Nanoinformatics: Emerging Databases and Available Tools
Abstract
:1. Introduction
2. Growth of Bioinformatics and Applications for Nanoinformatics
3. Databases
3.1. ISA-TAB-Nano
3.2. caNanoLab
3.3. Nanomaterial Registry
4. Text Mining
5. Molecular Modeling
5.1. Docking
5.2. Quantitative Structure-Activity Relationships (QSAR)
High-Throughput Screening Data Analysis Tools (HDAT)
5.3. MD Simulations
6. Imaging
7. Computational Design of Biomolecular Nanostructures
8. Specific Challenges and Opportunities in Nanoinformatics
9. Conclusions
Acknowledgments
Conflicts of Interest
- Author ContributionsS.P. obtained and analyzed the data and wrote the manuscript; S.C. supervised the work and wrote the manuscript.
References
- Albanese, A.; Tang, P.S.; Chan, W.C. The effect of nanoparticle size, shape, and surface chemistry on biological systems. Annu. Rev. Biomed. Eng 2012, 14, 1–16. [Google Scholar]
- Toumey, C. Why not zwergo-technology? Nat. Nanotechnol 2011, 6, 393–394. [Google Scholar]
- Kim, B.Y.; Rutka, J.T.; Chan, W.C. Nanomedicine. N. Engl. J. Med 2010, 363, 2434–2443. [Google Scholar]
- Makarucha, A.J.; Todorova, N.; Yarovsky, I. Nanomaterials in biological environment: A review of computer modelling studies. Eur. Biophys. J 2011, 40, 103–115. [Google Scholar]
- Nie, S.; Xing, Y.; Kim, G.J.; Simons, J.W. Nanotechnology applications in cancer. Annu. Rev. Biomed. Eng 2007, 9, 257–288. [Google Scholar]
- Yokel, R.A.; Macphail, R.C. Engineered nanomaterials: Exposures, hazards, and risk prevention. J. Occup. Med. Toxicol 2011, 6, 7. [Google Scholar]
- Love, S.A.; Maurer-Jones, M.A.; Thompson, J.W.; Lin, Y.S.; Haynes, C.L. Assessing nanoparticle toxicity. Annu. Rev. Anal. Chem 2012, 5, 181–205. [Google Scholar]
- Bharali, D.J.; Khalil, M.; Gurbuz, M.; Simone, T.M.; Mousa, S.A. Nanoparticles and cancer therapy: A concise review with emphasis on dendrimers. Int. J. Nanomed 2009, 4, 1–7. [Google Scholar]
- Buzea, C.; Pacheco, I.I.; Robbie, K. Nanomaterials and nanoparticles: Sources and toxicity. Biointerphases 2007, 2, 17–71. [Google Scholar]
- De la Iglesia, D.; Garcia-Remesal, M.; de la Calle, G.; Kulikowski, C.; Sanz, F.; Maojo, V. The impact of computer science in molecular medicine: Enabling high-throughput research. Curr. Top. Med. Chem 2013, 13, 526–575. [Google Scholar]
- Yang, S.T.; Liu, Y.; Wang, Y.W.; Cao, A. Biosafety and bioapplication of nanomaterials by designing protein-nanoparticle interactions. Small 2013, 9, 1635–1653. [Google Scholar]
- Nanoinformatics. Available online: http://nanoinformatics.org/nanoinformatics/index.php/Main_Page accessed on 14 March 2014.
- Maojo, V.; Fritts, M.; de la Iglesia, D.; Cachau, R.E.; Garcia-Remesal, M.; Mitchell, J.A.; Kulikowski, C. Nanoinformatics: A new area of research in nanomedicine. Int. J. Nanomed 2012, 7, 3867–3890. [Google Scholar]
- NNI Signature Initiative. Available online: http://www.nano.gov/sites/default/files/nki_nsi_white_paper_-_may_14_2012_secured.pdf accessed on 14 March 2014.
- The ACTION-Grid White Paper on Nanoinformatics. Available online: http://www.action-grid.eu/documents/Final%20White%20Paper%20-%20Nano.pdf accessed on 14 March 2014.
- Ostraat, M.L.; Mills, K.C.; Guzan, K.A.; Murry, D. The Nanomaterial Registry: Facilitating the sharing and analysis of data in the diverse nanomaterial community. Int. J. Nanomed 2013, 8 Suppl 1, 7–13. [Google Scholar]
- Sharon, G.; George, W.H.; Stephanie, A.M.; Michal, L.; Mervi, H.; Juli, D.K. Cananolab: Data sharing to expedite the use of nanotechnology in biomedicine. Comput. Sci. Discov 2013, 6, 014010. [Google Scholar]
- De la Iglesia, D.; Cachau, R.E.; García-Remesal, M.; Maojo, V. Nanoinformatics knowledge infrastructures: Bringing efficient information management to nanomedical research. Comput. Sci. Discov 2013, 6, 014011. [Google Scholar]
- Thomas, D.G.; Gaheen, S.; Harper, S.L.; Fritts, M.; Klaessig, F.; Hahn-Dantona, E.; Paik, D.; Pan, S.; Stafford, G.A.; Freund, E.T.; et al. ISA-TAB-Nano: A specification for sharing nanomaterial research data in spreadsheet-based format. BMC Biotechnol 2013, 13, 2. [Google Scholar]
- Inkscape Is a Professional Vector Graphics. Available online: http://www.inkscape.org/en/ accessed on 14 March 2014.
- GIMP—The GNU Image Manipulation Program. Available online: http://www.gimp.org/ accessed on 14 March 2014.
- Genbank. Available online: https://www.ncbi.nlm.nih.gov/genbank/ accessed on 14 March 2014.
- Protein Data Bank. Available online: http://www.rcsb.org/pdb/home/home.do accessed on 14 March 2014.
- Brown, P.O.; Eisen, M.B.; Varmus, H.E. Why PLoS became a publisher. PLoS Biol 2003, 1, e36. [Google Scholar]
- Fox, J.A.; Butland, S.L.; McMillan, S.; Campbell, G.; Ouellette, B.F. The bioinformatics links directory: A compilation of molecular biology web servers. Nucleic Acids Res 2005, 33, W3–W24. [Google Scholar]
- Web Server Issue. Available online: http://nar.oxfordjournals.org/content/41/W1.toc accessed on 14 March 2014.
- González-Nilo, F.; Pérez-Acle, T.; Guínez-Molinos, S.; Geraldo, D.A.; Sandoval, C.; Yévenes, A.; Santos, L.S.; Laurie, V.F.; Mendoza, H.; Cachau, R.E. Nanoinformatics: An emerging area of information technology at the intersection of bioinformatics, computational chemistry and nanobiotechnology. Biol. Res 2011, 44, 43–51. [Google Scholar]
- Nanomaterial-Biological Interactions Knowledgebase. Available online: http://nbi.oregonstate.edu/ accessed on 14 March 2014.
- Internano—Resources for Nanomanufacturing. Available online: http://www.internano.org/ accessed on 14 March 2014.
- Nano-EHS Database Analysis Tool. Available online: http://icon.rice.edu/report.cfm accessed on 14 March 2014.
- Nanoparticle Information Library. Available online: http://nanoparticlelibrary.net/ accessed on 14 March 2014.
- Nanomaterial Registry. Available online: https://www.nanomaterialregistry.org/ accessed on 14 March 2014.
- National Toxicology Program Database. Available online: http://tools.niehs.nih.gov/ntp_tox/ accessed on 14 March 2014.
