2.1. Effects of γ Irradiation on Mortality and Reproduction of Adult Mites
The goal of irradiation is not to cause severe mortality, but to prevent development or reproduction because 100% acute mortality required very high doses of irradiation that could not be tolerated by most fresh commodities [
14]. In the present study, with the results shown in
Table 1, the mortality increased significantly (
p < 0.05) after the irradiation treatment from day 1 to day 9, and the number of eggs laid by the irradiated adults decreased significantly; furthermore, these eggs did not hatch to produce the next generation. Consistent with our data, Castro
et al. (2004) reported that the mortality of
Brevipalpus chilensis increased to 46.6% when the irradiation dose was increased to 350 Gy; additionally, a decrease in the capacity for oviposition occurred when the irradiation dose was increased to 300 Gy, and
B. chilensis was not viable [
15]. Moreover, our previous research suggested that irradiation with 400 Gy from Co
60γ was an effective postharvest technique to mitigate the risk of
P. citri on citrus fruit [
6]. We investigated the possibility of using irradiation as a quarantine treatment for
P. citri; because of parthenogenesis in
P. citri females, a suitable dose of radiation without effects on the citrus fruit was measured by F
1 sterility.
Table 1.
F1 fecundity (no. of eggs), percent of egg hatch and mortality of adult P. citri irradiated with Co60γ-rays.
Table 1.
F1 fecundity (no. of eggs), percent of egg hatch and mortality of adult P. citri irradiated with Co60γ-rays.
| Treatment Dose (Gy) | 0 | 1 | 2 | 5 | 7 | 9 | 14 | Mean No. of Eggs | F1 Hatched Eggs |
|---|
| 0 | 0.67a | 2.67a | 6.00a | 10.00a | 14.00a | 46.67a | 92.67a | 1133 | 94.6 |
| 400 | 2.00a | 12.00b | 22.67b | 42.67b | 64.67b | 84.00b | 96.67a | 375 | 0 |
2.2. Effects of γ Irradiation on Adult DNA and Egg Development
The fragmentation of DNA is a common phenomenon with apoptotic cell death, which is conserved in different species, including
Drosophila melanogaster and
Bombyx mori [
16,
17]. The results of the DNA fragmentation, as determined by agarose gel electrophoresis, are illustrated in
Figure 1. A significant fragmentation of DNA was observed in the samples irradiated with more than 400 Gy of γ rays, and the DNA fragments, which varied in size between 250 and 2500 base pairs, were clearly visible after the electrophoresis. Apoptosis is well known to be an orderly cellular process of suicide in response to various stimuli [
18]. It has been reported that ionizing radiation could cause cellular response in relation to the genes that mediated the complex regulatory pathways including cell cycle, apoptosis, and DNA repair [
19]. Irradiation causes double-stranded breaks in the DNA, which results in three well-studied cellular responses: the regulation of the cell cycle by checkpoints, DNA repair and apoptosis [
20]. Our results showed that the 400 Gy of γ rays caused serious damage to the chromosomes of
