X-ray-Induced Changes in the Expression of Inflammation-Related Genes in Human Peripheral Blood
Abstract
:1. Introduction
2. Result and Discussion
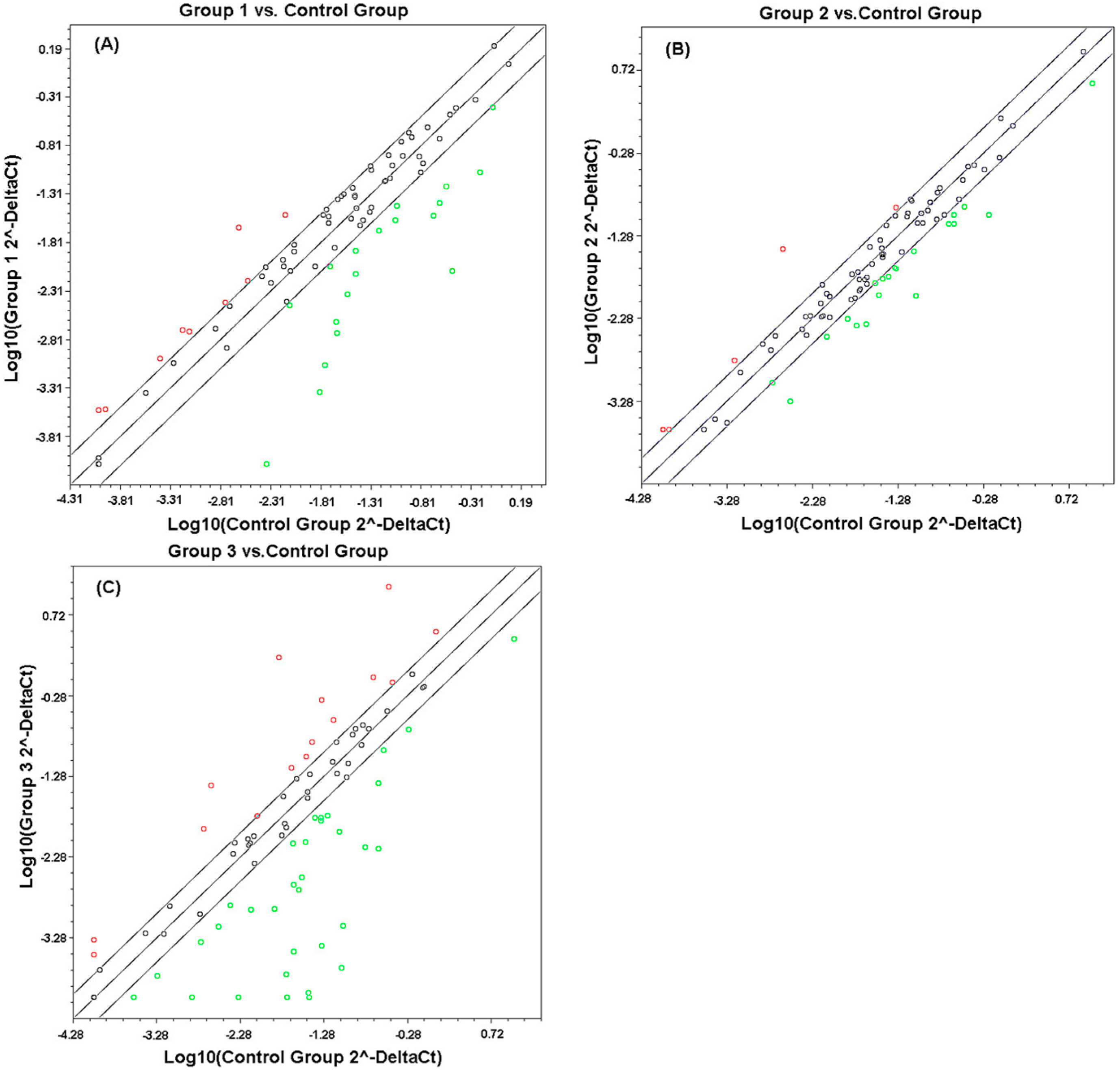
2.1. Comparison of Transcriptional Profiles of Human Peripheral Blood Cells after Exposure to Different X-ray Doses
| Expression Changes | Gene Number | ||
|---|---|---|---|
| 0.5 Gy | 3 Gy | 10 Gy | |
| Up-regulated | 9 | 8 | 16 |
| Down-regulated | 19 | 19 | 32 |
| Total | 28 | 27 | 48 |
| Symbol | GenBank | Description | Fold Regulation | ||
|---|---|---|---|---|---|
| 0.5 Gy | 3 Gy | 10 Gy | |||
| CCL2 | NM_002982 | Chemokine (C–C motif) ligand 2 | −39.5871 | −2.6739 | 37.1193 |
| CXCL1 | NM_001511 | Chemokine (C–X–C motif) ligand 1 (melanoma growth stimulating activity, alpha) | −7.0153 | −2.8083 | 4.2787 |
| NLRX1 | NM_024618 | NLR family member X1 | 2.1804 | −5.4679 | −4.0013 |
| MEFV | NM_000243 | Mediterranean fever | −6.3207 | −2.0551 | −9.6336 |
| CXCL2 | NM_002089 | Chemokine (C–X–C motif) ligand 2 | −5.9388 | −2.5035 | −35.3923 |
| P2RX7 | NM_002562 | Purinergic receptor P2X, ligand-gated ion channel, 7 | −9.4334 | −4.9114 | −2.8395 |
| PTGS2 | NM_000963 | Prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) | −3.2288 | −8.6517 | −382.4951 |
| TNF | NM_000594 | Tumor necrosis factor | −4.7188 | −2.1824 | −359.0338 |
| IFNB1 | NM_002176 | Interferon, beta 1, fibroblast | 3.0783 | 2.5595 | 3.4564 |
| TNFSF4 | NM_003326 | Tumor necrosis factor (ligand) superfamily, member 4 | 9.3354 | 15.2667 | 17.2687 |
| IL1B | NM_000576 | Interleukin 1, beta | −7.3595 | −6.4265 | |
| IL6 | NM_000600 | Interleukin 6 (interferon, beta 2) | −20.1839 | −3.9760 | |
| NLRP4 | NM_134444 | NLR family, pyrin domain containing 4 | 2.6747 | 2.1798 | |
