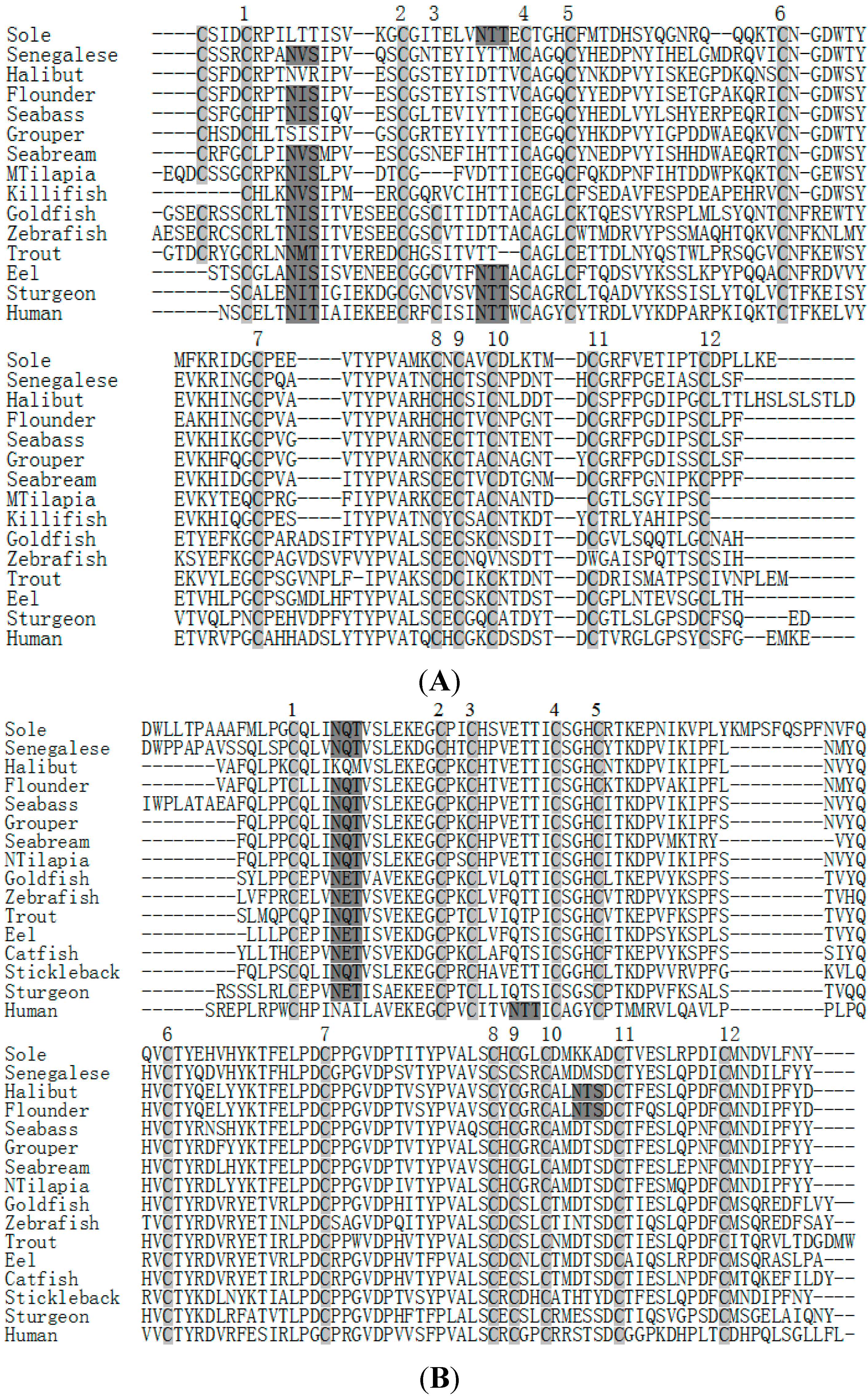
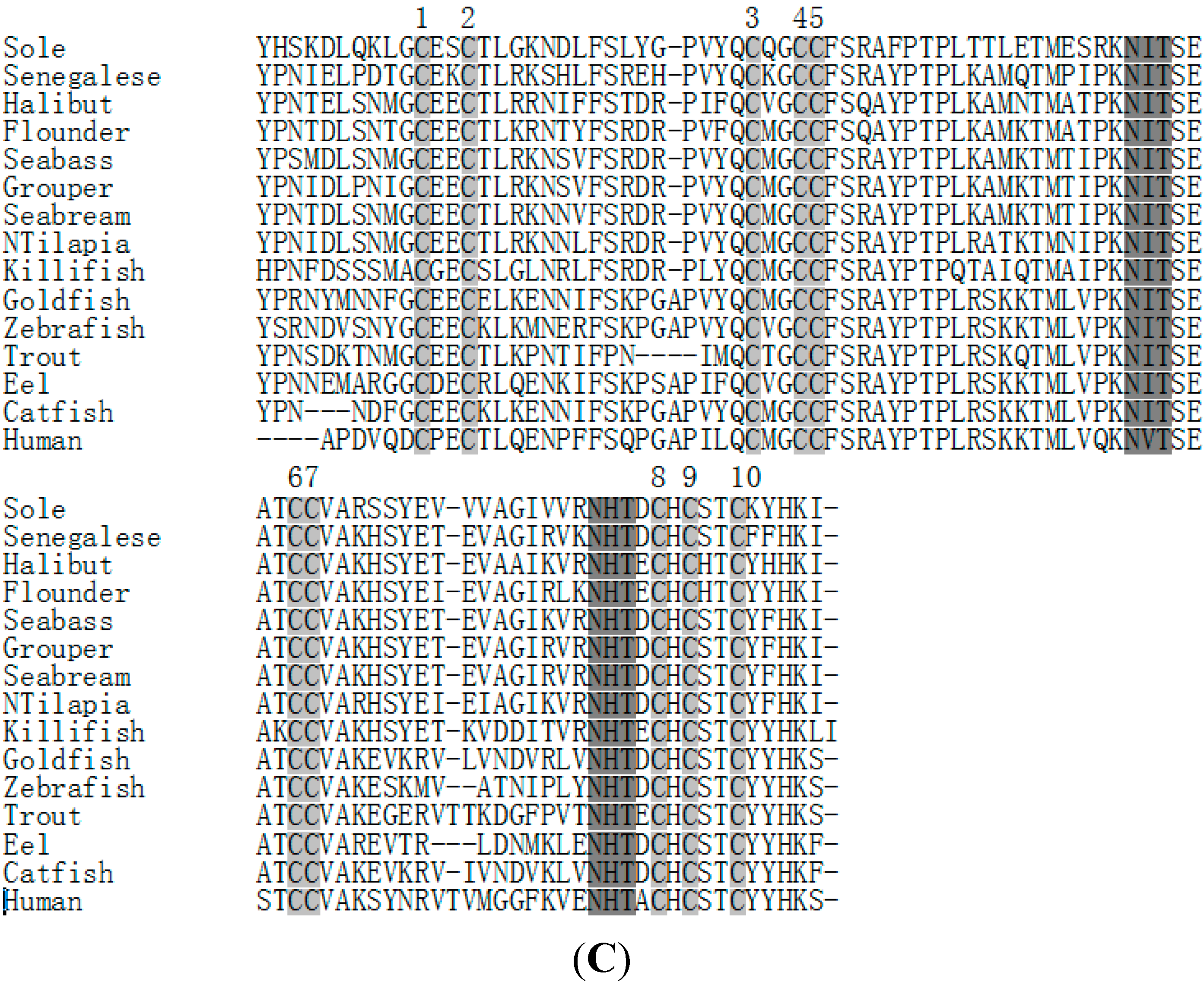
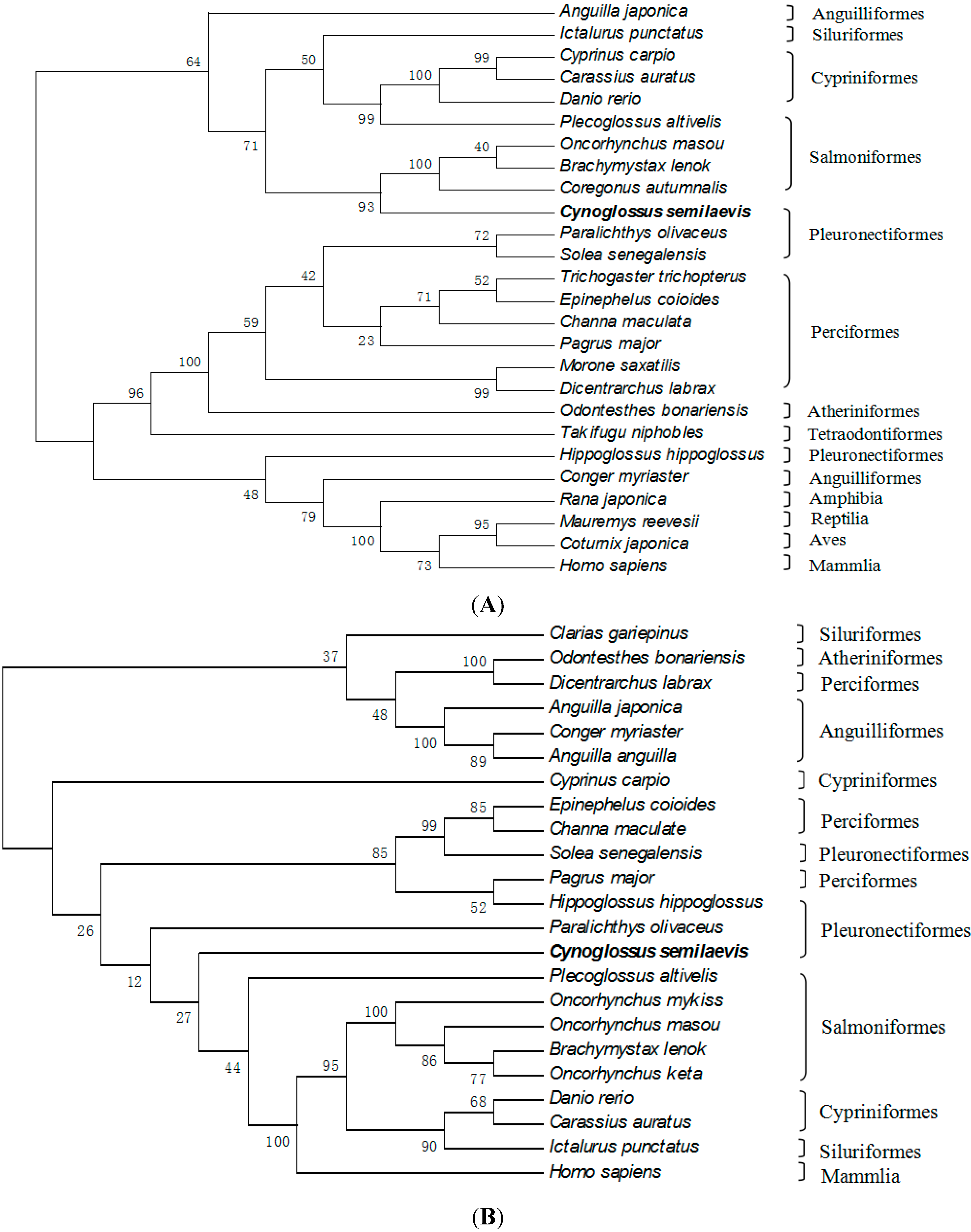
2.3. Amino Acid Sequence Analysis of Tongue Sole GTH Subunits

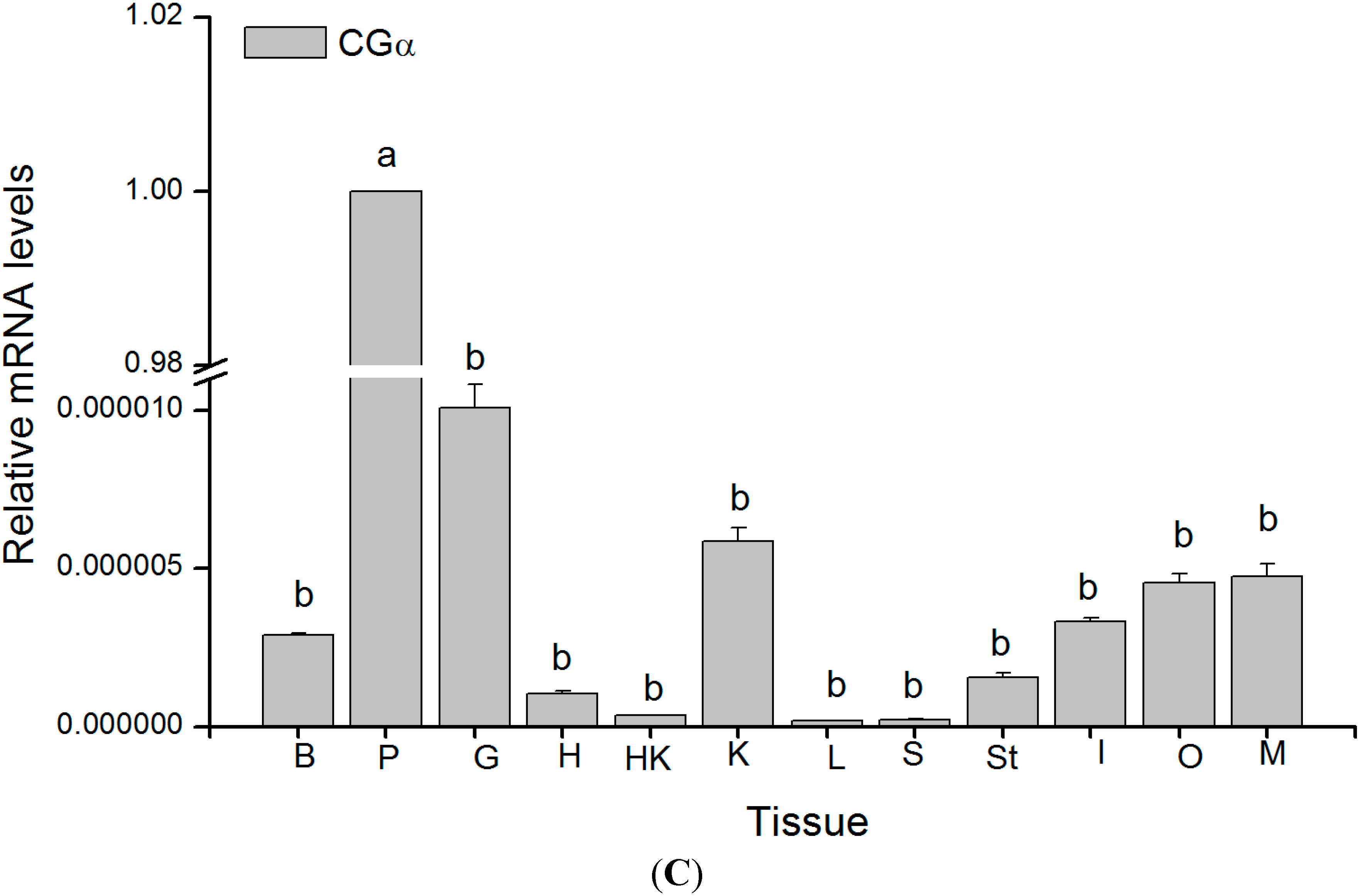
2.4. Tissue Distribution of GTH Subunits in Maturing Female Tongue Sole

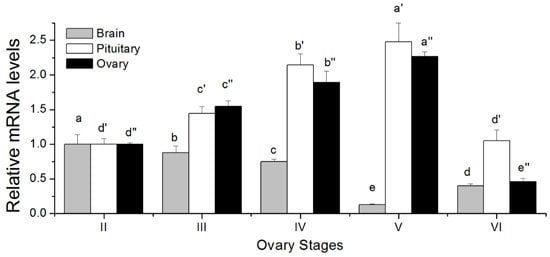
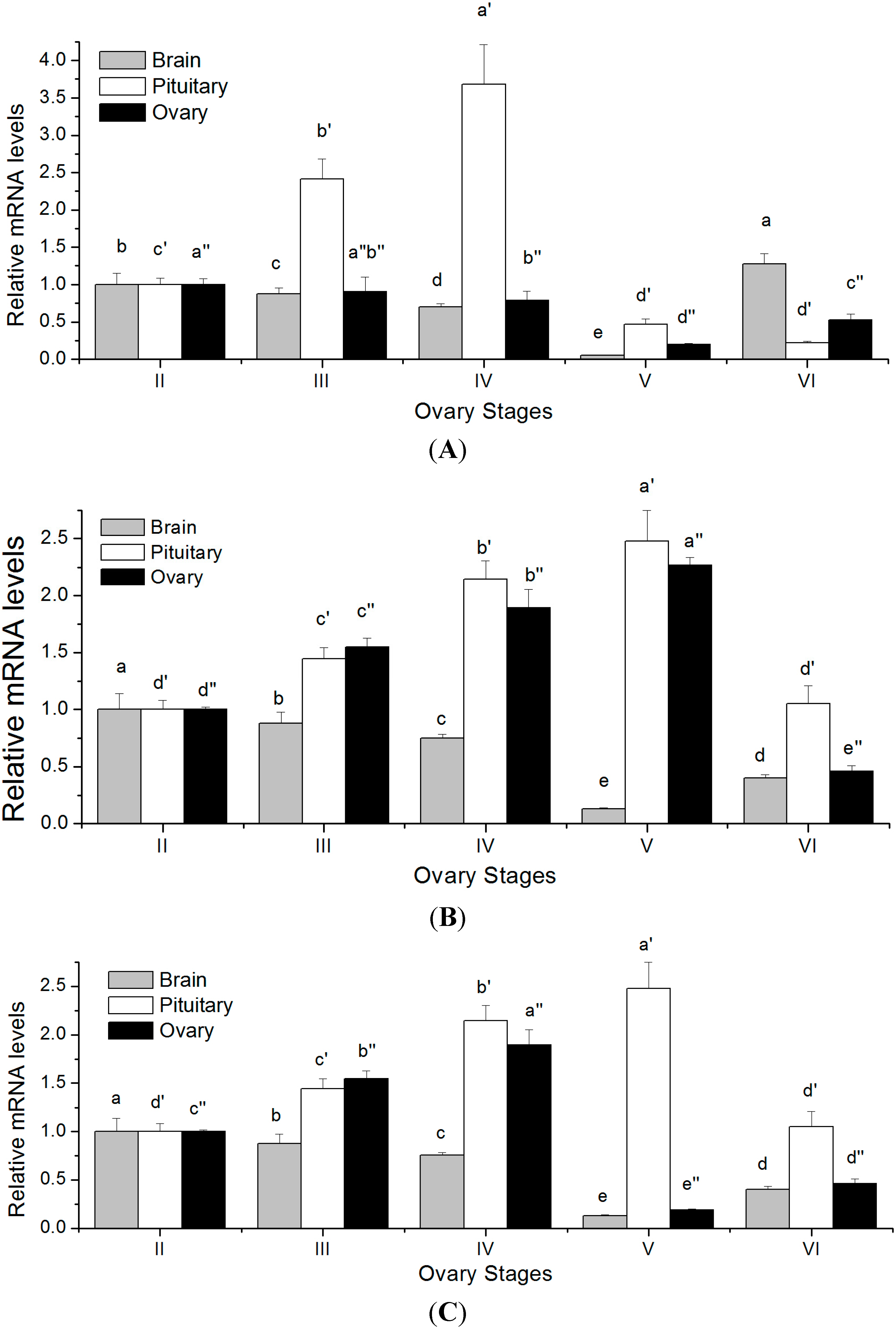
2.5. Changes in GTH Subunits Transcript Levels in Brain, Pituitary and Ovary at Different Gonadal Stages
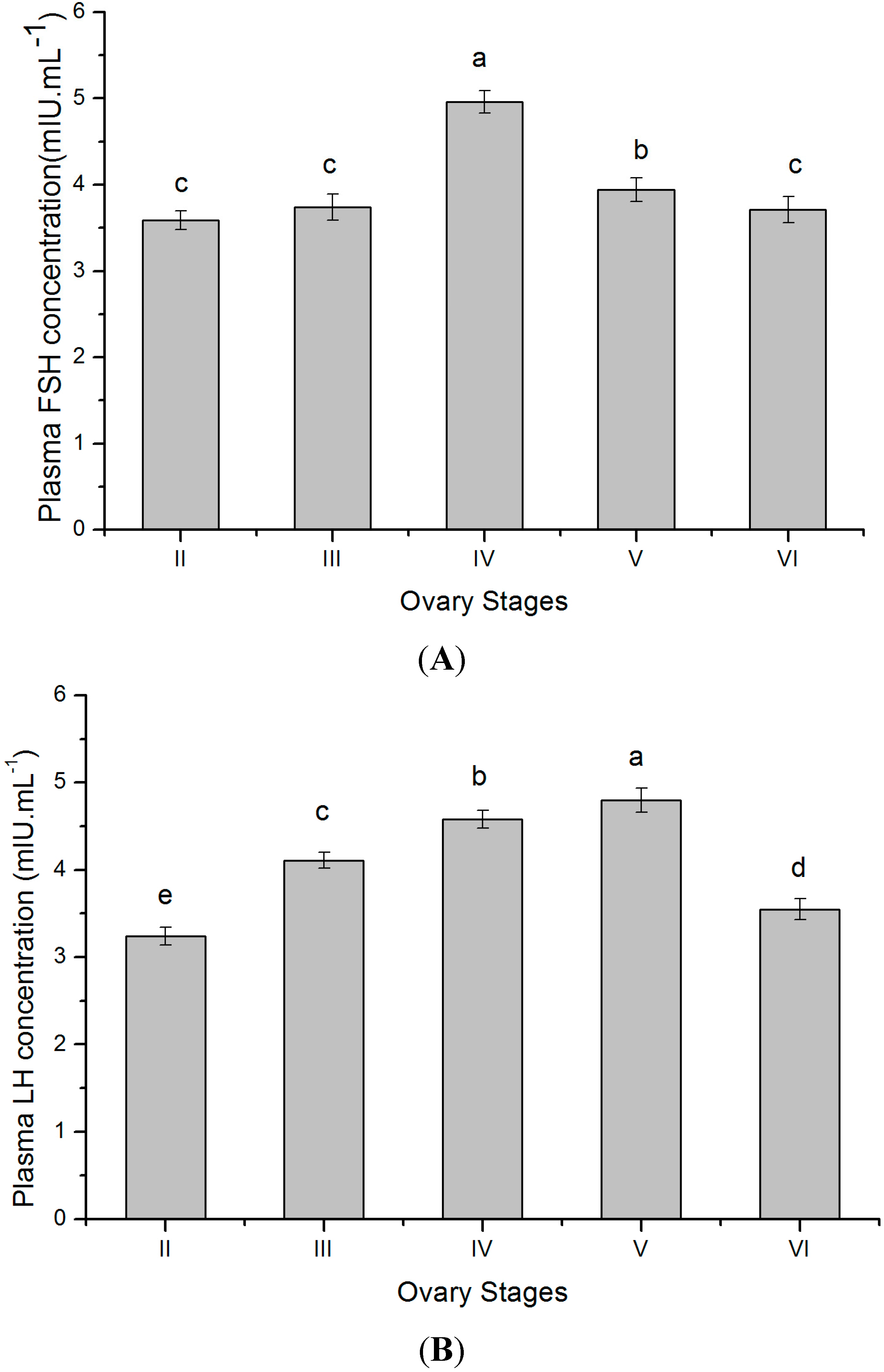
2.6. Plasma Gonadotropins Levels at Different Gonadal Stages
2.7. Discussion
3. Experimental Section
3.1. Experimental Fish and Sample Collection
3.2. Histological Analysis
3.3. Total RNA Extraction and Reverse Transcription (RT)
3.4. Isolation and PCR Amplification of FSHβ, LHβ and CGα cDNA Fragments
| Name | Sequence (5'→3') | Usage | Size (bp) |
|---|---|---|---|
| FSHβF | AGCAGAGGATGMAGCTGGT | Degenerate primer | 241 |
| FSHβR | TGGCCACAGGGTAGGTSAC | ||
| FSHβGSP1 | GCCACAGGGTAGGTCACTTCCTCTGGAC | 5' RACE primer | 321 |
| FSHβNGSP1 | GGTCCAGTCTCCGTTGCACGTCTTCTGC | Nested 5' RACE primer | 268 |
| FSHβGSP2 | GTGCTGGCGATGGTTTGTCCTGGGAAGG | 3' RACE primer | 409 |
| FSHβF1 | TGATGGGTGTCCAGAGGAAG | qRT-PCR primer | 95 |
| FSHβR1 | CAACAAACCGTCCACAGTCC | ||
| LHβF | CCATYTGCAGCGGYCAC | Degenerate primer | 162 |
| LHβR | GCAGCTCAMAGCCACMGG | ||
| LHβGSP1 | GCTCAAAGCCACAGGGTACGTGATGGTG | 5' RACE primer | 399 |
| LHβGSP2 | AAGATGCCGTCGTTTCAGTCACCCTTCA | 3' RACE primer | 353 |
| LHβF1 | AGACGGTGTCTCTGGAGAAAGAAG | qRT-PCR primer | 105 |
| LHβR1 | ACGGCACCTTGATGTTTGGT | ||
| CGαF | TAGYTGATTCTTACCCCARCAT | Degenerate primer | 270 |
| CGαR | TGCAGTGRCAGTCTGTGTGGTT | ||
| CGαGSP1 | CACCACAATACCAGCCACCACTACCTCA | 5' RACE primer | 340 |
| CGαNGSP1 | GCACGTCGCCTCTGAAGTGATGTTCTTT | Nested 5' RACE primer | 292 |
| CGαGSP2 | GTCTACCAGTGCCAGGGCTGCTGCTTCT | 3' RACE primer | 441 |
| CGαF1 | TTCCCCACTCCTCTAACGACA | qRT-PCR primer | 116 |
| CGαR1 | ACCACAATACCAGCCACCACTAC | ||
| 18S F | GGTCTGTGATGCCCTTAGATGTC | Internal control | 113 |
| 18S R | AGTGGGGTTCAGCGGGTTAC |
3.5. Rapid Amplification of cDNA 3' and 5' Ends (3' and 5' RACE)
3.6. Sequence Analysis and Phylogenetic Analysis
3.7. Quantitative Real-Time PCR
3.8. Radioimmunoassay
3.9. Statistical Analysis
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Yaron, Z.; Gur, G.; Melamed, P.; Rosenfeld, H.; Elizur, A.; Levavi-Sivan, B. Regulation of fish gonadotropins. Int. Rev. Cytol. 2003, 225, 131–185. [Google Scholar] [PubMed]
- Levavi-Sivan, B.; Bogerd, J.; Mañanós, E.L.; Gómez, A.; Lareyre, J.J. Perspectives on fish gonadotropins and their receptors. Gen. Comp. Endocrinol. 2010, 165, 412–437. [Google Scholar] [CrossRef] [PubMed]