P. citri and that the chromosome fragments might lead to delays in cell division or may even cause some cells to stop dividing or die.
Figure 1.
DNA fragmentation was examined with agarose gel electrophoresis. 1 represents the irradiated mites, and 2 represents the normal mites.
Figure 1.
DNA fragmentation was examined with agarose gel electrophoresis. 1 represents the irradiated mites, and 2 represents the normal mites.
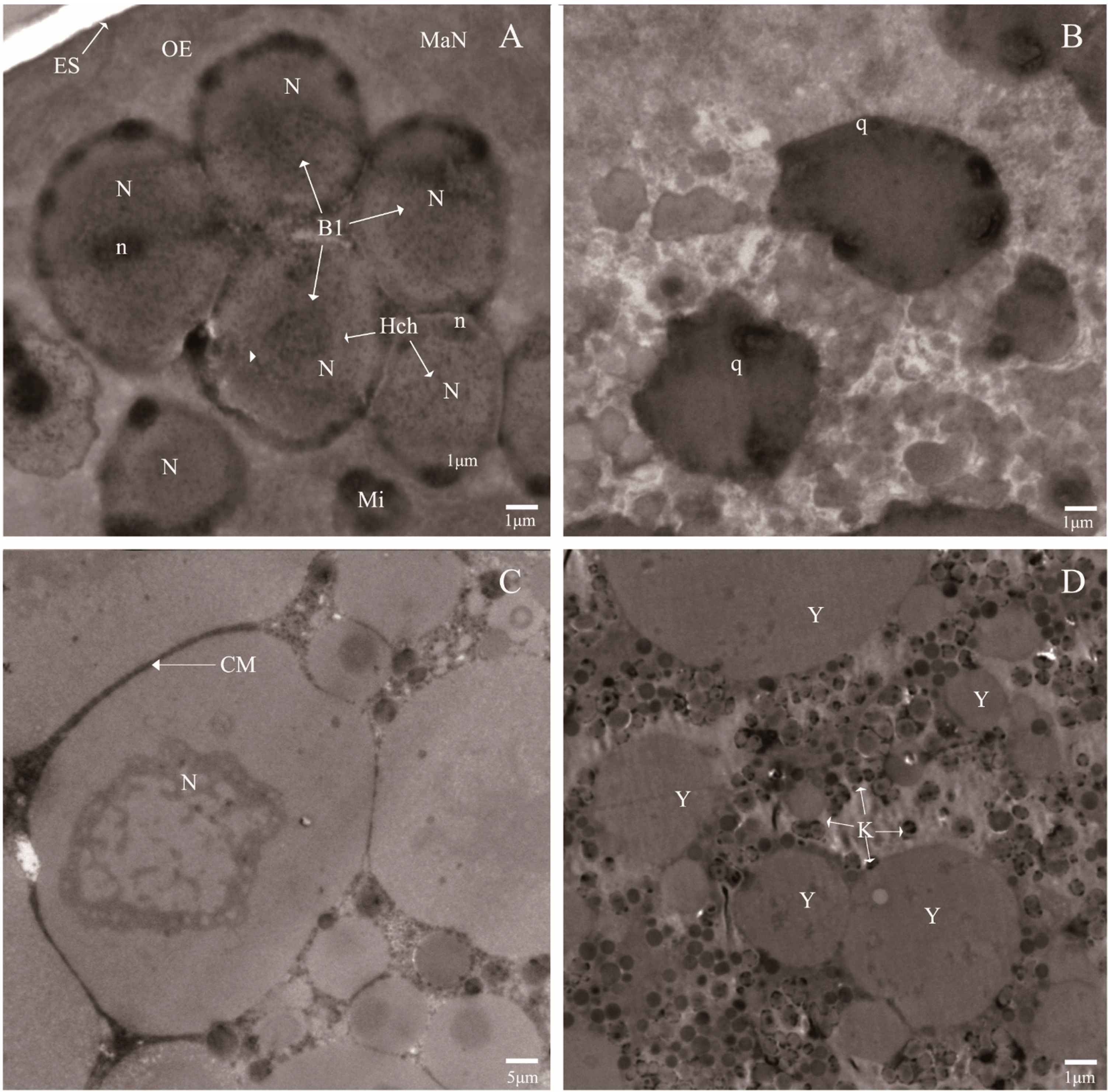
Compared with the normal eggs, the ultra-structural investigation showed that variations occurred in the eggs laid by the irradiated adults. As shown in
Figure 2B,D, the plasma membrane was destroyed, which resulted in cytoplasmic leakage and the overflowed cytoplasm forming the gathered yolk, and the nuclei contain compact granular masses, which may be formed by chromatin associated with the nuclear envelope. However, the normal development of eggs were shown well-organized morphology and cell membrane system. Moreover, the macromere (MaN) was found at the position of newly formed OE (outer envelope), and the other blastomere nuclei well aligned with the micromere (Mi) and possessed different particle sizes (
Figure 2A,C). It was reported that high-dose irradiation easily causes complete egg sterility [
21]. Our data were consistent with this observation and demonstrated that high-dose irradiation damaged the endomembrane system of the eggs of
P. citri, particularly the plasma membrane, which led to the condensed nucleoli and the gathered yolk.
Apoptosis is the process of programmed cell death that depends on the expression of many gene products, such as caspases [
22]. Although this study did not examine the characteristics of apoptosis in preimplantation embryos, the traits that were exhibited were fully along with the process of programmed cell death. A previous study confirmed that some types of cell deaths were found in the preimplantation embryos, particularly during the postcompaction stages, and some types of blastomere DNA fragmentation and embryonic cell cycle arrest were agreed with an autophagic mode of cell death; thus, the regulation of this phenomenon by a subset of genetic and biochemical events that controlled apoptosis was supported by this study [
23].
Figure 2.
Ultra-structural features of the eggs laid by the irradiated mites (B,D) and the normal mites (A,C). Blastomeres (Bl); Nuclei (N); Outer envelope (OE); Micromere (Mi); Egg shell (ES); Nucleolus (n); macromere (MaN); Cell membrane (CM); Apoptotic body (q); Heterochromatin islands (Hch); Degenerating nucleoli (k); and Gathered yolk (Y).
Figure 2.
Ultra-structural features of the eggs laid by the irradiated mites (B,D) and the normal mites (A,C). Blastomeres (Bl); Nuclei (N); Outer envelope (OE); Micromere (Mi); Egg shell (ES); Nucleolus (n); macromere (MaN); Cell membrane (CM); Apoptotic body (q); Heterochromatin islands (Hch); Degenerating nucleoli (k); and Gathered yolk (Y).