- Nano-HUB Database. Available online: http://nanohub.org/resources/databases accessed on 14 March 2014.
- National Center for Biomedical Ontology. Available online: http://www.bioontology.org/ accessed on 14 March 2014.
- ISA-TAB-Nano. Available online: https://wiki.nci.nih.gov/display/ICR/ISA-TAB-Nano accessed on 14 March 2014.
- caNanoLab. Available online: https://cananolab.nci.nih.gov/caNanoLab/ accessed on 14 March 2014.
- Toxicology Data Network. Available online: http://toxnet.nlm.nih.gov/ accessed on 14 March 2014.
- Nanotechnology Characterization Laboratory. Available online: http://ncl.cancer.gov/objectives_ncl_obj5.asp accessed on 14 March 2014.
- The Collaboratory for Structural Nanobiology. Available online: http://csn.ncifcrf.gov/Advanced_Structure_Analysis/HOME.html accessed on 14 March 2014.
- Toxicology Literature Online. Available online: http://toxnet.nlm.nih.gov/cgi-bin/sis/htmlgen?TOXLINE accessed on 14 March 2014.
- OECD Database on Research into the Safety of Manufactured Nanomaterials. Available online: http://webnet.oecd.org/NANOMATERIALS/Pagelet/Front/Default.aspx accessed on 14 March 2014.
- Schuurman, N.; Leszczynski, A. Ontologies for bioinformatics. Bioinform. Biol. Insights 2008, 2, 187–200. [Google Scholar]
- Bard, J.B.; Rhee, S.Y. Ontologies in biology: Design, applications and future challenges. Nat. Rev. Genet 2004, 5, 213–222. [Google Scholar]
- Thomas, D.G.; Klaessig, F.; Harper, S.L.; Fritts, M.; Hoover, M.D.; Gaheen, S.; Stokes, T.H.; Reznik-Zellen, R.; Freund, E.T.; Klemm, J.D.; et al. Informatics and standards for nanomedicine technology. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol 2011, 3, 511–532. [Google Scholar]
- NanoParticle Ontology for Cancer Nanotechnology Research. Available online: http://www.nano-ontology.org/ accessed on 14 March 2014.
- BioPortal. Available online: http://bioportal.bioontology.org/ accessed on 14 March 2014.
- The Open Biological and Biomedical Ontologies. Available online: http://www.obofoundry.org/ accessed on 14 March 2014.
- caOBR—A Tool for Indexing Cancer Informatics Resources. Available online: http://www.bioontology.org/caOBR accessed on 14 March 2014.
- The Database and Ontology of Chemical Entities of Biological Interest. Available online: http://www.ebi.ac.uk/chebi/ accessed on 14 March 2014.
- The Gene Ontology. Available online: http://www.geneontology.org/ accessed on 14 March 2014.
- The Foundational Model of Anatomy Ontology. Available online: http://sig.biostr.washington.edu/projects/fm/AboutFM.html accessed on 14 March 2014.
- Zebrafish Anatomy and Development Ontology. Available online: http://www.obofoundry.org/cgi-bin/detail.cgi?id=zebrafish_anatomy accessed on 14 March 2014.
- Ontology Lookup Service. Available online: http://www.ebi.ac.uk/ontology-lookup/browse.do?ontName=REX accessed on 14 March 2014.
- Phenotypic Quality Ontology. Available online: http://bioportal.bioontology.org/ontologies/PATO accessed on 14 March 2014.
- Cohen, K.B.; Hunter, L. Getting started in text mining. PLoS Comput. Biol 2008, 4, e20. [Google Scholar]
- Rzhetsky, A.; Seringhaus, M.; Gerstein, M.B. Getting started in text mining: Part two. PLoS Comput. Biol 2009, 5, e1000411. [Google Scholar]
- Grulke, C.M.; Goldsmith, M.R.; Vallero, D.A. Toward a blended ontology: Applying knowledge systems to compare therapeutic and toxicological nanoscale domains. J. Biomed. Biotechnol 2012, 2012, 308381. [Google Scholar]
- Garcia-Remesal, M.; Garcia-Ruiz, A.; Perez-Rey, D.; de la Iglesia, D.; Maojo, V. Using nanoinformatics methods for automatically identifying relevant nanotoxicology entities from the literature. Biomed. Res. Int 2013, 2013, 410294. [Google Scholar]
- Liu, R.; France, B.; George, S.; Rallo, R.; Zhang, H.; Xia, T.; Nel, A.E.; Bradley, K.; Cohen, Y. Association rule mining of cellular responses induced by metal and metal oxide nanoparticles. Analyst 2014, 139, 943–953. [Google Scholar]
- Google Scholar. Available online: http://scholar.google.com accessed on 14 March 2014.
- Gopubmed. Available online: http://www.gopubmed.org/ accessed on 14 March 2014.
- Textpresso. Available online: http://www.textpresso.org/ accessed on 14 March 2014.
- BioRAT. Available online: http://bioinf.cs.ucl.ac.uk/software_downloads/biorat/ accessed on 14 March 2014.
- iHOP. Available online: http://www.ihop-net.org/UniPub/iHOP/ accessed on 14 March 2014.
- Lewar, E.G. Computational Chemistry: Introduction to the Theory and Applications of Molecular and Quantum Mechanics, 2nd ed; Springer: New York, NY, USA, 2010. [Google Scholar]
- Molecular Mechanics and Dynamics. Available online: http://employees.csbsju.edu/hjakubowski/classes/ch331/protstructure/mechdynam2.html accessed on 14 March 2014.
- PyMOL. Available online: http://www.pymol.org/ accessed on 14 March 2014.
- RasMol. Available online: http://www.bernstein-plus-sons.com/software/rasmol/ accessed on 14 March 2014.
- Raster3D. Available online: http://skuld.bmsc.washington.edu/raster3d/raster3d.html accessed on 14 March 2014.
- UCSF Chimera. Available online: http://www.cgl.ucsf.edu/chimera/index.html accessed on 14 March 2014.
- Visual Molecular Dynamics. Available online: http://www.ks.uiuc.edu/Research/vmd/ accessed on 14 March 2014.
- Cn3D Is a Macromolecular Structure Viewer. Available online: http://www.ncbi.nlm.nih.gov/Structure/CN3D/cn3d.shtml accessed on 14 March 2014.
- BALL—Biochemical Algorithms Library. Available online: http://www.ball-project.org/ accessed on 14 March 2014.
- Jmol. Available online: http://jmol.sourceforge.net/ accessed on 14 March 2014.
- ICM-Browser. Available online: http://www.molsoft.com/icm_browser.html accessed on 14 March 2014.
- Chemkit. Available online: http://sourceforge.net/projects/chemkit/files/chemkit-0.1.tar.gz/download accessed on 14 March 2014.
- OpenStructure. Available online: http://www.openstructure.org/ accessed on 14 March 2014.
- VEGA, Z.Z. Available online: http://www.vegazz.net/ accessed on 14 March 2014.
- Molekel. Available online: http://molekel.cscs.ch/wiki/pmwiki.php accessed on 14 March 2014.
- Avogadro. Available online: http://avogadro.cc/wiki/Main_Page accessed on 14 March 2014.
- Xeo. Available online: http://sourceforge.net/projects/xeo/ accessed on 14 March 2014.
- ChemSketch. Available online: http://www.acdlabs.com/resources/freeware/chemsketch/ accessed on 14 March 2014.
