ThWRKY4 from Tamarix hispida Can Form Homodimers and Heterodimers and Is Involved in Abiotic Stress Responses
Abstract
:1. Introduction
2. Results and Discussion
2.1. Analysis of the Hetero- and Homo-Dimers of ThWRKY4
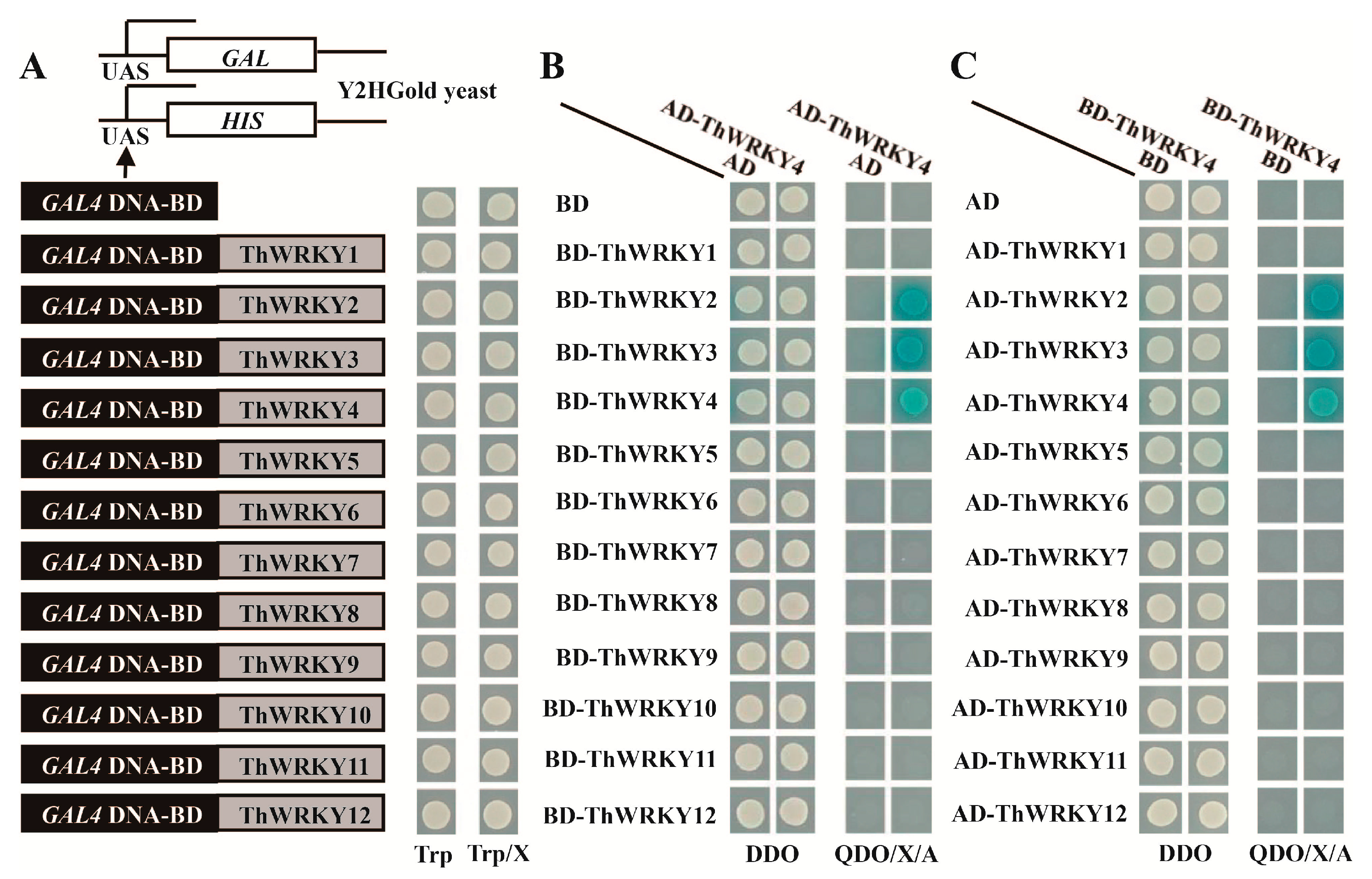
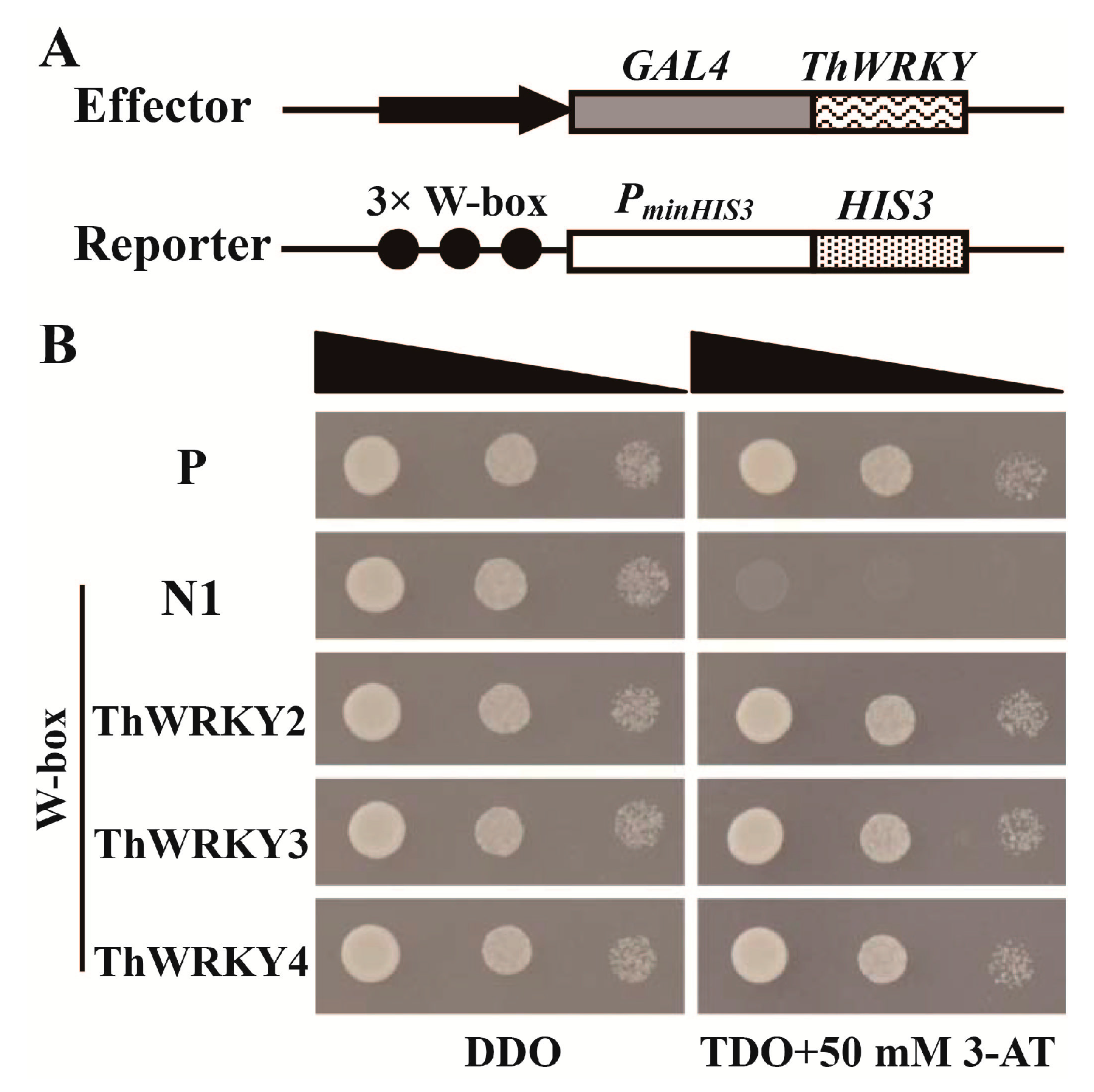
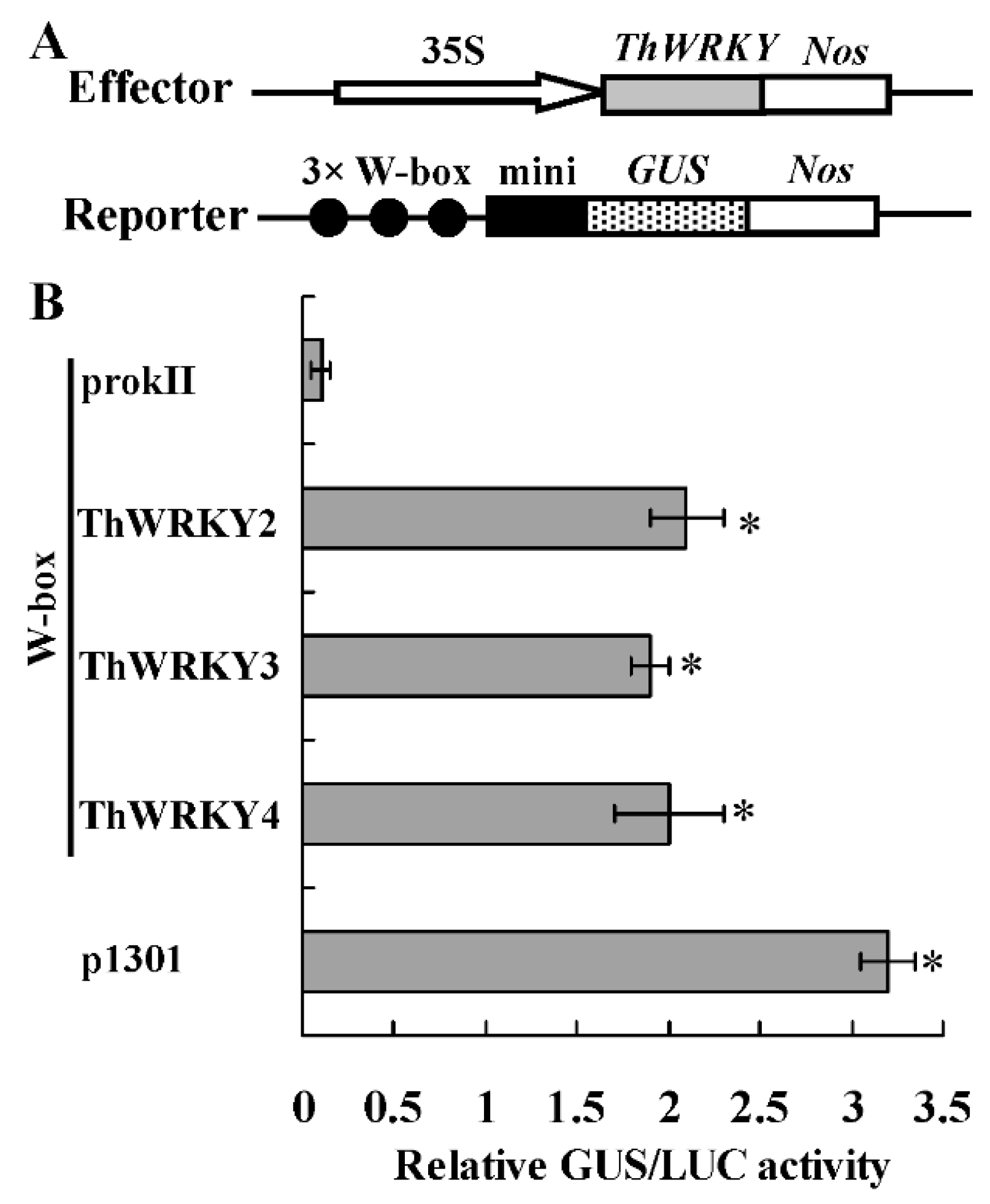
2.2. Analysis of the Binding of ThWRKY2 and ThWRKY3 to the W-Box Motif
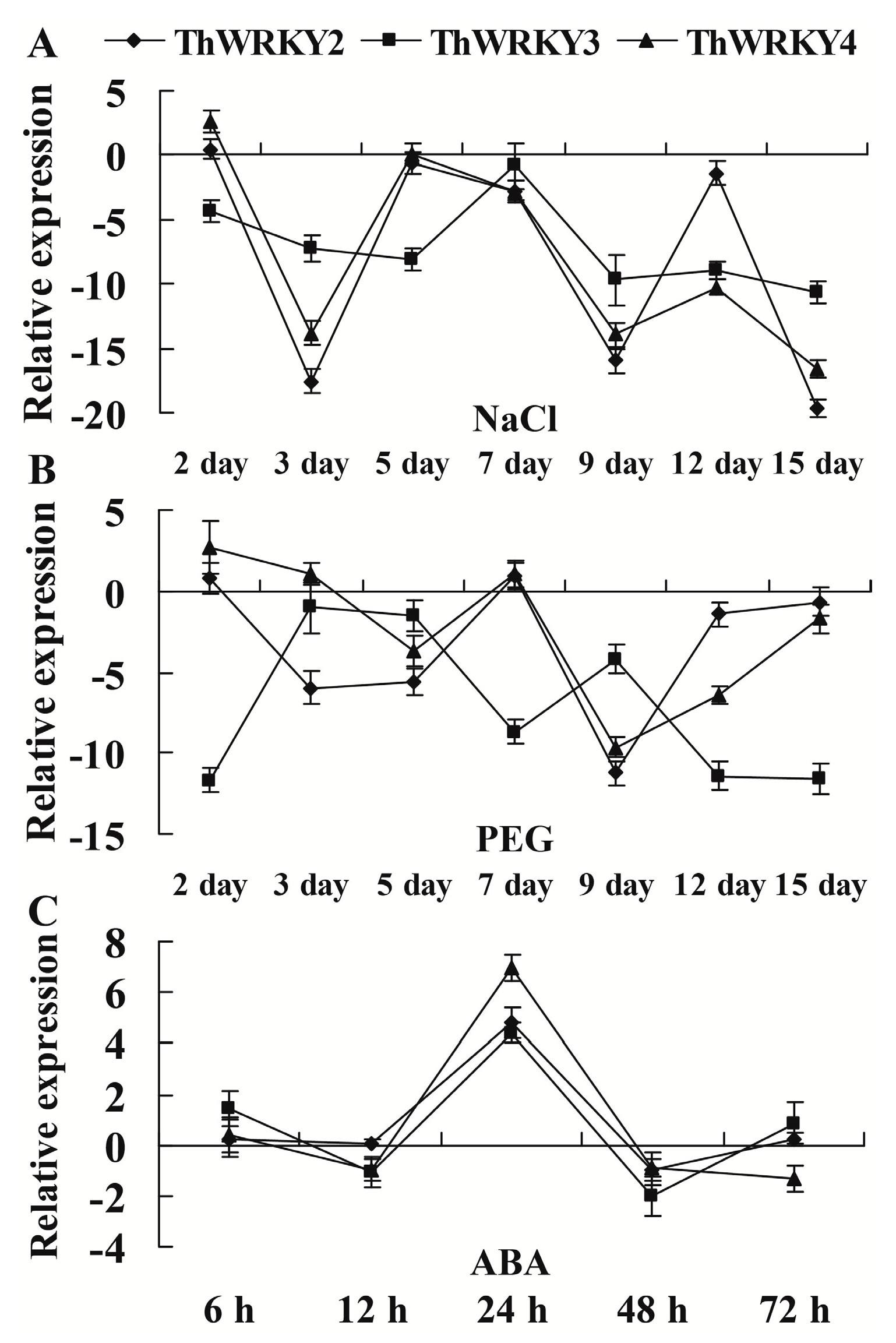
2.3. Expression of ThWRKY2, 3, and 4 in Response to Abiotic Stresses
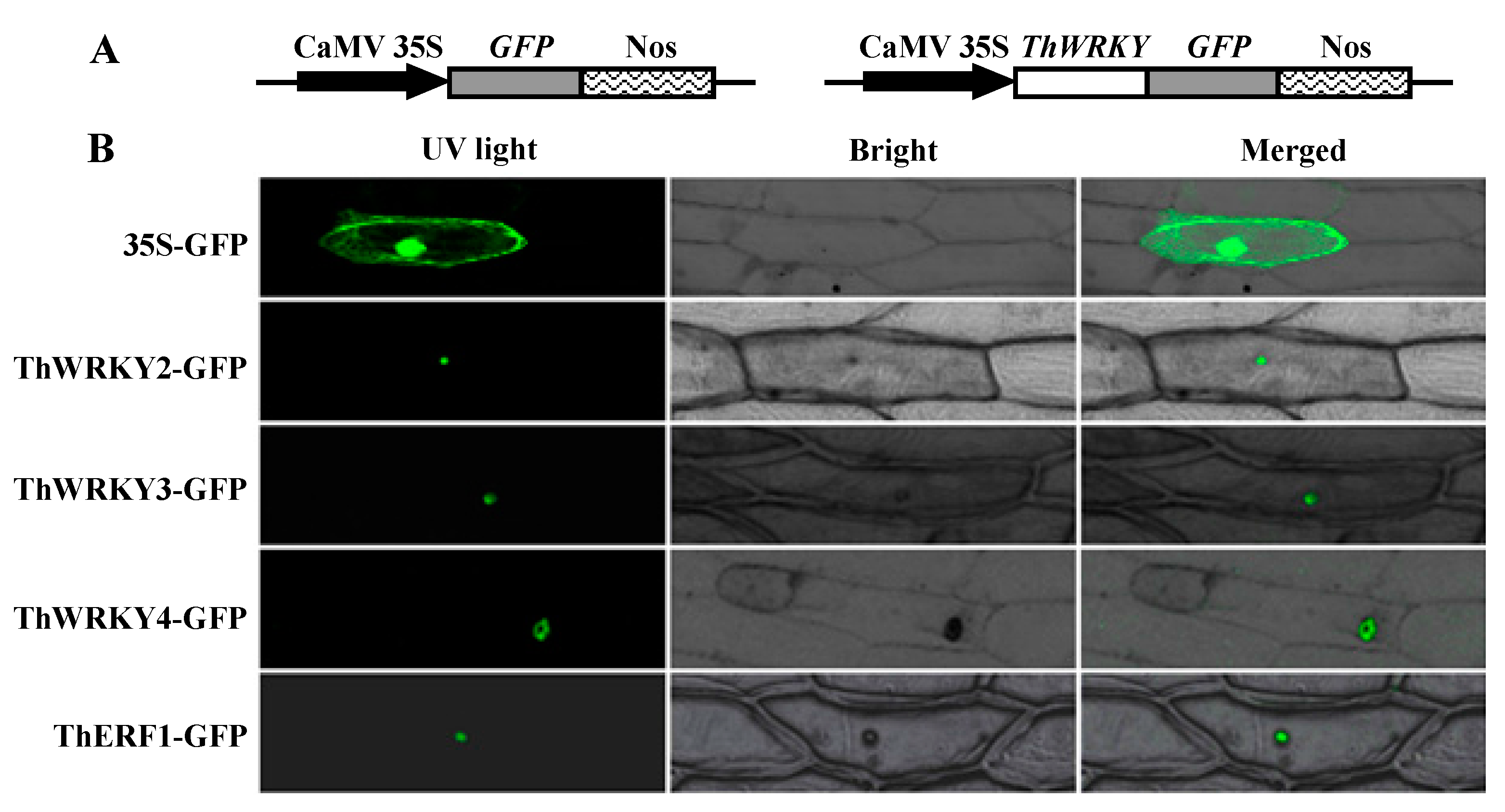
2.4. Subcellular Localization of ThWRKY2, ThWRKY3, and ThWRKY4
3. Experimental Section
3.1. Plant Materials and Growth Conditions
3.2. Cloning the ThWRKY Gene Sequences
3.3. Construction of the Yeast Expression Vector and Yeast Two-Hybrid Analysis
3.4. Yeast One-Hybrid Assays
3.5. Transient Expression Assays
3.6. Real-Time RT-PCR Analyses
3.7. Subcellular Localization Analysis
3.8. Statistical Analysis
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Rushton, P.J.; Somssich, I.E.; Ringler, P.; Shen, Q.J. WRKY transcription factors. Trends Plant Sci. 2010, 15, 247–258. [Google Scholar] [CrossRef] [PubMed]
- Eulgem, T.; Rushton, P.J.; Robatzek, S.; Somssich, I.E. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000, 5, 199–206. [Google Scholar] [CrossRef]