2.3. An Overview of the Transcriptome of P. citri
The total number of the Illumina sequencing short reads was 78,795,170 (
Table 2, NCBI SRA No. SRX800412), and 60,619 contigs were assembled with a mean length of 636 bp. 54,595 unigenes were further assembled by using the paired-end reads and Trinity, with a mean length of 604 bp. The assembly generated a large number of contigs, including 10,708 contigs were >1000 bp, and 8644 unigenes were >1000 bp. Of note, the assembly and annotation of different
P. citri cDNA libraries resulted in different data outputs. For example, a report on the transcriptomes of resistant and susceptible strains of
P. citri found that the number of unigenes was 34,159 and 30,466 and that the mean length of the unigenes was 489 and 536 bp, respectively [
3]. All of the assembled and annotated contigs were deposited in the NCBI with the BioProject ID: SUB773748.
Table 2.
Summary for P. citri transciptome.
Table 2.
Summary for P. citri transciptome.
| | Total Number | Total Nucleotides (nt) | Average Length (bp) | N50 |
|---|
| Reads | 78,795,170 | 7,879,517,000 | - | - |
| Contigs | 60,619 | 38,562,819 | 636 | 932 |
| Unigenes | 54,595 | 33,003,960 | 604 | 868 |
Based on searching the unigenes using BLASTx against Nr, Swiss-Port, COG and KEGG, 29,904 unigenes were annotated with a ratio of 54.77% in all the unigenes. Furthermore, the unigene sequences were evaluated by the Clusters of Orthologous Groups (COG) classification. Of the 28,872 Nr hits, 11,356 sequences were shown a COG classification (
Figure S1). Among the 25 COG categories, the cluster for “General function prediction only” (3695; 32.54%) was the largest group, followed by “Carbohydrate transport and metabolism” (2134; 18.79%), “Transcription” (1931; 17.00%), “Translation, ribosomal structure and biogenesis” (1832; 16.13%), and “Posttranslational modification, protein turnover, chaperones” (1630; 14.35%). The categories “extracellular structures” (25; 0.22%) and “nuclear structure” (13; 0.11%) had the fewest corresponding genes. Apoptosis is generally considered as a process of programmed cell death, which associates with the environmentally determined elimination of cells [
24]. In this study, the sequences associated with pathways that could be identified are presented in
Table 3. The cell cycle and apoptosis play important roles in the genetic networks that explain the radiation induced sterility, such as the p53 signaling pathway, which was investigated with 84 unigenes in relation to the high-dose radiation on
P. citri (
Table 3 and
Table S1). As reported, the p53 pathway played a critical role in regulating the response to DNA damage, which led to micronucleus formation in chronically irradiated cells [
25].
Table 3.
Selected pathways are enriched and involved in the pathways of apoptosis, cell death and the cell cycle induced by the irradiation of P. citri.
Table 3.
Selected pathways are enriched and involved in the pathways of apoptosis, cell death and the cell cycle induced by the irradiation of P. citri.