- Hu, X.; Shelver, W.H. Docking studies of matrix metalloproteinase inhibitors: Zinc parameter optimization to improve the binding free energy prediction. J. Mol. Graph. Model 2003, 22, 115–126. [Google Scholar]
- Morris, G.M.; Huey, R.; Olson, A.J. Using autoDock for ligand-receptor docking. Curr. Protoc. Bioinform 2008. [Google Scholar] [CrossRef]
- Yang, S.T.; Wang, H.; Guo, L.; Gao, Y.; Liu, Y.; Cao, A. Interaction of fullerenol with lysozyme investigated by experimental and computational approaches. Nanotechnology 2008, 19, 395101. [Google Scholar]
- Calvaresi, M.; Zerbetto, F. Baiting Proteins with C60. ACS Nano 2010, 4, 2283–2299. [Google Scholar]
- AutoDock 4. Available online: http://autodock.scripps.edu/ accessed on 14 March 2014.
- AutoDock Vina. Available online: http://vina.scripps.edu/ accessed on 14 March 2014.
- Dock. Available online: http://dock.compbio.ucsf.edu/ accessed on 14 March 2014.
- GOLD. Available online: http://www.ccdc.cam.ac.uk/Solutions/GoldSuite/Pages/GOLD.aspx accessed on 14 March 2014.
- FlexX. Available online: http://www.biosolveit.de/FlexX/ accessed on 14 March 2014.
- Surflex-Dock. Available online: http://www.tripos.com/index.php?family=modules,SimplePage,,,&page=surflex_dock&s=0 accessed on 14 March 2014.
- Tropsha, A. Best Practices for QSAR model development, validation, and exploitation. Mol. Inf 2010, 29, 476–488. [Google Scholar]
- Mays, C.; Benfenati, E.; Pardoe, S. Use and perceived benefits and barriers of QSAR models for reach: Findings from a questionnaire to stakeholders. Chem. Cent. J 2012, 6, 159. [Google Scholar]
- Toropov, A.A.; Toropova, A.P.; Benfenati, E.; Leszczynska, D.; Leszczynski, J. Smiles-based optimal descriptors: QSAR analysis of fullerene-based HIV-1 pr inhibitors by means of balance of correlations. J. Comput. Chem 2010, 31, 381–392. [Google Scholar]
- Fourches, D.; Pu, D.; Tassa, C.; Weissleder, R.; Shaw, S.Y.; Mumper, R.J.; Tropsha, A. Quantitative nanostructure—Activity relationship modeling. ACS Nano 2010, 4, 5703–5712. [Google Scholar]
- Fourches, D.; Pu, D.; Tropsha, A. Exploring quantitative nanostructure-activity relationships (QNAR) modeling as a tool for predicting biological effects of manufactured nanoparticles. Comb. Chem. High Throughput Screen 2011, 14, 217–225. [Google Scholar]
- Puzyn, T.; Rasulev, B.; Gajewicz, A.; Hu, X.; Dasari, T.P.; Michalkova, A.; Hwang, H.M.; Toropov, A.; Leszczynska, D.; Leszczynski, J. Using nano-QSAR to predict the cytotoxicity of metal oxide nanoparticles. Nat. Nano 2011, 6, 175–178. [Google Scholar]
- Liu, R.; Rallo, R.; George, S.; Ji, Z.; Nair, S.; Nel, A.E.; Cohen, Y. Classification NanoSAR development for cytotoxicity of metal oxide nanoparticles. Small 2011, 7, 1118–1126. [Google Scholar]
- Poater, A.; Saliner, A.G.; Solà, M.; Cavallo, L.; Worth, A.P. Computational methods to predict the reactivity of nanoparticles through structure–property relationships. Expert Opin. Drug Deliv 2010, 7, 295–305. [Google Scholar]
- Winkler, D.A.; Mombelli, E.; Pietroiusti, A.; Tran, L.; Worth, A.; Fadeel, B.; McCall, M.J. Applying quantitative structure-activity relationship approaches to nanotoxicology: Current status and future potential. Toxicology 2013, 313, 15–23. [Google Scholar]
- Epa, V.C.; Burden, F.R.; Tassa, C.; Weissleder, R.; Shaw, S.; Winkler, D.A. Modeling biological activities of nanoparticles. Nano Lett 2012, 12, 5808–5812. [Google Scholar]
- ChemProp. Available online: http://www.ufz.de/index.php?en=10684 accessed on 14 March 2014.
- OECD. Available online: http://www.oecd.org/env/ehs/ accessed on 14 March 2014.
- RmSquare. Available online: http://aptsoftware.co.in/rmsquare/ accessed on 14 March 2014.
- T.E.S.T. Available online: http://www.epa.gov/nrmrl/std/qsar/qsar.html accessed on 14 March 2014.
- Virtual Computational Chemical Laboratory. Available online: http://www.vcclab.org/ accessed on 14 March 2014.
- CORAL. Available online: http://www.insilico.eu/coral/ accessed on 14 March 2014.
- Rong, L.; Taimur, H.; Robert, R.; Yoram, C. HDAT: Web-based high-throughput screening data analysis tools. Comput. Sci. Discov 2013, 6, 014006. [Google Scholar]
- High Throughput Screening Data Analysis Tools (HDAT). Available online: http://nanoinfo.org/hdat/ accessed on 14 March 2014.
- Assisted Model Building with Energy Refinement. Available online: http://ambermd.org/ accessed on 14 March 2014.
- NAMD—Scalable Molecular Dynamics. Available online: http://www.ks.uiuc.edu/Research/namd/ accessed on 14 March 2014.
- Gromacs. Available online: http://www.gromacs.org/ accessed on 14 March 2014.
- Chemistry at HARvard Macromolecular Mechanics. Available online: http://www.charmm.org/ accessed on 14 March 2014.
- Desmond. Available online: http://www.deshawresearch.com/resources_desmond.html accessed on 14 March 2014.
- LAMMPS Molecular Dynamics Simulator. Available online: http://lammps.sandia.gov/ accessed on 14 March 2014.
- TINKER—Software Tools for Molecular Design. Available online: http://dasher.wustl.edu/tinker/ accessed on 14 March 2014.
- MDynaMix: A Molecular Dynamics Program. Available online: http://www.mmk.su.se/~sasha/mdynamix/ accessed on 14 March 2014.
- CP2K—Open Source Molecular Dynamics. Available online: http://www.cp2k.org/ accessed on 14 March 2014.
- ABINIT. Available online: http://www.abinit.org/ accessed on 14 March 2014.
- Advanced Concepts in Electronic Structure. Available online: http://www.qtp.ufl.edu/ACES/ accessed on 14 March 2014.
- BigDFT. Available online: http://bigdft.org/Wiki/index.php?title=BigDFT_website accessed on 14 March 2014.
- Quantum Monte Carlo. Available online: http://vallico.net/casinoqmc/ accessed on 14 March 2014.
- CASTEP. Available online: http://www.castep.org/ accessed on 14 March 2014.
- The Columbus Quantum Chemistry Programs. Available online: http://www.univie.ac.at/columbus/ accessed on 14 March 2014.
- Coupled-Cluster Techniques for Computational Chemistry. Available online: http://www.cfour.de/ accessed on 14 March 2014.
- Conquest: Linear Scaling DFT. Available online: http://www.order-n.org/ accessed on 14 March 2014.
- CPMD. Available online: http://www.cpmd.org/ accessed on 14 March 2014.