| TNFSF11 | NM_003701 | Tumor necrosis factor (ligand) superfamily, member 11 | 2.9932 | 2.5231 | |
| CASP5 | NM_004347 | Caspase 5, apoptosis-related cysteine peptidase | −12.5221 | −63.2637 | |
| CCL7 | NM_006273 | Chemokine (C–C motif) ligand 7 | −33.7867 | 103.5741 | |
| IRF2 | NM_002199 | Interferon regulatory factor 2 | −2.8683 | −3.4050 | |
| MAPK13 | NM_002754 | Mitogen-activated protein kinase 13 | −2.7257 | −313.9804 | |
| NLRP1 | NM_033004 | NLR family, pyrin domain containing 1 | −2.3960 | −119.3437 | |
| NLRP3 | NM_183395 | NLR family, pyrin domain containing 3 | −2.1744 | −197.9183 | |
| PEA15 | NM_003768 | Phosphoprotein enriched in astrocytes 15 | −4.6696 | −2.4521 | |
| CARD18 | NM_021571 | Caspase recruitment domain family, member 18 | 2.5595 | 5.3355 | |
| SUGT1 | NM_006704 | SGT1, suppressor of G2 allele of SKP1 ( S. cerevisiae) | 2.3183 | 9.3633 | |
| CIITA | NM_000246 | Class II, major histocompatibility complex, transactivator | −2.6392 | −11.2611 | |
| IKBKG | NM_003639 | Inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma | −2.2396 | −2.9962 | |
| IRAK1 | NM_001569 | Interleukin-1 receptor-associated kinase 1 | −3.1312 | −3.8438 | |
| IRF1 | NM_002198 | Interferon regulatory factor 1 | −3.2151 | −5.4590 | |
| MAPK3 | NM_002746 | Mitogen-activated protein kinase 3 | −2.3435 | −119.1749 | |
| NFKBIB | NM_002503 | Nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta | −2.4474 | −2.5170 | |
| NLRC5 | NM_032206 | NLR family, CARD domain containing 5 | −2.3274 | −7.4314 | |
| NOD1 | NM_006092 | Nucleotide-binding oligomerization domain containing 1 | −2.0327 | −3.8188 | |
| CD40LG | NM_000074 | CD40 ligand | 4.3937 | ||
| IL12A | NM_000882 | Interleukin 12A (natural killer cell stimulatory factor 1, cytotoxic lymphocyte maturation factor 1, p35) | 2.5043 | ||