- Ko, H.; Park, W.; Kim, D.; Kobayashi, M.; Sohn, Y.C. Biological activities of recombinant Manchurian trout FSH and LH: Their receptor specificity, steroidogenic and vitellogenic potencies. J. Mol. Endocrinol. 2007, 38, 99–111. [Google Scholar] [CrossRef] [PubMed]
- Kitano, H.; Irie, S.; Ohta, K.; Hirai, T.; Yamaguchi, A.; Matsuyama, M. Molecular cloning of two gonadotropin receptors and their distinct mRNA expression profiles in daily oogenesis of the wrasse P. sieboldi. Gen. Comp. Endocrinol. 2011, 172, 268–276. [Google Scholar] [CrossRef] [PubMed]
- Schulz, R.W.; Miura, T. Spermatogenesis and its endocrine regulation. Fish Physiol. Biochem. 2002, 26, 43–56. [Google Scholar] [CrossRef]
- Swanson, P.; Dickey, J.T.; Campbell, B. Biochemistry and physiology of fish gonadotropins. Fish Physiol. Biochem. 2003, 28, 53–59. [Google Scholar] [CrossRef]
- Weltzien, F.A.; Andersson, E.; Andersen, O.; Shalchian-Tabrizi, K.; Norberg, B. The brain-pituitary-gonad axis in male teleosts, with special emphasis on flatfish (Pleuronectiformes). Comp. Biochem. Physiol. A 2004, 137, 447–477. [Google Scholar] [CrossRef]
- Lubzens, E.; Young, G.; Bobe, J.; Cerdà, J. Oogenesis in teleosts: How fish eggs are formed. Gen. Comp. Endocrinol. 2010, 165, 367–389. [Google Scholar] [CrossRef] [PubMed]
- Gomez, J.M.; Weil, C.; Ollitrault, M.; le Bail, P.Y.; Breton, B.; Gac, F.L. Growth hormone (GH) and gonadotropin subunit gene expression and pituitary and plasma changes during spermatogenesis and oogenesis in rainbow trout (Oncorhynchus mykiss). Gen. Comp. Endocrinol. 1999, 113, 413–428. [Google Scholar] [CrossRef] [PubMed]
- Elisio, M.; Chalde, T.; Miranda, L.A. Seasonal changes and endocrine regulation of pejerrey (Odontesthes bonariensis) oogenesis in the wild. Comp. Biochem. Physiol. A 2014, 175, 102–109. [Google Scholar] [CrossRef]
- Jackson, K.; Goldberg, D.; Ofir, M.; Abraham, M.; Degani, G. Blue gourami (Trichogaster trichopterus) gonadotropic β subunits (I and II) cDNA sequences and expression during oogenesis. J. Mol. Endocrinol. 1999, 23, 177–187. [Google Scholar] [CrossRef] [PubMed]
- Sohn, Y.C.; Yoshiura, Y.; Kobayashi, M.; Aida, K. Seasonal changes in mRNA levels of gonadotropin and thyrotropin subunits in the goldfish, Carassius auratus. Gen. Comp. Endocrinol. 1999, 113, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Kajimura, S.; Yoshiura, Y.; Suzuki, M.; Aida, K. cDNA cloning of two gonadotropin β subunits (GTH-Iβ and -IIβ) and their expression profiles during gametogenesis in the Japanese flounder (Paralichthys olivaceus). Gen. Comp. Endocrinol. 2001, 122, 117–129. [Google Scholar] [CrossRef] [PubMed]
- Meiri, I.; Knibb, W.R.; Zohar, Y.; Elizur, A. Temporal profile of β follicle-stimulating hormone, β luteinizing hormone, and growth hormone gene expression in the protandrous hermaphrodite, gilthead seabream, Sparus aurata. Gen. Comp. Endocrinol. 2004, 137, 288–299. [Google Scholar] [CrossRef] [PubMed]