- A Computational Tool for Solid State Chemistry and Physics. Available online: http://www.crystal.unito.it/index.php accessed on 14 March 2014.
- Dacapo Is a Total Energy Program Based on Density Functional Theory. Available online: https://wiki.fysik.dtu.dk/dacapo accessed on 14 March 2014.
- Dalton. Available online: http://daltonprogram.org/ accessed on 14 March 2014.
- Density Functional Based Tight Binding. Available online: http://www.dftb-plus.info/ accessed on 14 March 2014.
- DIRAC—Program for Atomic and Molecular Direct Iterative Relativistic All-electron Calculations. Available online: http://www.diracprogram.org/doku.php accessed on 14 March 2014.
- The Elk FP-LAPW Code. Available online: http://elk.sourceforge.net/ accessed on 14 March 2014.
- ErgoSCF Is a Quantum Chemistry Program for Large-Scale Self-Consistent Field Calculations. Available online: http://www.ergoscf.org/ accessed on 14 March 2014.
- ERKALE Is a Quantum Chemistry Program Used to Solve the Electronic Structure of Atoms, Molecules and Molecular Clusters. Available online: https://code.google.com/p/erkale/ accessed on 14 March 2014.
- FLEUR—The Julich FLAPW Code Family. Available online: http://www.flapw.de/pm/index.php accessed on 14 March 2014.
- FreeON Is a Suite of Programs for Linear Scaling Quantum Chemistry. Available online: https://www.freeon.org/index.php/Main_Page accessed on 14 March 2014.
- The General Atomic and Molecular Electronic Structure System (GAMESS) Is a General ab initio Quantum Chemistry Package. Available online: http://www.msg.ameslab.gov/gamess/index.html accessed on 14 March 2014.
- Grid-Based Projector-Augmented Wave Method. Available online: https://wiki.fysik.dtu.dk/gpaw/ accessed on 14 March 2014.
- Gaussian. Available online: http://www.gaussian.com/ accessed on 14 March 2014.
- Joint Density Functional Theory. Available online: http://sourceforge.net/p/jdftx/wiki/Home/ accessed on 14 March 2014.
- Multiresolution Adaptive Numerical Environment for Scientific Simulation. Available online: http://code.google.com/p/m-a-d-n-e-s-s/ accessed on 14 March 2014.
- A Practical Quantum Chemistry Tool for Modeling Biological Systems and Co-Crystals. Available online: http://cacheresearch.com/mopac/Mopac2002manual/ accessed on 14 March 2014.
- The Massively Parallel Quantum Chemistry Program. Available online: http://www.mpqc.org/ accessed on 14 March 2014.
- NWChem—Delivering High-Performance Computational Chemistry. Available online: http://www.nwchem-sw.org/index.php/Main_Page accessed on 14 March 2014.
- Octopus Is a Scientific Program Aimed at the Ab Initio Virtual Experimentation. Available online: http://www.tddft.org/programs/octopus/wiki/index.php/Main_Page accessed on 14 March 2014.
- ONTEP Is a Linear-Scaling Code for Quantum-Mechanical Calculations Based on Density-Functional Theory. Available online: http://www2.tcm.phy.cam.ac.uk/onetep/ accessed on 14 March 2014.
- OpenAtom Is a Highly Scalable and Portable Parallel Application for Molecular Dynamics Simulations at the Quantum Level. Available online: http://charm.cs.uiuc.edu/OpenAtom/ accessed on 14 March 2014.
- Open Source Package for Material Explorer. Available online: http://www.openmx-square.org/ accessed on 14 March 2014.
- The ORCA Program Is to Carry out Geometry Optimizations and to Predict a Large Number of Spectroscopic Parameters at Different Levels of Theory. Available online: http://www.cec.mpg.de/forum/ accessed on 14 March 2014.
- PSI4—Ab Initio Quantum Chemistry Program. Available online: http://www.psicode.org/ accessed on 14 March 2014.
- PyQuante—Python Quantum Chemistry. Available online: http://pyquante.sourceforge.net/ accessed on 14 March 2014.
- Quantum Espresso Is an Integrated Suite of Open-Source Computer Codes for Electronic-Structure Calculations and Materials Modeling at the Nanoscale. Available online: http://www.quantum-espresso.org/ accessed on 14 March 2014.
- Vienna Ab Initio Simulation Package. Available online: http://www.vasp.at/ accessed on 14 March 2014.
- Yambo Is a FORTRAN/C Code for Many-Body Calculations in Solid State and Molecular Physics. Available online: http://www.yambo-code.org/ accessed on 14 March 2014.
- Dodson, G.G.; Lane, D.P.; Verma, C.S. Molecular simulations of protein dynamics: New windows on mechanisms in biology. EMBO Rep 2008, 9, 144–150. [Google Scholar]
- Dror, R.O.; Dirks, R.M.; Grossman, J.P.; Xu, H.; Shaw, D.E. Biomolecular simulation: A computational microscope for molecular biology. Annu. Rev. Biophys 2012, 41, 429–452. [Google Scholar]
- Yanamala, N.; Kagan, V.E.; Shvedova, A.A. Molecular modeling in structural nano-toxicology: Interactions of nano-particles with nano-machinery of cells. Adv. Drug Deliv Rev 2013, 65, 2070–2077. [Google Scholar]
- Micro-Manager Open Source Microscopy Software. Available online: http://www.micro-manager.org/wiki/Micro-Manager accessed on 14 March 2014.
- OMERO. Available online: http://www.openmicroscopy.org/site/products/omero accessed on 14 March 2014.
- Bisque Database. Available online: http://www.bioimage.ucsb.edu/bisque accessed on 14 March 2014.
- Bio-Formats Is a Standalone Java Library for Reading and Writing Life Sciences Image file Formats. Available online: http://loci.wisc.edu/software/bio-formats accessed on 14 March 2014.
- Image Processing and Analysis in Java. Available online: http://rsb.info.nih.gov/ij/ accessed on 14 March 2014.
- Fiji Is an Image Processing Package. Available online: http://fiji.sc/Fiji accessed on 14 March 2014.
- Quantitative Tool for Studying Complex and Dynamic Biological Microenvironments from 4D/5D Microscopy Data. Available online: http://farsight-toolkit.org/wiki/Main_Page accessed on 14 March 2014.
- Icy—An Open Community Platform for Bioimage Informatics. Available online: http://icy.bioimageanalysis.org/ accessed on 14 March 2014.
- Cellprofiler—Cell Image Analysis Software. Available online: http://www.cellprofiler.org/ accessed on 14 March 2014.
- Micropiolot—Microscope Automation. Available online: http://www.embl.de/services/core_facilities/almf/services/high-throughput-microscopy/microscope-automation/micropilot/index.html accessed on 14 March 2014.
- Cell-ID Is an Open-Source Cell-Finding, Tracking, and Analysis Package. Available online: http://www.molsci.org/protocols/software.html accessed on 14 March 2014.
- BioView3D Is an Open Source and Cross-Platform Application Intended for Biologists to Visualize 3D Stack Imagery. Available online: http://www.bioimage.ucsb.edu/downloads/BioView3D accessed on 14 March 2014.
- Vaa3D Is a Swiss Army Knife for Bioimage Visualization & Analysis. Available online: http://penglab.janelia.org/proj/vaa3d/Vaa3D/About_Vaa3D.html accessed on 14 March 2014.