| MAPK12 | NM_002969 | Mitogen-activated protein kinase 12 | 2.5661 | ||
| MOK | NM_014226 | Renal tumor antigen | 2.1817 | ||
| IL12B | NM_002187 | Interleukin 12B (natural killer cell stimulatory factor 2, cytotoxic lymphocyte maturation factor 2, p40) | −55.6021 | ||
| IL18 | NM_001562 | Interleukin 18 (interferon-gamma-inducing factor) | −2.2094 | ||
| NFKBIA | NM_020529 | Nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha | −2.0706 | ||
| IL33 | NM_033439 | Interleukin 33 | 2.5595 | ||
| NLRP5 | NM_153447 | NLR family, pyrin domain containing 5 | 2.5595 | ||
| BCL2L1 | NM_138578 | Bcl2-like 1 | −2.4431 | ||
| AIM2 | NM_004833 | Absent in melanoma 2 | 2.0252 | ||
| HSP90AA1 | NM_001017963 | Heat shock protein 90 kDa alpha (cytosolic), class A member 1 | 2.2279 | ||
| HSP90B1 | NM_003299 | Heat shock protein 90 kDa beta (Grp94), member 1 | 2.8132 | ||
| IFNG | NM_000619 | Interferon, gamma | 6.1451 | ||
| MAP3K7 | NM_003188 | Mitogen-activated protein kinase kinase kinase 7 | 2.8662 | ||
| MYD88 | NM_002468 | Myeloid differentiation primary response gene (88) | 3.8568 | ||
| NLRC4 | NM_021209 | NLR family, CARD domain containing 4 | 3.6631 | ||
| PANX1 | NM_015368 | Pannexin 1 | 2.0607 | ||
| RIPK2 | NM_003821 | Receptor-interacting serine-threonine kinase 2 | 3.1423 | ||
| CARD6 | NM_032587 | Caspase recruitment domain family, member 6 | −5.9438 | ||
| CASP1 | NM_033292 | Caspase 1, apoptosis-related cysteine peptidase (interleukin 1, beta, convertase) | −23.2608 | ||
| FADD | NM_003824 | Fas (TNFRSF6)-associated via death domain | −100.3419 | ||
| MAPK11 | NM_002751 | Mitogen-activated protein kinase 11 | −2.9619 | ||
| MAPK9 | NM_002752 | Mitogen-activated protein kinase 9 | −12.8590 | ||
| NLRP12 | NM_033297 | NLR family, pyrin domain containing 12 | −14.3709 | ||