- Hellqvist, A.; Schmitz, M.; Mayer, I.; Borg, B. Seasonal changes in expression of LH-β and FSH-β in male and female three-spined stickleback, Gasterosteus aculeatus. Gen. Comp. Endocrinol. 2006, 145, 263–269. [Google Scholar] [CrossRef] [PubMed]
- Nyuji, M.; Selvaraj, S.; Kitano, H.; Ohga, H.; Yamaguchi, A.; Matasuyama, M. Changes in the expression of pituitary gonadotropin subunits during reproductive cycle of multiple spawning female chub mackerel Scomber japonicus. Fish Physiol. Biochem. 2012, 38, 883–897. [Google Scholar] [CrossRef] [PubMed]
- Aizen, J.; Kasuto, H.; Levavi-Sivan, B. Development of specific enzyme-linked immu-nosorbent assay for determining LH and FSH levels in tilapia, using recombinant go-nadotropins. Gen. Comp. Endocrinol. 2007, 153, 323–332. [Google Scholar] [CrossRef] [PubMed]
- Parhar, I.S.; Soga, T.; Ogawa, S.; Sakuma, Y. FSH and LH-β subunits in the preoptic nucleus: Ontogenic expression in teleost. Gen. Comp. Endocrinol. 2003, 132, 369–378. [Google Scholar] [CrossRef] [PubMed]
- Wong, T.T.; Zohar, Y. Novel expression of gonadotropin subunit genes in oocytes of the gilthead seabream (Sparus aurata). Endocrinology 2004, 145, 5210–5220. [Google Scholar] [CrossRef] [PubMed]
- Zohar, Y.; Muñoz-Cueto, J.A.; Elizur, A.; Kah, O. Neuroendocrinology of reproduction in teleost fish. Gen. Comp. Endocrinol. 2010, 165, 438–455. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.Y.; Zhang, Q.Q.; Qi, J.; Chen, Y.J.; Zhong, Q.W.; Li, C.M.; Yu, Y.; Li, S.; Wang, Z.G. Identification of differential genes in the ovary relative to the testis and their expression patterns in half-smooth tongue sole (Cynoglossus semilaevis). J. Genet. Genomics 2010, 37, 137–145. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.L.; Zhang, G.J.; Shao, C.W.; Huang, Q.F.; Liu, G.; Zhang, P.; Song, W.T.; An, N.; Chalopin, D.; Volff, J.; et al. Whole-genome sequence of a flatfish provides insights into ZW sex chromosome evolution and adaptation to a benthic lifestyle. Nat. Genet. 2014, 46, 253–260. [Google Scholar] [CrossRef] [PubMed]
- Shao, C.W.; Li, Q.Y.; Chen, S.L.; Zhang, P.; Lian, J.M.; Hu, Q.M.; Sun, B.; Jin, L.J.; Liu, S.S.; Wang, Z.J.; et al. Epigenetic modification and inheritance in sexual reversal of fish. Genome Res. 2014, 24, 604–615. [Google Scholar] [CrossRef] [PubMed]
- Shao, C.W.; Chen, S.L.; Scheuring, C.F.; Xu, J.Y.; Sha, Z.X.; Dong, X.L.; Zhang, H.B. Construction of two BAC libraries from half-smooth tongue sole Cynoglossus semilaevis and identification of clones containing candidate sex-determination genes. Mar. Biotechnol. 2010, 12, 558–568. [Google Scholar] [CrossRef] [PubMed]