- The IMOD Is a Set of Image Processing, Modeling and Display Programs. Available online: http://bio3d.colorado.edu/imod/ accessed on 14 March 2014.
- iCluster SubCellular Localisation Image Visualiser. Available online: http://icluster.imb.uq.edu.au/ accessed on 14 March 2014.
- ImageSurfer Is free 3D Imaging Software to Visualize and Analyze Multi-Channel Volumes. Available online: http://imagesurfer.cs.unc.edu/ accessed on 14 March 2014.
- Rosen, J.E.; Yoffe, S.; Meerasa, A.; Verma, M.; Gu, F.X. Nanotechnology and diagnostic imaging: New advances in contrast agent technology. J. Nanomed. Nanotechnol 2011, 2, 115. [Google Scholar]
- Kostarelos, K.; Bianco, A.; Prato, M. Promises, facts and challenges for carbon nanotubes in imaging and therapeutics. Nat. Nano 2009, 4, 627–633. [Google Scholar]
- Hamilton, N.A. Open source tools for fluorescent imaging. Methods Enzymol 2012, 504, 393–417. [Google Scholar]
- Sanderson, K. Bioengineering: What to make with DNA origami. Nature 2010, 464, 158–159. [Google Scholar]
- Zhao, Y.X.; Shaw, A.; Zeng, X.; Benson, E.; Nystrom, A.M.; Hogberg, B. DNA origami delivery system for cancer therapy with tunable release properties. ACS Nano 2012, 6, 8684–8691. [Google Scholar]
- Tiamat Is a Graphical User Interface Program for the Design of DNA Nanostructures and Sequences. Available online: http://yanlab.asu.edu/Resources.html accessed on 14 March 2014.
- Computer-Aided Engineering for DNA Origami. Available online: http://cando-dna-origami.org/ accessed on 14 March 2014.
- DNA Origami Design Software. Available online: http://www.cdna.dk/default.asp?getreq=%2Findex.php%2Fsoftware.html accessed on 14 March 2014.
- Cadnano Simplifies and Enhances the Process of Designing Three-Dimensional DNA Origami Nanostructures. Available online: http://cadnano.org/ accessed on 14 March 2014.
- Uniquimer3D Is a User-Friendly Software System for Structural DNA Nanotechnology. Available online: http://ihome.ust.hk/~keymix/ accessed on 14 March 2014.
- Contub Is a Set of Tools Dedicated to the Construction of Complex Carbon Nanotube Structures for Use in Computational Chemistry. Available online: http://www.ugr.es/~gmdm/java/contub/contub.html accessed on 14 March 2014.
- A Program for Creating Fullerene Structures and for Performing Topological Analyses. Available online: http://ctcp.massey.ac.nz/index.php?group=&page=fullerenes&menu=fullerenes accessed on 14 March 2014.
- CS Catalyst. Available online: http://www.camsoft.co.kr/chemnews/catalyst/5/51/bucky.html accessed on 14 March 2014.
- Ninithi Is a Free Modeling Software Used in Nanotechnology. Available online: http://sourceforge.net/projects/ninithi/files/ accessed on 14 March 2014.
- TubeGen Is a Web Accessible Nanotube Structure Generator. Available online: http://turin.nss.udel.edu/research/tubegenonline.html accessed on 14 March 2014.
- Nanotube Coordinate Generator with a Viewer for Windows. Available online: http://www.photon.t.u-tokyo.ac.jp/~maruyama/wrapping3/wrapping.html accessed on 14 March 2014.
- Van Noorden, R. Tensions grow as data-mining discussions fall apart. Nature 2013, 498, 14–15. [Google Scholar]
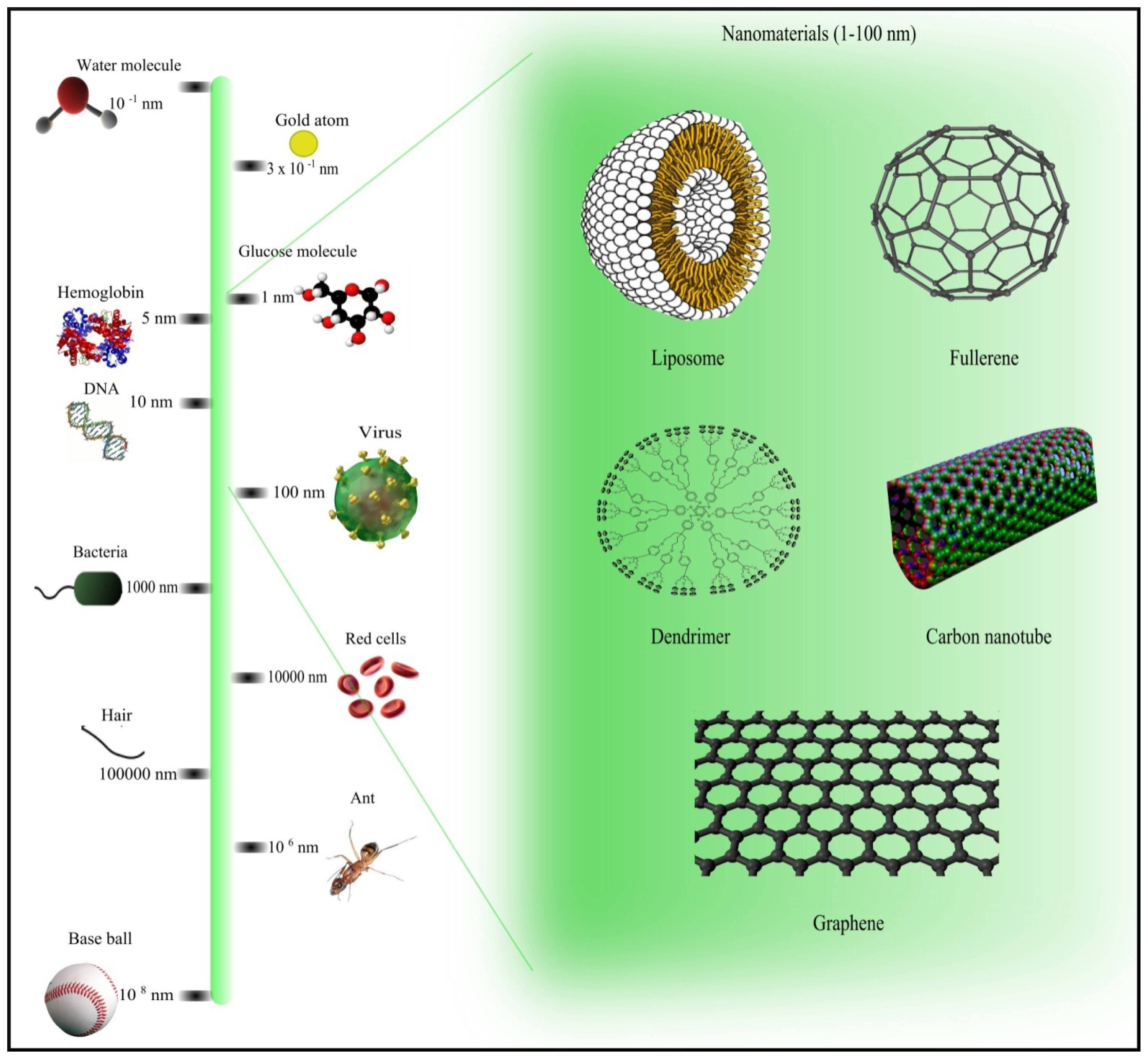
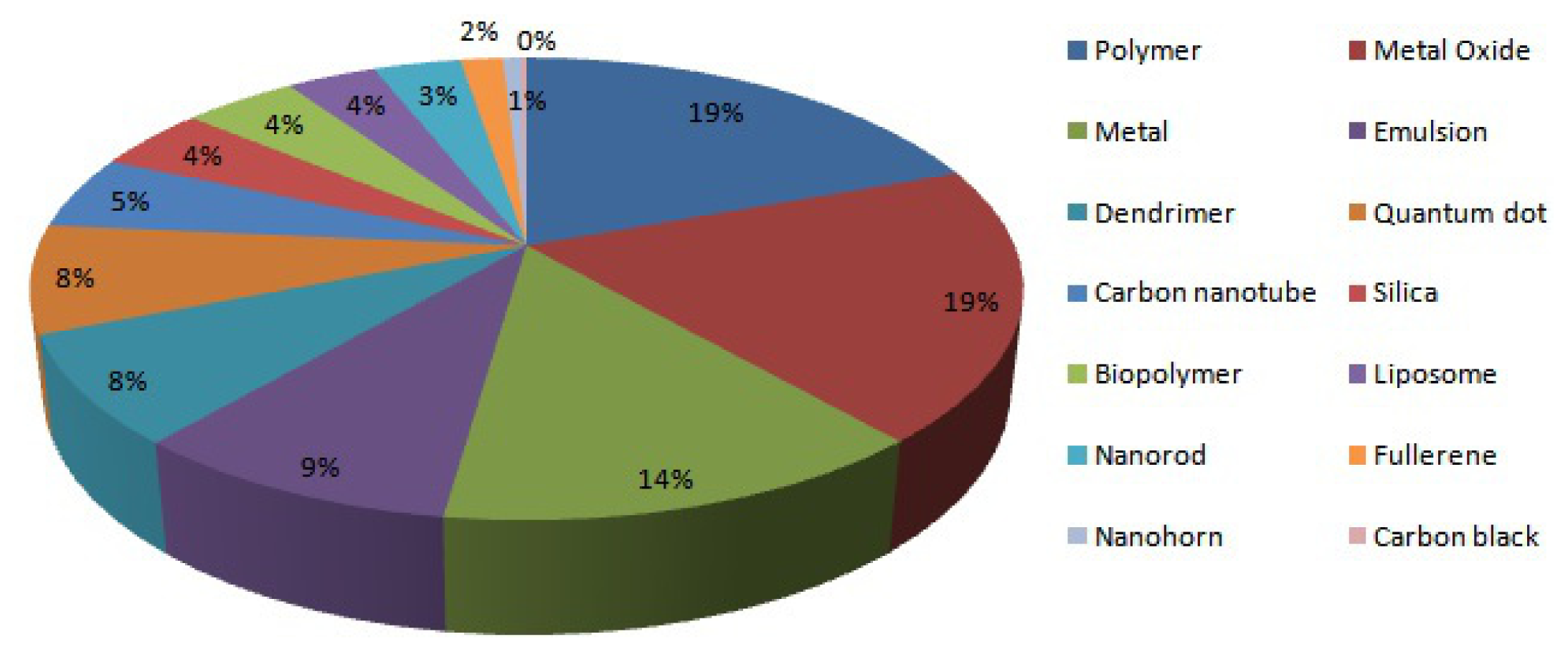
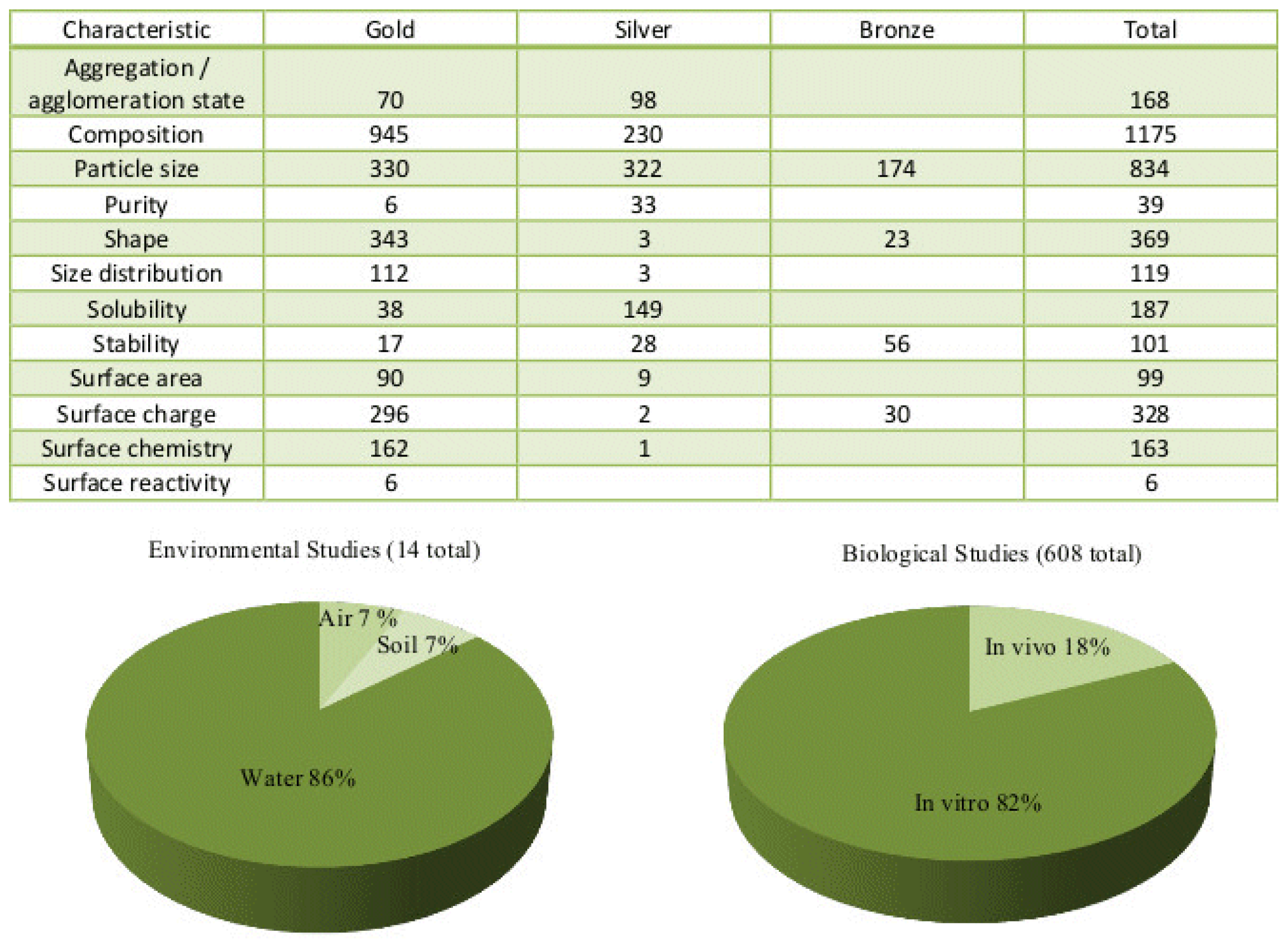
| No. | Name | Description | Url |
|---|---|---|---|
| 1 | Nanomaterial Biological Interactions Knowledgebase | The knowledgebase serves as a repository for annotated data on nanomaterial characterization (purity, size, shape, charge, composition, functionalization, and agglomeration state), synthesis methods, and nanomaterial-biological interactions. | [28] |
| 2 | InterNano | InterNano supports the information needs of the nanomanufacturing community by bringing together resources related to advances in applications, devices, metrology, and materials that will facilitate the commercial development and/or marketable application of nanotechnology. | [29] |
| 3 | Nano-EHS Database Analysis Tool | This web tool provides a quick and thorough synopsis of the Environment, Health and Safety Database. | [30] |
| 4 | Nanoparticle Information Library | The goal of the NIL is to help occupational health professionals, industrial users, worker groups, and researchers organize and share information on nanomaterials, including their health and safety-associated properties. | [31] |