| Pathway | All Genes with Pathway | Pathway ID |
|---|
| Annotation (16,065) |
|---|
| P53 signaling pathway | 84 (0.52%) | ko04115 |
| Apoptosis | 108 (0.67%) | ko04210 |
| Regulation of autophagy | 39 (0.24%) | ko04140 |
| MAPK signaling pathway | 434 (2.7%) | ko04010 |
| Calcium signaling pathway | 296 (1.84%) | ko04020 |
| Protein processing in endoplasmic reticulum | 488 (3.04%) | ko04141 |
| Lysosome | 474 (2.95%) | ko04142 |
| Cell cycle | 296 (1.84%) | ko04110 |
| Wnt signaling pathway | 158 (1.61%) | ko04310 |
| Chemokine signaling pathway | 243 (1.51%) | ko04062 |
| Neurotrophin signaling pathway | 222 (1.38%) | ko03015 |
When the unigenes were blasted against the NCBI nonredundant (nr) database using BLASTX with a cutoff e-value of 10
−5. Of the 54,595 unigenes, 28,872 (52.9%) returned at least one match with the
e-value >10
−5. As there is lack of genome and EST information for
P. citri, 47.1% of the unigenes could not be matched to any known genes in the Nr (Non-redundant) database (Available online:
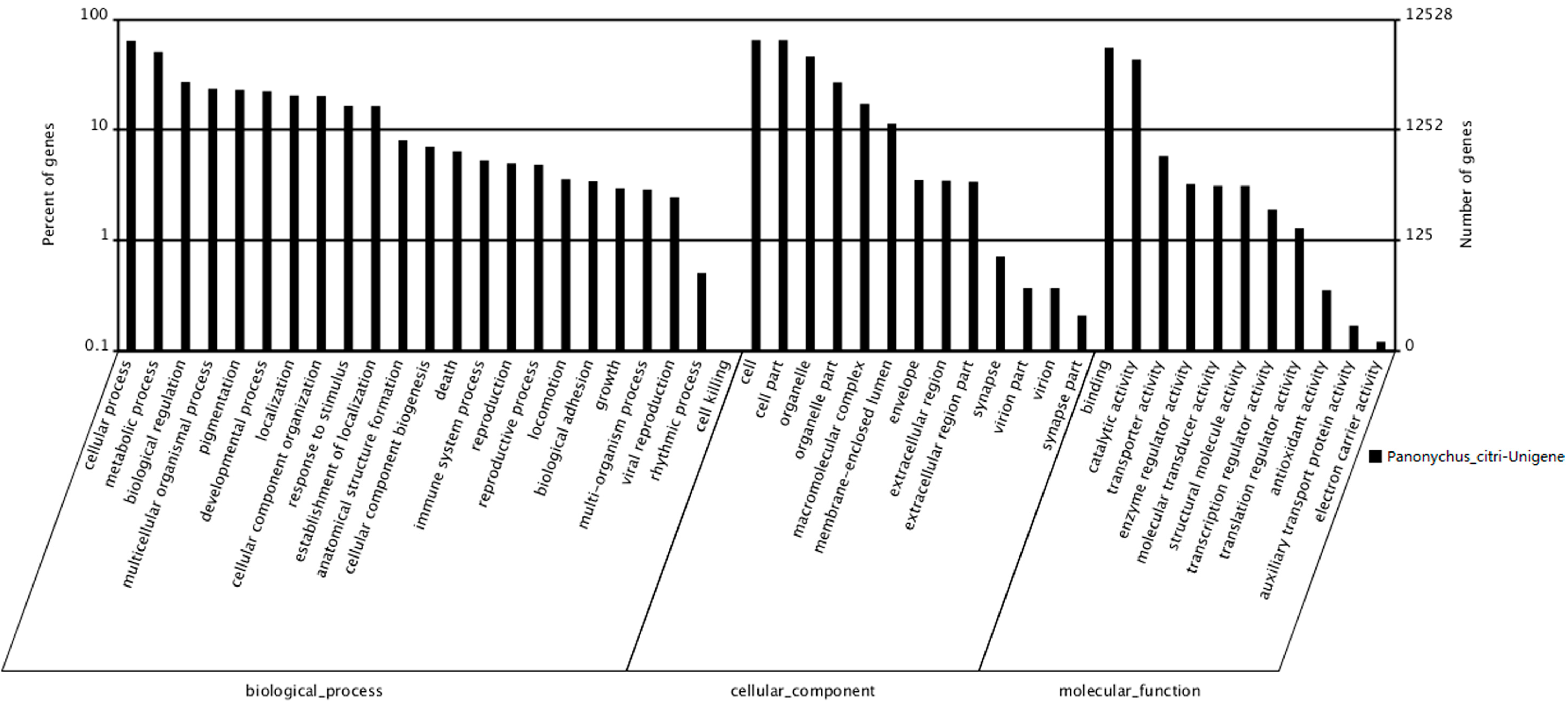
http://blast.ncbi.nlm.nih.gov/Blast.cgi). According to sequence homology, 12,528 unigenes were categorized into 47 Gene Ontology (GO) terms (
Figure 3). The results showed that among the three categories, the terms cellular progress and metabolic progress occurred most frequently in the ontology of biological processes, whereas the terms cell, cell part and organelle and the terms binding and catalytic activity occurred most frequently in the ontology of cellular components and molecular functions, respectively. The response to stimulus, death, reproduction and viral reproduction were highly represented groups in the biological process category. In the “cellular process” category, 8015 unigenes were annotated, which suggested that our study identified novel genes involved in the cell cycle. The cell and organelle parts were the dominant groups in the “cellular component function” category. Though a high percentage of the genes in the “molecular function” category were from the “binding” (55.27%) and the “catalytic activity” (43.37%) groups. In the “cell killing” groups, only a few (10; 0.08%) unigenes were annotated, which suggested that the 400 Gy of irradiation caused damage to the cell before death (
Table 3). Compared with a similar study on the
P. citri transcriptome, the number of unigenes annotated against nr, KEGG, COG and GO were 32,535, 22,725, 15,412 and 10,421, respectively in which the COG annotations were involved in 25 molecular families, including such as general function prediction, translation, ribosomal structure and biogenesis [
23].
Figure 3.
Gene Ontology classifications of assembled unigenes. A total of 12,528 unigenes were categorized into three main categories: biological process, cellular component and molecular function.
Figure 3.
Gene Ontology classifications of assembled unigenes. A total of 12,528 unigenes were categorized into three main categories: biological process, cellular component and molecular function.