| NLRP6 | NM_138329 | NLR family, pyrin domain containing 6 | −9.6825 | ||
| PSTPIP1 | NM_003978 | Proline-serine-threonine phosphatase-interacting protein 1 | −3.3141 | ||
| PYDC1 | NM_152901 | PYD (pyrin domain) containing 1 | −2.9089 | ||
| TIRAP | NM_001039661 | Toll-interleukin 1 receptor (TIR) domain containing adaptor protein | −3.0328 | ||
| TNFSF14 | NM_003807 | Tumor necrosis factor (ligand) superfamily, member 14 | −51.3506 | ||
| TXNIP | NM_006472 | Thioredoxin-interacting protein | −2.7110 | ||
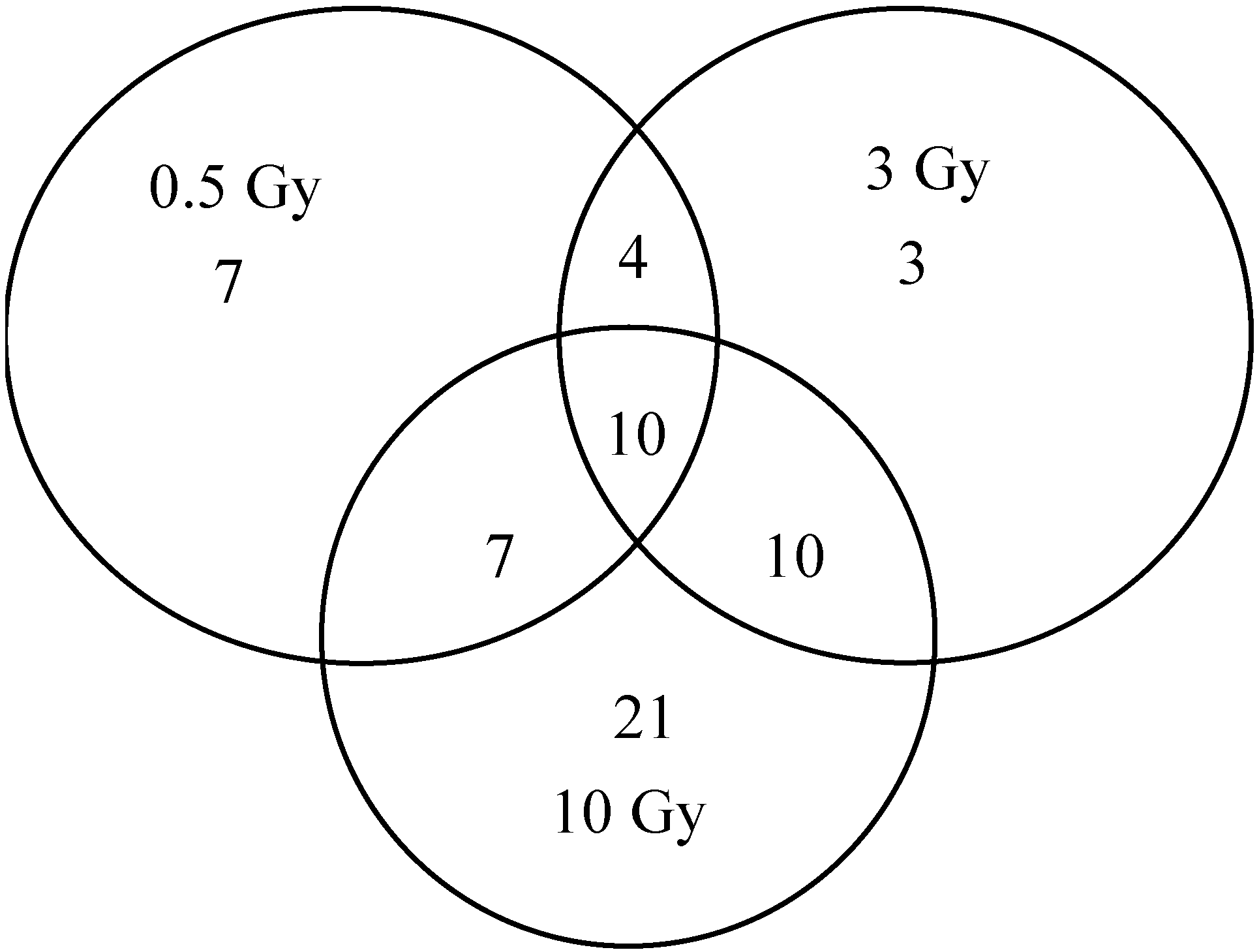

2.2. Comparison of qPCR Array with Other Studies
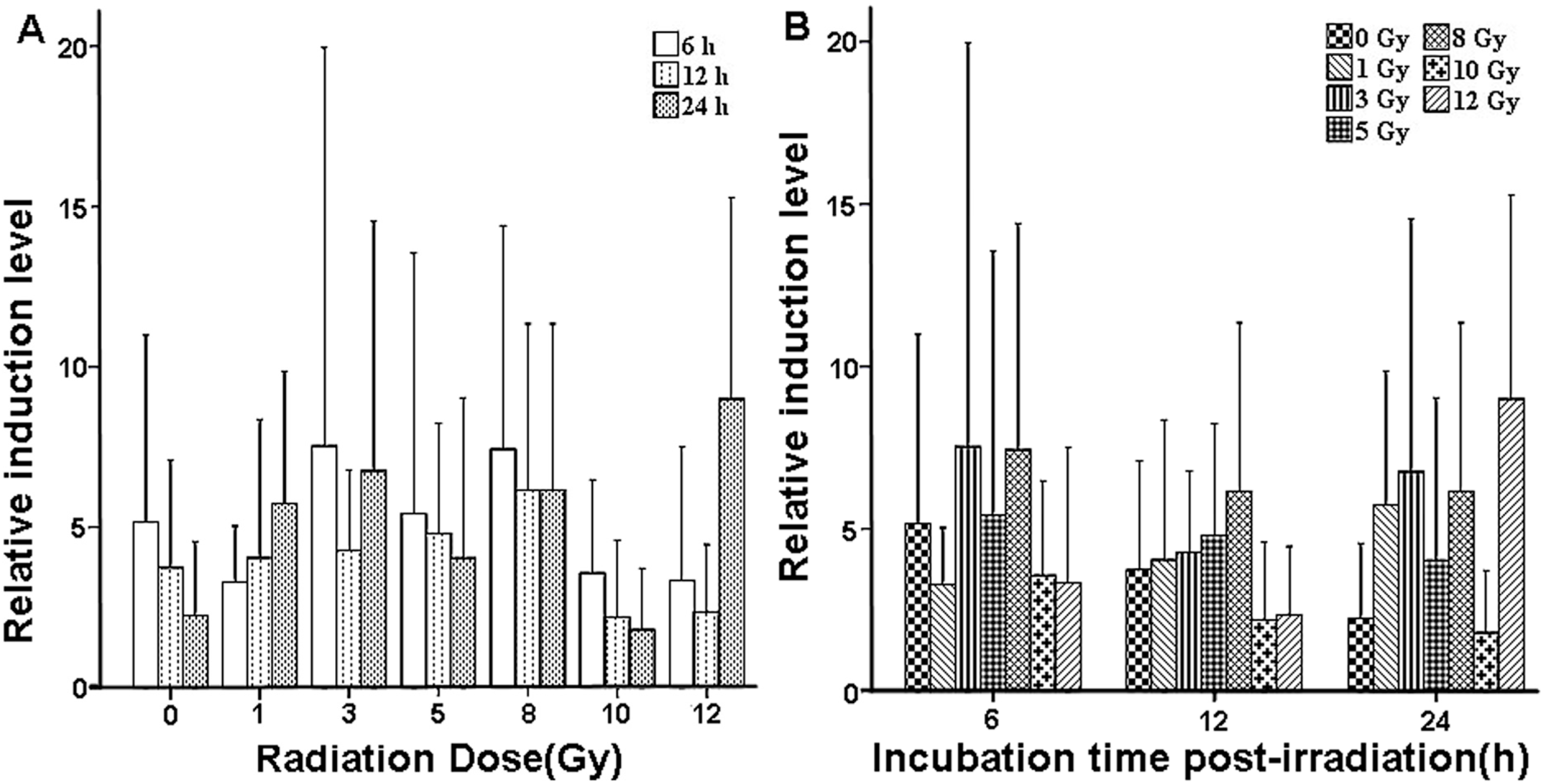
2.3. Validation of X-ray-Induced Expression Changes of CCL2 in Human Peripheral Blood
| Dose (Gy) | Relative Induction Level Relative Induction | ||
|---|---|---|---|
| 6 h | 12 h | 24 h | |
| 0 | 5.1547 ± 5.8280 | 3.7394 ± 3.3493 | 2.2400 ± 2.3159 |
| 1 | 3.1912 ± 1.7462 | 4.0470 ± 4.3098 | 5.7423 ± 4.1118 |
| 3 | 7.5279 ± 12.4202 | 4.2638 ± 2.5147 | 6.7529 ± 7.7754 |
| 5 | 5.4198 ± 8.1194 | 4.8103 ± 3.4379 | 4.0393 ± 4.9922 |
| 8 | 7.4343 ± 6.9659 | 6.1453 ± 5.1910 | 6.1574 ± 5.1804 |
| 10 | 3.5730 ± 2.9049 | 2.2096 ± 2.3899 | 1.8030 ± 1.9142 |
| 12 | 3.3331 ± 4.1677 | 2.3643 ± 2.0885 | 8.9944 ± 6.2734 |
3. Materials and Methods
3.1. Acquisition of Samples from Healthy Donors and Ethics Statement
3.2. In Vitro Irradiation of qPCR Array Analysis Samples and RNA Extraction
3.3. Inflammation-Related Gene Expression Measurement Using qPCR Array
3.4. In Vitro Irradiation of qRT-PCR Assay Samples and Subsequent Cell Isolation
3.5. Chemokine (C–C Motif) Ligand 2 (CCL2) Expression Measurement Using qRT-PCR
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Amundson, S.A.; Bittner, M.; Meltzer, P.; Trent, J.; Fornace, A.J. Biological indicators for the identification of ionizing radiation exposure in humans. Expert Rev. Mol. Diagn. 2001, 1, 211–219. [Google Scholar] [CrossRef]