- Rushton, D.L.; Tripathi, P.; Rabara, R.C.; Lin, J.; Ringler, P.; Boken, A.K.; Langum, T.J.; Smidt, L.; Boomsma, D.D.; Emme, N.J.; et al. WRKY transcription factors: Key components in abscisic acid signalling. Plant Biotechnol. J. 2012, 10, 2–11. [Google Scholar] [CrossRef] [PubMed]
- Ling, J.; Jiang, W.J.; Zhang, Y.; Yu, H.J.; Mao, Z.C.; Gu, X.F.; Huang, S.W.; Xie, B.Y. Genome-wide analysis of WRKY gene family in Cucumis sativus. BMC Genom. 2011, 12, 471. [Google Scholar] [CrossRef] [PubMed]
- Ma, D.; Pu, G.; Lei, C.; Ma, L.; Wang, H.; Guo, Y.; Chen, J.; Du, Z.; Wang, Z.; Li, G.; et al. Isolation and characterization of AaWRKY1, an Artemisia annua transcription factor that regulates the amorpha-4, 11-diene synthase gene, a key gene of artemisinin biosynthesis. Plant Cell Physiol. 2009, 50, 2146–2161. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhu, Y.; Wang, L.L.; Liu, X.; Liu, Y.X.; Phillips, J.; Deng, X. A WRKY transcription factor participates in dehydration tolerance in Boea hygrometrica by binding to the W-box elements of the galactinol synthase (BhGolS1) promoter. Planta 2009, 230, 1155–1166. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Xu, C.; Wang, C.; Wang, Y. Characterization of a eukaryotic translation initiation factor 5A homolog from Tamarix androssowii involved in plant abiotic stress tolerance. BMC Plant Biol. 2012, 12, 118. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Liu, G.; Meng, X.; Liu, Y.; Ji, X.; Li, Y.; Nie, X.; Wang, Y. A WRKY gene from Tamarix hispida, ThWRKY4, mediates abiotic stress responses by modulating reactive oxygen species and expression of stress-responsive genes. Plant Mol Biol. 2013, 82, 303–320. [Google Scholar] [CrossRef] [PubMed]