2.4. Profiles of Transcript Abundance through Time Intervals
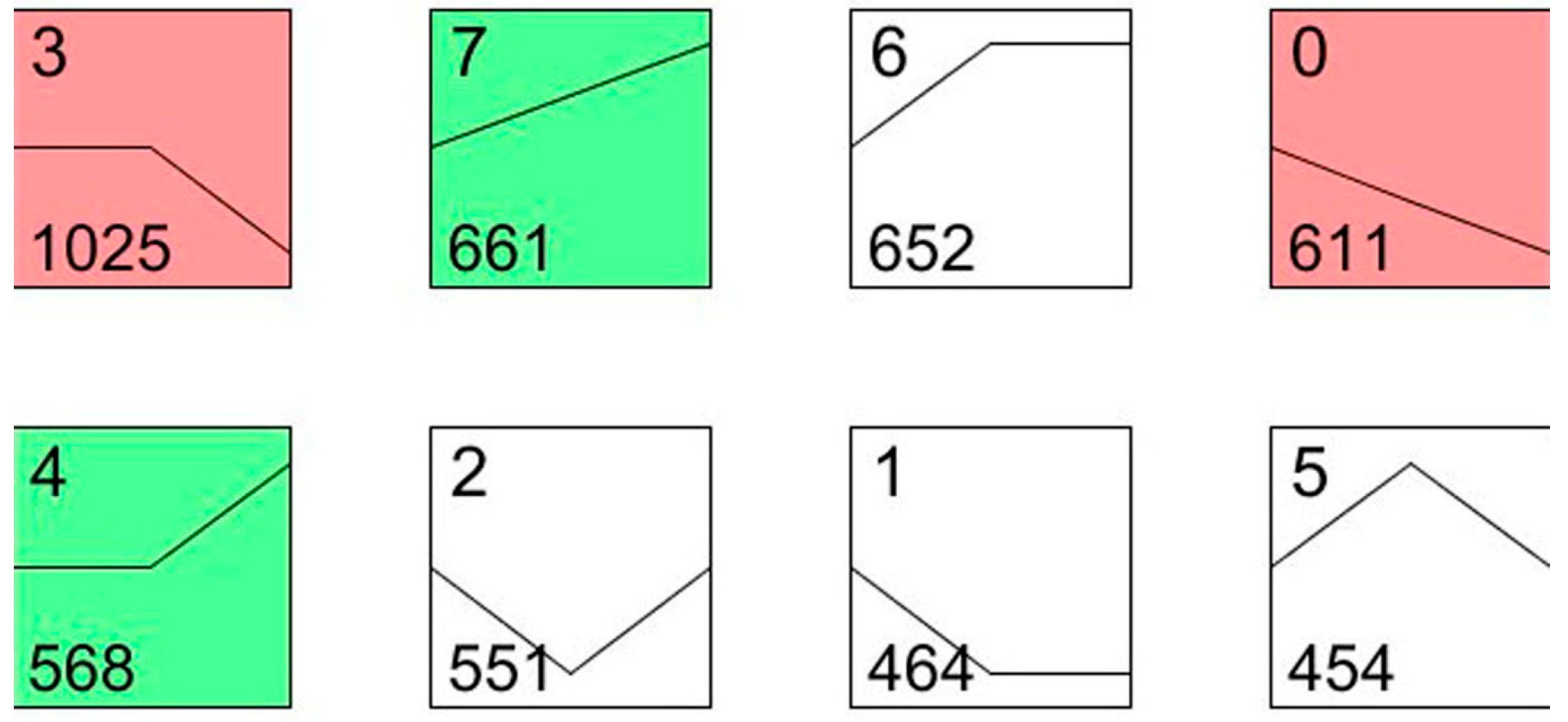
To understand the dynamic biological process of transcript abundance at two and five days after the irradiation treatment, the expression patterns of all differentially displayed genes were further analyzed using the expression level of the patterns considered as a common reference. According to this analysis, the 4986 differentially expressed genes were grouped into eight profiles using STEM (Short Time-series Expression Miner).
As shown in
Figure 4, profile 0 and profile 7 showed a monotonic decrease and increase, respectively, among the three time intervals. The genes that coded for chitinase (Unigene002740), cytochrome P450 (Unigene0012520) and ubiquitin hydrolase (Unigene0020738) were included in profile 0, and the gene that coded for the apoptosis regulator BAX (Unigene0053037) was included in profile 7. The most represented pattern was a mass increase after the irradiation from day 2 to day 5 (1025 genes, profile 3), which included the transcripts involved in ubiquinol-cytochrome c (Unigene0015549), p38 MAP kinase (Unigene0013647), and cytochrome P450 (Unigene0026480). The transcripts within profile 4 presented a curve in which the expression of 568 genes increased from day 2 to day 5, including caspase 7 (Unigene0023555) and autophagy-related protein 4 (Unigene0024949). Most of these genes are involved in apoptosis, cell death and the cell cycle. For example, caspase 7 was reported to be involved in the cleavage of claspin during apoptosis, which inhibited the Chk1 pathway [
26]. Cytochrome P450 is a large family of genes in insects, which plays important roles in insecticide resistance and hormone metabolism. In our study, these P450 genes, such as Unigene0026480 and Unigene0012520, were expected to be involved in the
P. citri defense against the environmental stress caused by the irradiation treatment.
Figure 4.