| 5 | Nanomaterials Registry | The Registry was established to provide researchers with a convenient data management and sharing plan. | [32] |
| 6 | National Toxicology Program Database | The goal of this database is to develop and apply modern toxicology and molecular biology tools to identify substances in the environment that may affect human health. | [33] |
| 7 | Nano-HUB database | Nano-HUB offers a searchable online database of nanoBIO tools. | [34] |
| 8 | National Center for Biomedical Ontology Bioportal | A major focus involves the use of biomedical ontologies to aid in the management and analysis of data derived from complex experiments. | [35] |
| 9 | ISA-TAB-Nano | The ISA-TAB-Nano standard specifies the format for representing and sharing information about nanomaterials, small molecules, and biological specimens along with their assay characterization data (including metadata and summary data) using spreadsheet or TAB-delimited files. | [36] |
| 10 | caNanoLab | caNanoLab is a data-sharing portal designed to facilitate information sharing in the biomedical nanotechnology research community to expedite and validate the use of nanotechnology in biomedicine. | [37] |
| 11 | Toxicology Data Network | Includes databases on toxicology, hazardous chemicals, environmental health, and toxic releases. | [38] |
| 12 | Nanotechnology Characterization Laboratory | The Nanotechnology Characterization Laboratory performs and standardizes the preclinical characterization of nanomaterials intended for cancer therapeutics and diagnostics developed by researchers from academia, government, and industry. | [39] |
| 13 | Collaboratory for Structural Nanobiology | A nanoinformatics service dedicated to the collection, curation, and correlation of structural, physicochemical, and biological and biomedical data. | [40] |
| 14 | Toxicology Literature Online | Provides references from the toxicology literature. | [41] |
| 15 | OECD | Database on research into the safety of manufactured nanomaterials. | [42] |
| No. | Name | Description | Url |
|---|---|---|---|
| 1 | NanoParticle Ontology | The NPO was developed to provide knowledge regarding the description, preparation, and characterization of nanomaterials in cancer nanotechnology research. | [46] |
| 2 | BioPortal | BioPortal provides access to commonly used biomedical ontologies and tools for their analysis. | [47] |
| 3 | The Open Biological and Biomedical Ontologies | A collaborative experiment involving developers of science-based ontologies who are establishing a set of principles for ontology development with the goal of creating a suite of orthogonal interoperable reference ontologies in the biomedical domain. | [48] |
| 4 | Cancer Open Biomedical Resource | A tool for indexing cancer nanotechnology informatics knowledge with open biomedical resources. | [49] |
| 5 | Chemical Entities of Biological Interest | A database and ontology of chemical entities of biological interest. | [50] |
| 6 | Gene Ontology | The Gene Ontology project is a major bioinformatics initiative with the aim of standardizing the representation of gene and gene product attributes across species and databases. | [51] |
| 7 | Foundational Model of Anatomy ontology | The Foundational Model of Anatomy Ontology is an evolving computer-based knowledge source for biomedical informatics. | [52] |
| 8 | Zebrafish Anatomy and Development ontology | A structured, controlled vocabulary of the anatomy and development of the zebrafish. | [53] |
| 9 | Ontology Lookup Service | Provides a web service interface to query multiple ontologies from a single location with a unified output format. | [54] |
| 10 | Phenotype Quality ontology | Phenotype Quality provides ontology of phenotypic qualities intended for use in a number of applications, primarily defining composite phenotypes and phenotype annotation. | [55] |
| No. | Name | Operating System | License | Url |
|---|---|---|---|---|
| 1 | PyMOL | Windows/GNU Linux | Free and commercial | [68] |
| 2 | RasMol | Windows/GNU Linux | GNU General Public License | [69] |
| 3 | Raster3D | GNU Linux/Unix/Mac OS X | Artistic License | [70] |