- Miao, Y.; Zentgraf, U. A HECT E3 ubiquitin ligase negatively regulates Arabidopsis leaf senescence through degradation of the transcription factor WRKY53. Plant J. 2010, 63, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Liang, G.; Yang, S.; Yu, D. Arabidopsis WRKY57 functions as a node of convergence for jasmonic acid- and auxin-mediated signaling in jasmonic acid–induced leaf senescence. Plant Cell 2014, 26, 230–245. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Sun, X.; Liu, B.; Zhu, D.; Bai, X.; Cai, H.; Ji, W.; Cao, L.; Wu, J.; Wang, M.; et al. Ectopic expression of a WRKY homolog from Glycine soja alters flowering time in Arabidopsis. PLoS ONE 2013, 8, e73295. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Chen, X.; Xie, K.; Xing, Q.; Wu, Y.; Li, J.; Du, C.; Sun, Z.; Guo, Z. Dlf1, a WRKY transcription factor, is involved in the control of flowering time and plant height in rice. PLoS ONE 2014, 9, e102529. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.S.; Kolevski, B.; Smyth, D.R. TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. Plant Cell 2002, 14, 1359–1375. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Yu, D. Arabidopsis WRKY2 transcription factor mediates seed germination and postgermination arrest of development by abscisic acid. BMC Plant Biol. 2009, 9, 96. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.Q.; Xu, Y.; Lu, Y.; Yu, H.X.; Gu, M.H.; Liu, Q.Q. The WRKY transcription factor OsWRKY78 regulates stem elongation and seed development in rice. Planta 2011, 234, 541–554. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.; Qamar, S.A.; Chen, Z.; Mengiste, T. Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J. 2006, 48, 592–605. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhang, L.; Li, D.; Wang, F.; Yu, D. WRKY8 transcription factor functions in the TMV-cg defense response by mediating both abscisic acid and ethylene signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 2013, 110, 1963–1971. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.Y.; Tian, A.G.; Zou, H.F.; Xie, Z.M.; Lei, G.; Huang, J.; Wang, C.M.; Wang, H.W.; Zhang, J.S.; Chen, S.Y. Soybean WRKY-type transcription factor genes, GmWRKY13, GmWRKY21, and GmWRKY54, confer differential tolerance to abiotic stresses in transgenic Arabidopsis plants. Plant Biotechnol. J. 2008, 6, 486–503. [Google Scholar] [CrossRef] [PubMed]
- Niu, C.F.; Wei, W.; Zhou, Q.Y.; Tian, A.G.; Hao, Y.J.; Zhang, W.K.; Ma, B.; Lin, Q.; Zhang, Z.B.; Zhang, J.S.; et al. Wheat WRKY genes TaWRKY2 and TaWRKY19 regulate abiotic stress tolerance in transgenic Arabidopsis plants. Plant Cell Environ. 2012, 35, 1156–1170. [Google Scholar] [CrossRef] [PubMed]