- Liao, X.L.; Xu, G.B.; Chen, S.L. Molecular method for sex identification of half-Smooth tongue sole (Cynoglossus semilaevis) using a novel sex-linked microsatellite marker. Int. J. Mol. Sci. 2014, 15, 12952–12958. [Google Scholar] [CrossRef] [PubMed]
- Ji, X.S.; Chen, S.L.; Jiang, Y.L.; Xu, T.J.; Yang, J.F.; Tian, Y.S. Growth differences and differential expression analysis of pituitary adenylate cyclase activating polypeptide (PACAP) and growth hormone-releasing hormone (GHRH) between the sexes in half-smooth tongue sole Cynoglossus semilaevis. Gen. Comp. Endocrinol. 2011, 170, 99–109. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Liu, S.F.; Zh, Z.M.; Lin, L.; Sun, Z.Z.; Liu, C.L.; Ma, H.; Su, Y.Q.; Tang, Q.S. Genomic structure, polymorphism and expression analysis of the growth hormone (GH) gene in female and male half-smooth tongue sole (Cynoglossus semilaevis). Gene 2012, 493, 92–104. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.F.; Wen, H.S.; Wang, Z.P.; He, F.; Zhang, J.R.; Chen, X.Y.; Jin, G.X.; Shi, B.; Shi, D.; Yang, Y.P.; et al. Cloning and expression of P450c17-I (17α-hydroxylase/17,20-lyase) in brain and ovary during gonad development in Cynoglossus semilaevis. Fish Physiol. Biochem. 2010, 36, 1001–1012. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Wang, X.L.; Sha, Z.X.; Yang, C.G.; Liu, S.S.; Wang, N.; Chen, S.L. Establishment and characterization of a testicular cell line from the half-smooth tongue sole, Cynoglossus semilaevis. Int. J. Biol. Sci. 2011, 7, 452–459. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.S.; Yi, Q.L.; Zhong, Q.W.; Li, C.M.; Muhammad, S.; Wang, X.B.; Wang, Z.G.; Qi, J.; Yu, H.Y.; Zhang, Q.Q. Molecular cloning, tissue distribution, and ontogeny of gonadotropin-releasing hormone III gene (GnRH-III) in half-smooth tongue sole (Cynoglossus semilaevis). Comp. Biochem. Physiol. B 2012, 163, 59–64. [Google Scholar] [CrossRef] [PubMed]
- Li, W.G.; Zhang, J.R.; Mu, W.J.; Wen, H.S. Cloning, characterization and expression of estrogen receptor β in the male half-smooth tongue sole, Cynoglossus semilaevis. Fish Physiol. Biochem. 2013, 39, 671–682. [Google Scholar] [CrossRef] [PubMed]
- Ulloa-Aguirre, A.; Timossi, C.; Damián-Matsumura, P.; Dias, J.A. Role of glycosylation in function of follicle-stimulating hormone. Endocrine 1999, 11, 205–215. [Google Scholar] [CrossRef] [PubMed]
- Weltzien, F.A.; Kobayashi, T.; Andersson, E.; Norberg, B.; Andersen, Ø. Molecular characterization and expression of FSHβ, LHβ, and common α-subunit in male Atlantic halibut (Hippoglossus hippoglossus). Gen. Comp. Endocrinol. 2003, 131, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Hearn, M.T.W.; Gomme, P.T. Molecular architecture and biorecognition processes of the cystine knot protein superfamily: Part I. The glycoprotein hormones. J. Mol. Recognit. 2000, 13, 223–278. [Google Scholar] [CrossRef] [PubMed]