| 4 | UCSF Chimera | Windows/GNU Linux/Mac OS X | Free for non-commercial use | [71] |
| 5 | VMD | Windows/GNU Linux/Mac OS X/Solaris | VMD License/Free to use | [72] |
| 6 | Cn3D | Windows/Mac OS X | Free to use | [73] |
| 7 | BALLView | Windows/GNU Linux/Unix/Mac OS X | GNU Lesser General Public License | [74] |
| 8 | Jmol | Windows/GNU Linux/Mac OS X | GNU Lesser General Public License | [75] |
| 9 | ICM-Browser | Windows, Mac, and GNU Linux | Commercial software | [76] |
| 10 | Chemkit | GNU Linux | BSD licenses (free to use) | [77] |
| 11 | OpenStructure | GNU Linux/Mac OS X | GNU Lesser General Public License | [78] |
| 12 | VEGA ZZ | Windows/GNU Linux/Mac OS X/Silicon Graphics/Amiga OS | Copyright (free for non-profit academic) | [79] |
| 13 | Molekel | Windows/GNU Linux/Mac OS X | GNU General Public License | [80] |
| 14 | Avogadro | Windows/GNU Linux/Mac OS X | GNU General Public License | [81] |
| 15 | Xeo | Windows/GNU Linux/Mac OS X | GNU General Public License | [82] |
| 16 | ChemSketch | GNU Linux/Mac OS X | Commercial software (free for academic and personal use) | [83] |
| No. | Name | Operating System | License | Url |
|---|---|---|---|---|
| 1 | Autodock 4 | Windows/GNU Linux/Mac OS X | GNU General Public License | [88] |
| 2 | AutoDockVina | Windows/GNU Linux/Mac OS X | Apache license | [89] |
| 3 | Dock | Windows/GNU Linux/Mac OS X | Academic software license | [90] |
| 4 | GOLD | Windows/GNU Linux | Commercial software | [91] |
| 5 | FlexX | Windows/GNU Linux | Commercial software | [92] |
| 6 | Surflex-dock | Windows/GNU Linux | Commercial software | [93] |
| No. | Name | Operating System | Url |
|---|---|---|---|
| 1 | ChemProp | Windows | [104] |
| 2 | OECD ToolBox | Windows/GNU Linux/Mac OS X | [105] |
| 3 | RmSquare | Web based application | [106] |
| 4 | T.E.S.T | Windows/GNU Linux/Mac OS X | [107] |
| 5 | Virtual Computational Chemistry Laboratory | Few online servers and commercial software | [108] |
| 6 | CORAL | Windows | [109] |
| No. | Name | Operating System | License | Url |
|---|---|---|---|---|
| Molecular Dynamic Simulations | ||||
| 1 | Amber | Unix-like systems | Academic/Non-profit/Government: $400 | [112] |
| 2 | NAMD | Windows/GNU Linux/Mac OS X/Solaris | Free for academics | [113] |
| 3 | Gromacs | Solaris, Linux, OS X, Windows by Cygwin, any other Unix variety | GNU Lesser General Public License for version | [114] |
| 4 | CHARMM | Unix-like | Academic research groups for a $600 licensing fee | [115] |
| 5 | Desmond | Linux/Windows | Academic and Commercial | [116] |
| 6 | LAMMPS | Windows/GNU Linux/Mac OS X | GNU General Public License | [117] |
| 7 | TINKER | Windows/GNU Linux/Mac OS X | Available free of charge | [118] |
| 8 | MDynaMix | - | GNU Lesser General Public License | [119] |
| 9 | CP2K | GNU Linux | GNU Lesser General Public License | [120] |
| DFT/Ab initio/Monte Carlo/Semi-empirical calculations | ||||
| 10 | ABINIT | Windows/GNU Linux/Mac OS X | GNU General Public License | [121] |
| 11 | ACES | GNU Linux/SGI | GNU General Public License | [122] |
| 12 | BigDFT | Cross-platform | GNU General Public License | [123] |
| 13 | CASINO | Linux related operating system/Windows by Cygwin | Charge-free license agreements for academics | [124] |
| 14 | CASTEP | - | Commercial software | [125] |
| 15 | COLUMBUS | GNU Linux | Obtained free of charge | [126] |
| 16 | CFOUR | GNU Linux | Charge-free license agreements for academics | [127] |
| 17 | CONQUEST | GNU Linux | GNU General Public License | [128] |
| 18 | CPMD | GNU Linux | CPMD License (free for non-profit) | [129] |
| 19 | CRYSTAL | Windows/GNU Linux/Mac OS X | Commercial software | [130] |
| 20 | DACAPO | GNU Linux | CAMP Open Software project | [131] |
| 21 | DALTON | GNU Linux | Charge-free license agreements for academics | [132] |
| 22 | DFTB+ | GNU Linux | Free for academic, educational, and non-profit research | [133] |
| 23 | DIRAC | Windows/GNU Linux/Mac OS X | Charge-free license agreements | [134] |
| 24 | Elk | GNU Linux | GNU General Public License | [135] |
| 25 | ErgoSCF | GNU Linux | GNU General Public License | [136] |
| 26 | ERKALE | Windows/GNU Linux | GNU General Public License | [137] |
| 27 | FLEUR | GNU Linux | Academic | [138] |