STEM (Short Time-series Expression Miner) clusters of expression profiles. The profiles are ordered based on the p-value significance of the number (at bottom-left corner) of genes assigned versus expected. The colored square frame denotes significant profiles (p-value ≤ 0.01). Each graph displays the mean pattern of expression (black lines) of the profile genes. The number of profiles in each cluster is at the top left corner of each STEM and the number of genes in the bottom left corner of the panels. The x-axis represents stages, and the y-axis represents log2-fold change in gene expression.
Figure 4.
STEM (Short Time-series Expression Miner) clusters of expression profiles. The profiles are ordered based on the p-value significance of the number (at bottom-left corner) of genes assigned versus expected. The colored square frame denotes significant profiles (p-value ≤ 0.01). Each graph displays the mean pattern of expression (black lines) of the profile genes. The number of profiles in each cluster is at the top left corner of each STEM and the number of genes in the bottom left corner of the panels. The x-axis represents stages, and the y-axis represents log2-fold change in gene expression.
2.5. Apoptosis Pathway Assignment by KEGG
Apoptosis is a type of programmed cell death, and many cells undergo such a programmed cell death during normal development or aging via a kind of homeostatic mechanism to keep cell populations in tissues. Apoptosis also serves as a defense responses in relation to the immune mechanism while cells were damaged by disease or noxious agents. It is not all cells have to die in response to multitudinous physiological and pathological stimulus. For example, irradiation or drugs used for pest control caused DNA damage in some cells, which led to apoptotic death through the MAPK signaling pathways or a p53-dependent pathway [
27].
In our study, 54,595 unigenes were annotated by KEGG, in which 23,092 unigenes were assigned to 275 KEGG pathways. The MAPK signaling pathways had the most representation among the unigenes (434 members, 2.7%), which was followed by apoptosis (108 members, 0.67%). These identified pathways provide a valuable resource to investigate specific genes, functional annotations and pathways for research on
P. citri damaged by irradiation. Notably, 1397 unigenes involved in apoptosis were confirmed including 11 pathways (
Table 3). The p53 is an important factor in the control of insect physiology and an important part of the apoptosis pathway [