- Lapthorn, A.J.; Harris, D.C.; Littlejohn, A.; Lustbader, J.W.; Canfield, R.E.; Machin, K.J.; Morgan, F.J.; Isaacs, N.W. Crystal structure of human chorionic gonadotropin. Nature 1994, 369, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Kim, S.; Karsi, A. Channel catfish follicle-stimulating hormone and luteinizing hormone: Complementary DNA cloning and expression during ovulation. Mar. Biotechnol. 2001, 3, 590–599. [Google Scholar] [CrossRef] [PubMed]
- Power, M.E.; Carolsfeld, J.; Wallis, G.P.; Sherwood, N.M. Isolation and characterization of a cDNA for gonadotropin II-β of Pacific herring, an ancient teleost. J. Fish Biol. 1997, 50, 315–323. [Google Scholar] [CrossRef]
- Zenkevics, H.; Vose, V.; Vosekalne, I.; Bucena, A. Partial deglycosylation of α subunit modifies sturgeon gonadotropin function. Acta Biochim. Pol. 2000, 47, 815–819. [Google Scholar] [PubMed]
- Gen, K.; Okuzawa, K.; Senthilkumaran, B.; Tanaka, H.; Moriyama, S.; Kagawa, H. Unique expression of gonadotropin-I and -II subunit genes in male and female red seabream (Pagrus major) during sexual maturation. Biol. Reprod. 2000, 63, 308–319. [Google Scholar] [CrossRef] [PubMed]
- Quérat, B.; Sellouk, A.; Salmon, C. Phylogenetic analysis of the vertebrate glycoprotein hormone family including new sequences of sturgeon (Acipenser baeri) β subunits of the two gonadotropins and the thyroid-stimulating hormone. Biol. Reprod. 2000, 63, 222–228. [Google Scholar] [CrossRef] [PubMed]
- Quérat, B.; Arai, Y.; Henry, A.; Akama, Y.; Longhurst, T.J.; Joss, J.M.P. Pituitary glycoprotein hormone β subunits in the Australian lungfish and estimation of the relative evolution rate of these subunits within vertebrates. Biol. Reprod. 2004, 70, 356–363. [Google Scholar] [CrossRef] [PubMed]
- Mateos, J.; Mañanós, E.; Martínez-Rodríguez, G.; Carrillo, M.; Querat, B.; Zanuy, S. Molecular characterization of sea bass gonadotropin subunits (α, FSHβ, and LHβ) and their expression during the reproductive cycle. Gen. Comp. Endocrinol. 2003, 133, 216–232. [Google Scholar] [CrossRef] [PubMed]
- Vischer, H.F.; Teves, A.C.C.; Ackermans, J.C.M.; van Dijk, W.; Schulz, R.W.; Bogerd, J. Cloning and spatiotemporal expression of the follicle-stimulating hormone β subunit complementary DNA in the African catfish (Clarias gariepinus). Biol. Reprod. 2003, 68, 1324–1332. [Google Scholar] [CrossRef] [PubMed]
- So, W.K.; Kwok, H.F.; Ge, W. Zebrafish gonadotropins and their receptors. II. Cloning and characterization of zebrafish follicle stimulating hormone and luteinizing hormone subunits—Their spatial-temporal expression patterns and receptor specificity. Biol. Reprod. 2005, 72, 1382–1396. [Google Scholar] [CrossRef] [PubMed]