- Scarpeci, T.E.; Zanor, M.I.; Mueller-Roeber, B.; Valle, E.M. Overexpression of AtWRKY30 enhances abiotic stress tolerance during early growth stages in Arabidopsis thaliana. Plant Mol Biol. 2013, 83, 265–277. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Chen, Z.; Liu, Y.; Zhang, H.; Zhang, M.; Liu, Q.; Hong, X.; Zhu, J.K.; Gong, Z. ABO3, a WRKY transcription factor, mediates plant responses to abscisic acid and drought tolerance in Arabidopsis. Plant J. 2010, 63, 417–429. [Google Scholar] [CrossRef] [PubMed]
- Antoni, R.; Rodriguez, L.; Gonzalez-Guzman, M.; Pizzio, G.A.; Rodriguez, P.L. News on ABA transport, protein degradation, and ABFs/WRKYs in ABA signaling. Curr. Opin. Plant Biol. 2011, 14, 547–553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Verk, M.C.; Bol, J.F.; Linthorst, H.J. WRKY transcription factors involved in activation of SA biosynthesis genes. BMC Plant Biol. 2011, 11, 89. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Zhang, Z.L.; Zou, X.; Yang, G.; Komatsu, S.; Shen, Q.J. Interactions of two abscisic acid induced WRKY genes in repressing gibberellin signaling in aleurone cells. Plant J. 2006, 46, 231–242. [Google Scholar] [CrossRef] [PubMed]
- Chi, Y.; Yang, Y.; Zhou, Y.; Zhou, J.; Fan, B.; Yu, J.Q.; Chen, Z. Protein–protein interactions in the regulation of WRKY transcription factors. Mol. Plant 2013, 6, 287–300. [Google Scholar] [CrossRef] [PubMed]
- Cormack, R.S.; Eulgem, T.; Rushton, P.J.; Kochner, P.; Hahlbrock, K.; Somssich, I.E. Leucine zipper-containing WRKY proteins widen the spectrum of immediate early elicitor-induced WRKY transcription factors in parsley. BBA-Gen. Subj. 2002, 1576, 92–100. [Google Scholar] [CrossRef]