- Blakely, W.F.; Salter, C.A.; Prasanna, P.G. Early-response biological dosimetry-recommended countermeasure enhancements for mass-casualty radiological incidents and terrorism. Health Phys. 2005, 89, 494–504. [Google Scholar] [CrossRef] [PubMed]
- Pellmar, T.C.; Rockwell, S. Priority list of research areas for radiological nuclear threat countermeasures. Radiat. Res. 2005, 163, 115–123. [Google Scholar] [PubMed]
- Rea, M.E.; Gougelet, R.M.; Nicolalde, R.J.; Geiling, J.A.; Swartz, H.M. Proposed triage categories for large-scale radiation incidents using high-accuracy biodosimetry methods. Health Phys. 2010, 98, 136–144. [Google Scholar] [CrossRef] [PubMed]
- Amundson, S.A.; Chen, D.J. Inverse dose-rate effect for mutation induction by γ-rays in human lymphoblasts. Int. J. Radiat. Biol. 1996, 69, 555–563. [Google Scholar] [CrossRef] [PubMed]
- Vilenchik, M.M.; Knudson, A.G. Inverse radiation dose-rate effects on somatic and germ-line mutations and DNA damage rates. Proc. Natl. Acad. Sci. USA 2000, 97, 5381–5386. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.R.; Mohankumar, M.N.; Hamza, V.Z.; Jeevanram, R.K. Dose-rate effect on the induction of HPRT mutants in human G0 lymphocytes exposed in vitro to gamma radiation. Radiat. Res. 2006, 165, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.; Wang, Y.; Li, T.; Shi, K.; Li, Z.; Ma, Y.; Li, F.; Luo, H.; Yang, Y.; Xu, C. Heat shock protein 90-mediated inactivation of nuclear factor-κB switches autophagy to apoptosis through becn1 transcriptional inhibition in selenite-induced NB4 cells. Mol. Biol. Cell 2011, 22, 1167–1180. [Google Scholar] [CrossRef]