- Mittelholzer, C.; Andersson, E.; Taranger, G.L.; Karlsen, Ø.; Norberg, B. Quantification of gonadotropin subunits GPα, FSHβ, and LHβ mRNA expression from Atlantic cod (Gadus morhua) throughout a reproductive cycle. Comp. Biochem. Physiol. B 2009, 153, 288–295. [Google Scholar] [CrossRef] [PubMed]
- Pandolfi, M.; Pozzi, A.G.; Cánepa, M.; Vissio, P.G.; Shimizu, A.; Maggese, M.C.; Lobo, G. Presence of β-follicle-stimulating hormone and β-luteinizing hormone transcripts in the brain of Cichlasoma dimerus (Perciformes: Cichlidae), effect of brain-derived gonadotropins on pituitary hormone release. Neuroendocrinology 2009, 89, 27–37. [Google Scholar] [CrossRef] [PubMed]
- Pakarainen, T.; Ahtiainen, P.; Zhang, F.P.; Rulli, S.; Poutanen, M.; Huhtaniemi, I. Extragonadal LH/hCG action-not yet time to rewrite textbooks. Mol. Cell. Endocrinol. 2007, 269, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Apaja, P.M.; Harju, K.T.; Aatsinki, J.T.; Petaja-Repo, U.E.; Rajaniemi, H.J. Identification and structural characterization of the neuronal luteinizing hormone receptor associated with sensory systems. J. Biol. Chem. 2004, 279, 1899–1906. [Google Scholar] [CrossRef] [PubMed]
- Elisio, M.; Soria, F.N.; Fernandino, J.I.; Strüssmann, C.A.; Somoza, G.M.; Miranda, L.A. Extrahypophyseal expression of gonadotropin subunits in pejerrey Odontesthes bonariensis and effects of high water temperatures on their expression. Gen. Comp. Endocrinol. 2012, 175, 329–336. [Google Scholar] [CrossRef] [PubMed]
- Tyler, C.R.; Pottinger, T.G.; Coward, K.; Beresford, N.; Maddix, S. Salmonid follicle-stimulating hormone (GTH-I) mediates vitellogenic development of oocytes in the rainbow trout, Oncorhynchus mykiss. Biol. Reprod. 1997, 57, 1238–1244. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Zhang, Y.; Huang, W.R.; Li, S.S.; Zhu, P.; Liu, Y.; Yin, S.W.; Liu, X.C.; Lin, H.R. Molecular characterization of marbled eel (Anguilla marmorata) gonadotropin subunits and their mRNA expression profiles during artificially induced gonadal development. Gen. Comp. Endocrinol. 2009, 162, 192–202. [Google Scholar] [CrossRef] [PubMed]
- Prat, F.; Sumpter, J.P.; Tyler, C.R. Validation of radioimmunoassay for two salmon gonadotropins (GTH-I and GTH-II) and their plasma concentrations throughout the reproductive cycle in male and female rainbow trout (Oncorhynchus mykiss). Biol. Reprod. 1996, 54, 1375–1382. [Google Scholar] [CrossRef] [PubMed]
- Nagae, M.; Todo, T.; Gen, K.; Kato, Y.; Young, G.; Adachi, S.; Yamauchi, K.J. Molecular cloning of the cDNAs encoding pituitary glycoprotein hormone α-and gonadotropin-II β-subunits of the Japanese eel, Anguilla japonica, and increase in their mRNAs during ovarian development induced by injection of chum salmon pituitary homogenate. J. Mol. Endocrinol. 1996, 16, 171–181. [Google Scholar] [CrossRef] [PubMed]