- Xu, X.; Chen, C.; Fan, B.; Chen, Z. Physical and functional interactions between pathogen-induced Arabidopsis WRKY18, WRKY40, and WRKY60 transcription factors. Plant Cell 2006, 18, 1310–1326. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.F.; Li, L.Q.; Xu, Q.; Kong, Y.H.; Wang, H.; Wu, W.H. The WRKY6 transcription factor modulates PHOSPHATE1 expression in response to low Pi stress in Arabidopsis. Plant Cell 2009, 21, 3554–3566. [Google Scholar] [CrossRef] [PubMed]
- Besseau, S.; Li, J.; Palva, E.T. WRKY54 and WRKY70 co-operate as negative regulators of leaf senescence in Arabidopsis thaliana. J. Exp. Bot. 2012, 63, 2667–2679. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Zhou, Y.; Yang, Y.; Chi, Y.J.; Zhou, J.; Chen, J.Y.; Wang, F.; Fan, B.; Shi, K.; Zhou, Y.H.; et al. Structural and functional analysis of VQ motif-containing proteins in Arabidopsis as interacting proteins of WRKY transcription factors. Plant Physiol. 2012, 159, 810–825. [Google Scholar] [CrossRef] [PubMed]
- Koo, S.C.; Moon, B.C.; Kim, J.K.; Kim, C.Y.; Sung, S.J.; Kim, M.C.; Cho, M.J.; Cheong, Y.H. OsBWMK1 mediates SA-dependent defense responses by activating the transcription factor OsWRKY33. Biochem. Biophys. Res. Commun. 2009, 387, 365–370. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; Liu, C.; Zhang, Y.; Meng, X.; Zhou, X.; Chu, C.; Wang, X. OsWRKY30 is activated by MAP kinases to confer drought tolerance in rice. Plant Mol. Biol. 2012, 80, 241–253. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.; Jiang, Y.; Ye, S.; Karim, A.; Ling, Z.; He, Y.; Yang, S.; Luo, K. PtrWRKY73, a salicylic acid-inducible poplar WRKY transcription factor, is involved in disease resistance in Arabidopsis thaliana. Plant Cell Rep. 2015, 34, 831–841. [Google Scholar] [CrossRef] [PubMed]
- Su, T.; Xu, Q.; Zhang, F.C.; Chen, Y.; Li, L.Q.; Wu, W.H.; Chen, Y.F. WRKY42 modulates phosphate homeostasis through regulating phosphate translocation and acquisition in Arabidopsis. Plant Physiol. 2015, 167, 1579–1591. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, P.; Rabara, R.C.; Rushton, P.J. A systems biology perspective on the role of WRKY transcription factors in drought responses in plants. Planta 2014, 239, 255–266. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, C.; Wang, D.; Wang, Y. Molecular characterization and transcript profiling of NAC genes in response to abiotic stress in Tamarix hispida. Tree Genet. Genomes 2014, 10, 157–171. [Google Scholar] [CrossRef]
- Wang, L.; Qin, L.; Liu, W.; Zhang, D.; Wang, Y. A novel ethylene-responsive factor from Tamarix hispida, ThERF1, is a GCC-box- and DRE-motif binding protein that negatively modulates abiotic stress tolerance in Arabidopsis. Physiol. Plant. 2014, 152, 84–97. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Jefferson, R.A.; Kavanagh, T.A.; Bevan, M.W. GUS fusions: β-Glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987, 6, 3901–3907. [Google Scholar] [PubMed]