- Wu, D.; Lee, Y.C.; Liu, H.C.; Yuan, R.Y.; Chiou, H.Y.; Hung, C.H.; Hu, C.J. Identification of TLR downstream pathways in stroke patients. Clin. Biochem. 2013, 46, 1058–1064. [Google Scholar] [CrossRef] [PubMed]
- Azab, S.S.; Salama, S.A.; Abdel-Naim, A.B.; Khalifa, A.E.; El-Demerdash, E.; Al-Hendy, A. 2-Methoxyestradiol and multidrug resistance: Can 2-methoxyestradiol chemosensitize resistant breast cancer cells? Breast Cancer Res. Treat. 2009, 113, 9–19. [Google Scholar]
- Fakhari, F.D.; Dittmer, D.P. Charting latency transcripts in Kaposi’s sarcoma-associated herpesvirus by whole-genome real-time quantitative PCR. J. Virol. 2002, 76, 6213–6223. [Google Scholar] [CrossRef]
- Dittmer, D.P. Transcription profile of Kaposi’s sarcoma-associated herpesvirus in primary Kaposi’s sarcoma lesions as determined by real-time PCR arrays. Cancer Res. 2003, 63, 2010–2015. [Google Scholar] [PubMed]
- Airoldi, I.; di Carlo, E.; Cocco, C.; Taverniti, G.; D’Antuono, T.; Ognio, E.; Watanabe, M.; Ribatti, D.; Pistoia, V. Endogenous IL-12 triggers an antiangiogenic program in melanoma cells. Proc. Natl. Acad. Sci. USA 2007, 104, 3996–4001. [Google Scholar] [CrossRef]
- Zhang, D.J.; Qu, J.; Xiong, M.; Qiao, Y.Y.; Wang, D.P.; Yu, J.Y.; Zhao, X.H. Effects of sea water immersion on the functional genes in vascular endothelial cells. Med. J. Chin. 2009, 34, 102–106. [Google Scholar]
- Gude, N.; Muraski, J.; Rubio, M.; Kajstura, J.; Schaefer, E.; Anversa, P.; Sussman, M.A. Akt promotes increased cardiomyocyte cycling and expansion of the cardiac progenitor cell population. Circ. Res. 2006, 99, 381–388. [Google Scholar] [CrossRef] [PubMed]
- Rangel-Moreno, J.; Hartson, L.; Navarro, C.; Gaxiola, M.; Selman, M.; Randall, T.D. Inducible bronchus-associated lymphoid tissue (iBALT) in patients with pulmonary complications of rheumatoid arthritis. J. Clin. Investig. 2006, 116, 3183–3194. [Google Scholar]