- Kagawa, H.; Tanaka, H.; Okuzawa, K.; Kobayashi, M. GTHII but not GTHI induces final maturation and the development of maturational competence of oocytes of red seabream in vitro. Gen. Comp. Endocrinol. 1998, 112, 80–88. [Google Scholar] [CrossRef]
- Mylonas, C.C.; Scott, A.P.; Zohar, Y. Plasma gonadotropin II, sex steroids, and thyroid hormones in wild striped bass (Morone saxatilis) during spermiation and final oocyte maturation. Gen. Comp. Endocrinol. 1997, 108, 223–236. [Google Scholar] [CrossRef] [PubMed]
- Yoshiura, Y.; Kobayashi, M.; Kato, Y.; Aida, K. Molecular cloning of the cDNAs encoding two gonadotropin β subunits (GTH-Iβ and -IIβ) from the goldfish, Carassius auratus. Gen. Comp. Endocrinol. 1997, 105, 379–389. [Google Scholar] [CrossRef] [PubMed]
- Cerdà, J.; Chauvigne, F.; Agulleiro, M.J.; Marin, E.; Halm, S.; Martínez-Rodríguez, G.; Prat, F. Molecular cloning of Senegalese sole (Solea senegalensis) follicle-stimulating hormone and luteinizing hormone subunits and expression pattern during spermatogenesis. Gen. Comp. Endocrinol. 2008, 156, 470–481. [Google Scholar] [CrossRef] [PubMed]
- Li, M.D.; Ford, J.J. A comprehensive evolutionary analysis based on nucleotide and amino acid sequences of the α- and β-subunits of glycoprotein hormone gene family. J. Endocrinol. 1998, 156, 529–542. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, K.; Kanamori, A.; Nagahama, Y.; Kawauchi, H. Development of salmon GTHI and GTHII radioimmunoassay. Gen. Comp. Endocrinol. 1988, 71, 459–467. [Google Scholar] [CrossRef] [PubMed]
- Swanson, P.; Bernard, M.; Nozaki, M.; Suzuki, K.; Kawauchi, H.; Dickhoff, W.W. Gonadotropin-I and Gonadotropin-II in Juvenile Coho Salmon. Fish Physiol. Biochem. 1989, 7, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Swanson, P.; Suzuki, K.; Kawauchi, H.; Dickhoff, W.W. Isolation and characterization of two coho salmon gonadotropins GTHI and GTHII. Biol. Reprod. 1991, 44, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Govoroun, M.; Chyb, J.; Breton, B. Immunological cross-reactivity between rainbow trout GTHI and GTHII and their α and β subunits: Application to the development of specific radioimmunoassay. Gen. Comp. Endocrinol. 1998, 111, 28–37. [Google Scholar] [CrossRef] [PubMed]
- Molés, G.; Gómez, A.; Rocha, A.; Carrillo, M.; Zanuy, S. Purification and characterization of follicle-stimulating hormone from pituitary glands of sea bass (Dicentrarchus labrax). Gen. Comp. Endocrinol. 2008, 158, 68–76. [Google Scholar] [CrossRef] [PubMed]