- Chang, S.; Puryear, J.; Cairney, J. A simple and efficient method for isolating RNA from pine trees. Plant Mol. Biol. Rep. 1993, 11, 113–116. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, L.; Zheng, L.; Zhang, C.; Wang, Y.; Lu, M.; Gao, C. ThWRKY4 from Tamarix hispida Can Form Homodimers and Heterodimers and Is Involved in Abiotic Stress Responses. Int. J. Mol. Sci. 2015, 16, 27097-27106. https://doi.org/10.3390/ijms161126009
Wang L, Zheng L, Zhang C, Wang Y, Lu M, Gao C. ThWRKY4 from Tamarix hispida Can Form Homodimers and Heterodimers and Is Involved in Abiotic Stress Responses. International Journal of Molecular Sciences. 2015; 16(11):27097-27106. https://doi.org/10.3390/ijms161126009
Chicago/Turabian StyleWang, Liuqiang, Lei Zheng, Chunrui Zhang, Yucheng Wang, Mengzhu Lu, and Caiqiu Gao. 2015. "ThWRKY4 from Tamarix hispida Can Form Homodimers and Heterodimers and Is Involved in Abiotic Stress Responses" International Journal of Molecular Sciences 16, no. 11: 27097-27106. https://doi.org/10.3390/ijms161126009
APA StyleWang, L., Zheng, L., Zhang, C., Wang, Y., Lu, M., & Gao, C. (2015). ThWRKY4 from Tamarix hispida Can Form Homodimers and Heterodimers and Is Involved in Abiotic Stress Responses. International Journal of Molecular Sciences, 16(11), 27097-27106. https://doi.org/10.3390/ijms161126009