27]. The induction of p53 occurs in response to a range of genotoxic or nongenotoxic stresses, which result in the biological outcomes of arrested growth or apoptosis. In the present study, the p53 signaling pathway was found in the KEGG pathway that involved 84 (0.52%) unigenes. Details of the p53 signaling pathway are shown in
Figure S2. This pathway will be given promising for further studies on the effects of irradiation-related processes on the pathway leading to sterility in
P. citri.
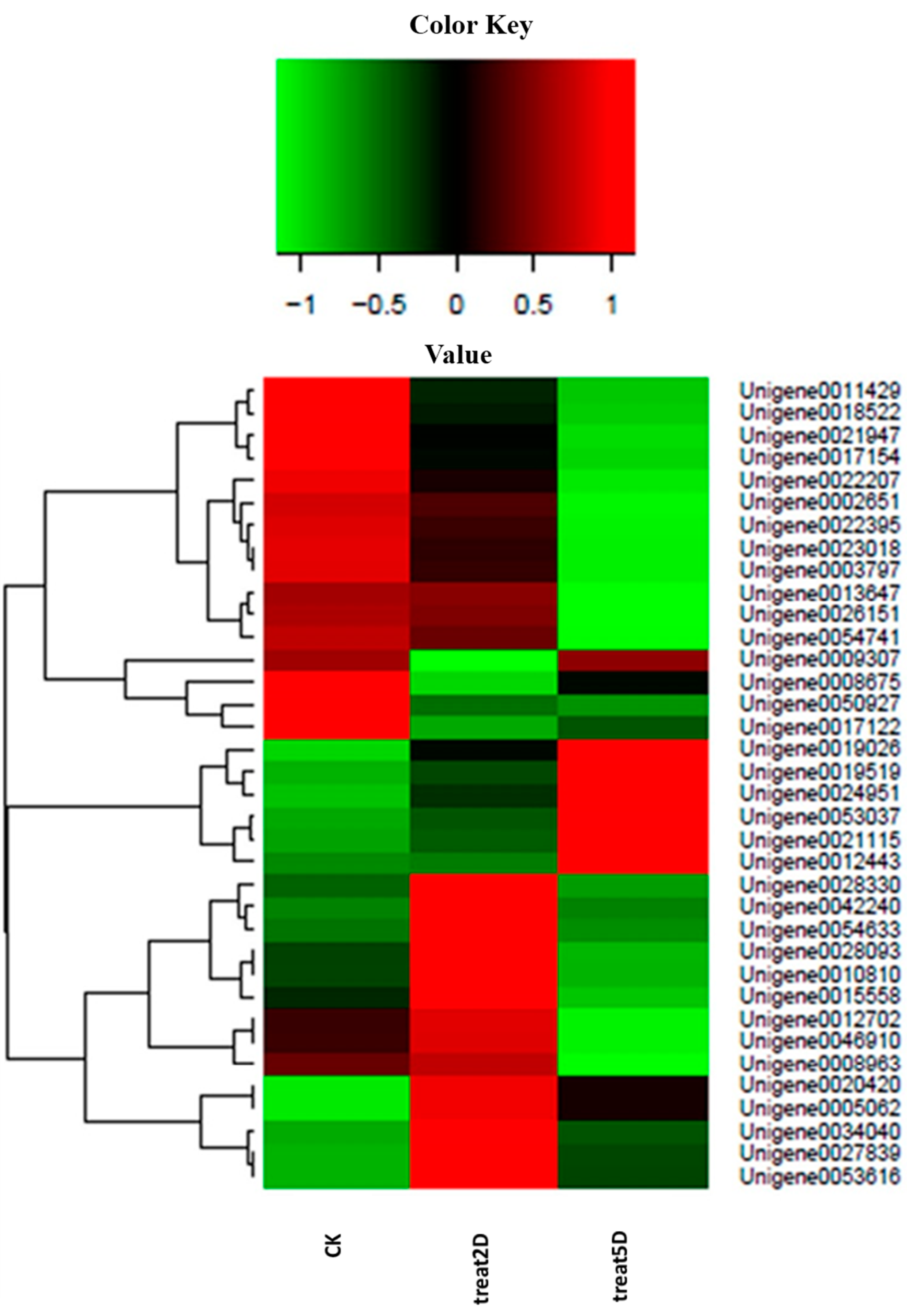
Figure 5.
Differential expression of genes related with terms in the P. citri STEM. Each column represents an experimental sample (e.g., CK, 2D and 5D), and each row represents a gene. Differences in expression are shown in different colors. Red indicates high expression, and green indicates low expression.
Figure 5.
Differential expression of genes related with terms in the P. citri STEM. Each column represents an experimental sample (e.g., CK, 2D and 5D), and each row represents a gene. Differences in expression are shown in different colors. Red indicates high expression, and green indicates low expression.
To improve our knowledge on the gene expression obtained from the
P. citri transcriptome, three DEG libraries were created to analyze the gene expression profiles; a large amount of genes showed significantly changed expression and involvement in the radiation-induced sterility. These genes were related to catalytic activity, autophagy, cellular response to stimuli, oxidoreductase activity, embryo development, hydrolase activity, and signal transduction. Furthermore, these genes had significantly different expression patterns among the three time intervals (
Figure 5). The assumption was that these genes might be induced independently of a p53 pathway at day 2 after irradiation, but the levels of expression increased at day 5, which might contribute to the delayed apoptotic response. For example, Unigene0042240 showed higher expression levels day 2 after irradiation compared with the control group and day 5 after irradiation; this result was homologous to MDM2 because an E3 ubiquitin ligase directed p53 ubiquitylation and degradation [
28]. Additionally, Unigene0020420 and Unigene0053037 showed higher levels of expression at day 2 and day 5 than in the CK, which were functional for stimuli (Unigene0053037 was homologous to Bax, which was activated in response to radiation) [
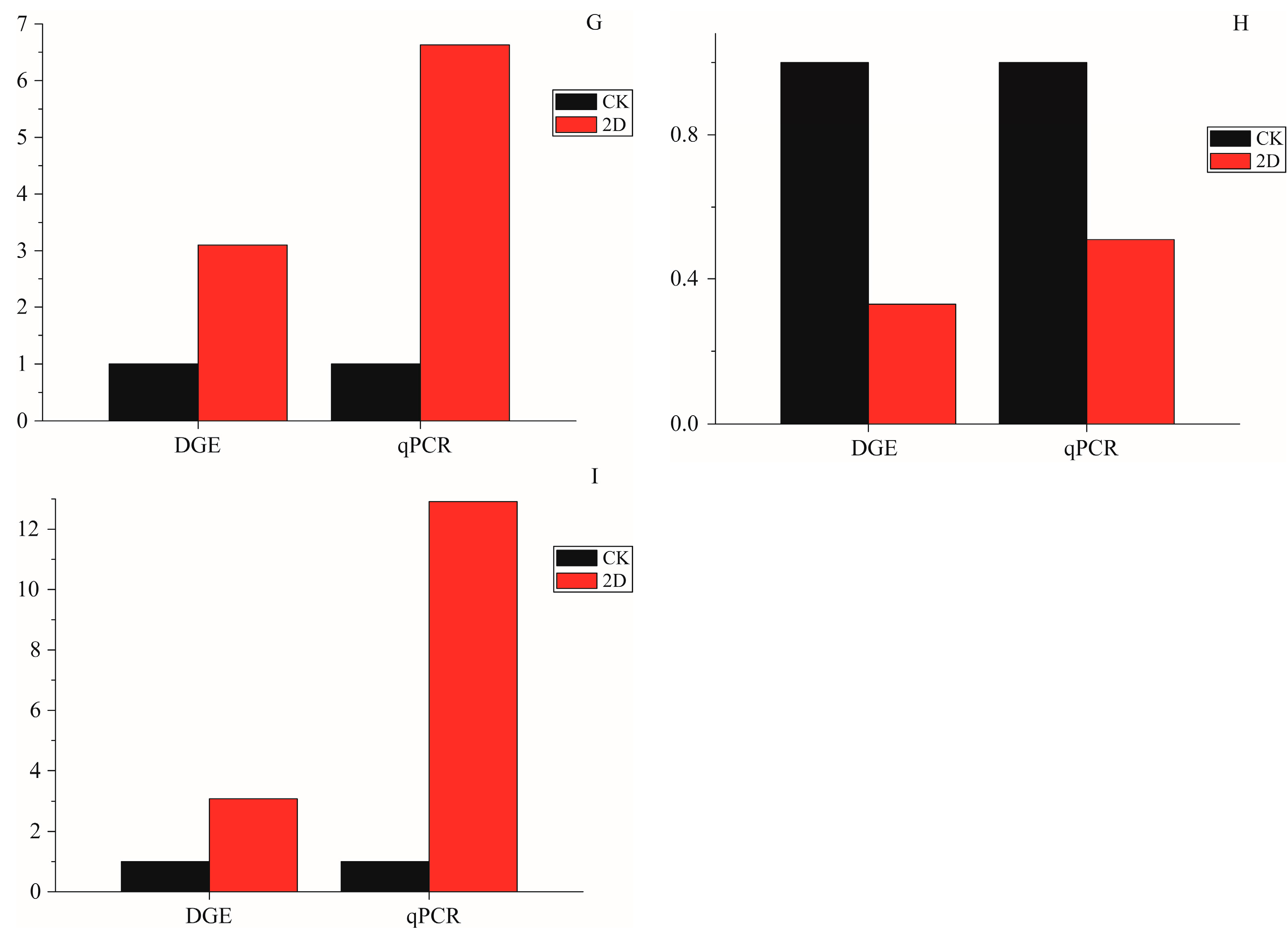