- Molés, G.; Gómez, A.; Carrillo, M.; Zanuy, S. Development of a homologous enzyme-linked immunosorbent assay for European sea bass FSH. Reproductive cycle plasma levels in both sexes and in yearling precocious and non-precocious males. Gen. Comp. Endocrinol. 2012, 176, 70–78. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, A.; Ohkubo, M.; Hamaguchi, M. Development of non-competitive enzyme-linked immunosorbent assays for mummichog Fundulus heteroclitus gonadotropins—Examining seasonal variations in plasma FSH and LH levels in both sexes. Gen. Comp. Endocrinol. 2012, 178, 463–469. [Google Scholar] [CrossRef] [PubMed]
- Breton, B.; Govoroun, M.; Mikolajczyk, T. GTHI and GTHII secretion profiles during the reproductive cycle in female rainbow trout: Relationship with pituitary responsiveness to GnRH-A stimulation. Gen. Comp. Endocrinol. 1998, 111, 38–50. [Google Scholar] [CrossRef] [PubMed]
- García-López, Á.; Couto, E.; Canario, A.V.M.; Sarasquete, C.; Martínez-Rodríguez, G. Ovarian development and plasma sex steroid levels in cultured female Senegalese sole Solea senegalensis. Comp. Biochem. Physiol. A 2007, 146, 342–354. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods. 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Tian, H.; Ru, S.G.; Wang, W.; Bing, X. Effects of monocrotophos on the reproductive axis in the female goldfish (Carassius auratus). Comp. Biochem. Physiol. C 2010, 152, 107–113. [Google Scholar]
- Shi, B.; Wen, H.S.; He, F.; Dong, S.L.; Ma, S.; Chen, C.F.; Chen, X.Y.; Zhang, J.R.; Jin, G.X. Single nucleotide polymorphisms within the estrogen receptor β gene are linked with reproductive indices in Japanese fiounder, Paralichthys olivaceus. Comp. Biochem. Physiol. B 2009, 154, 62–67. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, B.; Liu, X.; Xu, Y.; Wang, S. Molecular Characterization of Three Gonadotropin Subunits and Their Expression Patterns during Ovarian Maturation in Cynoglossus semilaevis. Int. J. Mol. Sci. 2015, 16, 2767-2793. https://doi.org/10.3390/ijms16022767
Shi B, Liu X, Xu Y, Wang S. Molecular Characterization of Three Gonadotropin Subunits and Their Expression Patterns during Ovarian Maturation in Cynoglossus semilaevis. International Journal of Molecular Sciences. 2015; 16(2):2767-2793. https://doi.org/10.3390/ijms16022767
Chicago/Turabian StyleShi, Bao, Xuezhou Liu, Yongjiang Xu, and Shanshan Wang. 2015. "Molecular Characterization of Three Gonadotropin Subunits and Their Expression Patterns during Ovarian Maturation in Cynoglossus semilaevis" International Journal of Molecular Sciences 16, no. 2: 2767-2793. https://doi.org/10.3390/ijms16022767
APA StyleShi, B., Liu, X., Xu, Y., & Wang, S. (2015). Molecular Characterization of Three Gonadotropin Subunits and Their Expression Patterns during Ovarian Maturation in Cynoglossus semilaevis. International Journal of Molecular Sciences, 16(2), 2767-2793. https://doi.org/10.3390/ijms16022767