| 28 | FreeON | GNU Linux | GNU General Public License | [139] |
| 29 | GAMESS | Windows/GNU Linux/Mac OS X | Charge free license agreements | [140] |
| 30 | GPAW | GNU Linux/Mac OS X | - | [141] |
| 31 | Gaussian | Windows/GNU Linux/Mac OS X | Commercial software | [142] |
| 32 | JDFTx | GNU Linux | GNU General Public License | [143] |
| 33 | MADNESS | Windows/GNU Linux/Mac OS X | GNU General Public License | [144] |
| 34 | MOPAC | Windows/GNU Linux/Mac OS X | Free and Commercial | [145] |
| 35 | MPQ | GNU Linux | GNU Lesser General Public License | [146] |
| 36 | NWChem | Linux, FreeBSD, Unix and like operating systems, Microsoft Windows, Mac OS X | Educational Community License | [147] |
| 37 | Octopus | GNU Linux | GNU General Public License | [148] |
| 38 | ONETEP | GNU Linux | Commercial software | [149] |
| 39 | OpenAtom | GNU Linux/Mac OS X | Free for academic | [150] |
| 40 | OpenMX | GNU Linux | GNU General Public License | [151] |
| 41 | ORCA | GNU Linux | Free for academic | [152] |
| 42 | PSI | GNU Linux/Windows, Mac OS X | GNU General Public License | [153] |
| 43 | PyQuante | - | BSD licenses (free for academic) | [154] |
| 44 | Quantum ESPRESSO | GNU Linux | Open source distribution | [155] |
| 45 | VASP | GNU Linux | Commercial software | [156] |
| 46 | Yambo Code | GNU Linux | GNU General Public License | [157] |
| No. | Name | Operating System | License | Url |
|---|---|---|---|---|
| 1 | Micro-manager | Windows and Mac | Copyright (distributed free of charge) | [161] |
| 2 | OMERO | Windows, Mac and Linux | GNU General Public License and few commercial licenses | [162] |
| 3 | Bisque | Windows, Mac and Linux | Free and open source | [163] |
| 4 | Bio-Formats | GNU Linux | GNU General Public License | [164] |
| 5 | ImageJ | Windows, Mac and GNU Linux (any java based) | Public domain | [165] |
| 6 | Fiji | Windows, Mac OS X, GNU Linux and Unix | GNU General Public License (the plugin interface is excluded from the license; some plugins have different licenses) | [166] |
| 7 | FARSIGHT TOOLKIT | Windows, Mac OS X and GNU Linux | Apache License, Version 2.0 | [167] |
| 8 | ICY | Windows, Mac OS X and GNU Linux | GNU General Public License | [168] |
| 9 | CellProfiler | Windows, Mac OS X, and GNU Linux | GNU General Public License and some portions are also BSD-licensed | [169] |
| 10 | Micro-Pilot | Windows | Free for academic users | [170] |
| 12 | Cell ID | Windows and GNU Linux | GNU Lesser General Public License | [171] |
| 13 | bioView3D | Windows, Mac OS X, and GNU Linux | Open source (free for non-commercial) | [172] |
| 14 | V3D | Windows, Mac and GNU Linux | Free for non-profit research | [173] |
| 15 | IMOD | Windows, Mac and GNU Linux | Free to use | [174] |
| 16 | iCluster | Windows, Mac OS X, and GNU Linux | GNU General Public License | [175] |
| 17 | Image Surfer | Windows and Mac | Free to use | [176] |
| No. | Name | Operating System | Url |
|---|---|---|---|
| 1 | Tiamat | Windows | [182] |
| 2 | caDNAno | Web-Accessible | [183] |
| 3 | SARSE | GNU Linux/Mac | [184] |
| 4 | cadnano | Windows, OSX and GNU Linux | [185] |
| 5 | Uniquimer3D | Windows | [186] |
| 6 | CoNTub | Windows, Mac OS X, GNU Linux and Unix | [187] |
| 7 | Fullerene | GNU Linux/Unix and Mac | [188] |
| 8 | CS Catalyst | Windows | [189] |
| 9 | Ninithi | Windows | [190] |
| 10 | TubeGen | Web-Accessible | [191] |
| 11 | Wrapping | Windows | [192] |
© 2014 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Panneerselvam, S.; Choi, S. Nanoinformatics: Emerging Databases and Available Tools. Int. J. Mol. Sci. 2014, 15, 7158-7182. https://doi.org/10.3390/ijms15057158
Panneerselvam S, Choi S. Nanoinformatics: Emerging Databases and Available Tools. International Journal of Molecular Sciences. 2014; 15(5):7158-7182. https://doi.org/10.3390/ijms15057158
Chicago/Turabian StylePanneerselvam, Suresh, and Sangdun Choi. 2014. "Nanoinformatics: Emerging Databases and Available Tools" International Journal of Molecular Sciences 15, no. 5: 7158-7182. https://doi.org/10.3390/ijms15057158
APA StylePanneerselvam, S., & Choi, S. (2014). Nanoinformatics: Emerging Databases and Available Tools. International Journal of Molecular Sciences, 15(5), 7158-7182. https://doi.org/10.3390/ijms15057158