29]. Other genes with significantly different levels of expression in the three time intervals were also annotated, such as Laminin subunit α (Unigene0026151), hydrolase activity (Unigene0008675 and 0017122), catalytic activity (Unigene0017154, 0003797, and -0054633), oxidoreductase activity (Unigene0012443 and 0019026), and autophagy (Unigene0021115 and 0024951). To verify a subset of the DGE data, the qRT-PCR analyses were performed. The expression of 9 genes that were annotated in apoptosis-related pathways were studied using qRT-PCR. The results showed a similar direction of expression induced by irradiation between the DGE and the qRT-PCR results (
Figure 6), and the qRT-PCR analysis confirmed the same tendency of expression detected by the DGE analysis.
Until now, in sterile P. citri, no study examined which genetic program was affected that regulated a vital process, such as cell division, differentiation or apoptosis. This study provides the first evidence that apoptosis operates and is induced by γ radiation in P. citri. Although the detailed functions of genes in relation to the genetic pathways of sterility in P. citri remain unknown, the current transcriptome analysis provides valuable gene sequence information on P. citri sterility, which may facilitate further investigations into the details of sterility caused by irradiation.
Figure 6.
The qPCR data for the expression pattern validation of selected genes. The x-axis indicates treatment, and the y-axis indicates relative expression level. The following selection of genes was tested, with their descriptions. (A) Apolipoprotein D (Unigene0009307); (B) Calmodulin-like (Unigene0019519); (C) Breakpoint cluster region protein (Unigene0022395); (D) Serine protease (Unigene0017154); (E) Intestinal-type alkaline phosphatase 1 (Unigene0015558); (F) Cu/Zn superoxide dismutase (Unigene0054633); (G) Microtubule-associated proteins (Unigene0021115); (H) DNA repair endonuclease XPF-like (Unigene0022207); and (I) autophagy-related 4 (Unigene0024951).
Figure 6.
The qPCR data for the expression pattern validation of selected genes. The x-axis indicates treatment, and the y-axis indicates relative expression level. The following selection of genes was tested, with their descriptions. (A) Apolipoprotein D (Unigene0009307); (B) Calmodulin-like (Unigene0019519); (C) Breakpoint cluster region protein (Unigene0022395); (D) Serine protease (Unigene0017154); (E) Intestinal-type alkaline phosphatase 1 (Unigene0015558); (F) Cu/Zn superoxide dismutase (Unigene0054633); (G) Microtubule-associated proteins (Unigene0021115); (H) DNA repair endonuclease XPF-like (Unigene0022207); and (I) autophagy-related 4 (Unigene0024951).