- Hansel, N.N.; Cheadle, C.; Diette, G.B.; Wright, J.; Thompson, K.M.; Barnes, K.C.; Georas, S.N. Analysis of CD4+ T-cell gene expression in allergic subjects using two different microarray platforms. Allergy 2008, 63, 366–369. [Google Scholar] [CrossRef]
- Rödel, F.; Frey, B.; Gaipl, U.; Keilholz, L.; Fournier, C.; Manda, K.; Schöllnberger, H.; Hildebrandt, G.; Rödel, C. Modulation of inflammatory immune reactions by low-dose ionizing radiation: Molecular mechanisms and clinical application. Curr. Med. Chem. 2012, 19, 1741–1750. [Google Scholar] [CrossRef] [PubMed]
- Amundson, S.A.; Do, K.T.; Shahab, S.; Bittner, M.; Meltzer, P.; Trent, J.; Fornace, A.J. Identification of potential mRNA biomarkers in peripheral blood lymphocytes for human exposure to ionizing radiation. Radiat. Res. 2000, 154, 342–346. [Google Scholar] [CrossRef] [PubMed]
- Johnston, C.J.; Williams, J.P.; Okunieff, P.; Finkelstein, J.N. Radiation-induced pulmonary fibrosis: Examination of chemokine and chemokine receptor families. Radiat. Res. 2002, 157, 256–265. [Google Scholar] [CrossRef]
- Bennett, L.; Palucka, A.K.; Arce, E.; Cantrell, V.; Borvak, J.; Banchereau, J.; Pascual, V. Interferon and granulopoiesis in systemic lupus erythematosus blood. J. Exp. Med. 2003, 197, 711–723. [Google Scholar] [CrossRef] [PubMed]
- Ponnampalam, A.P.; Weston, G.C.; Trajstman, A.C.; Susil, B.; Rogers, P.A. Molecular classification of human endometrial cycle stages by transcriptional profiling. Mol. Hum. Reprod. 2004, 10, 879–893. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.Q.; Kim, M.; Kim, J.H.; Kong, G.; Kang, K.S.; Kim, H.L.; Yoon, B.I.; Lee, M.O.; Lee, B.H. Differential gene expression and lipid metabolism in fatty liver induced by acute ethanol treatment in mice. Toxicol. Appl. Pharmacol. 2007, 223, 225–233. [Google Scholar] [CrossRef] [PubMed]
- Pacini, S; Ruggiero, M; Sardi, I; Aterini, S; Gulisano, F; Gulisano, M. Exposure to global system for mobile communication (GSM) cellular phone radiofrequecy alters gene expression, proliferation, and morphology of human skin fibroblasts. Oncol. Res. 2002, 13, 19–24. [Google Scholar]
- Chilakapati, J.; Wallace, K.; Ren, H.; Aterini, S.; Gulisano, F.; Gulisano, M. Genome-wide analysis of BEAS-2B cells exposed to trivalent arsenicals and dimethylthioarsinic acid. Toxicology 2010, 268, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Long, X.H.; Zhao, Z.; He, X.P.; Wang, H.P.; Xu, Q.Z.; An, J.; Bai, B.; Sui, J.L.; Zhou, P.K. Dose-dependent expression changes of early response genes to ionizing radiation in human lymphoblastoid cells. Int. J. Mol. Med. 2007, 19, 607–615. [Google Scholar] [PubMed]
- Lu, X.; Li, Y.W.; Feng, J.B.; Chen, D.Q.; Liu, Q.J. Differential gene expression profiling in lymphocyte line following exposure to different doses of 60Co γ-ray by gene chip. J. Radiat. Res. Radiat. Process. 2009, 27, 244–247. [Google Scholar]
- Wang, H.P.; Long, X.H.; Xu, Q.Z.; Bai, B.; Sui, J.L.; Zhou, P.K. Screening of radiation-induced genes in human lymphoblastoid cells with 20 cGy of γ-ray by gene chip. Radiat. Prot. 2006, 26, 151–156. [Google Scholar]
- Long, X.H.; Xu, Q.Z.; He, X.P.; An, J.; Sui, J.L.; Bai, B.; Zhou, P.K. Comparison of transcription profiles between 2 Gy and 10 Gy irradiated human lymphoblastoid cells. Chin. J. Radiol. Med. Prot. 2006, 26, 110–113. [Google Scholar]
- Li, K.; Liu, W.; Hou, D.J.; Qiao, J.W.; Ma, Y.; Jia, X.M.; Li, J.Q. Gene transcriptional profiles in human lymphoblastoid cells with low and high doses of irradiation. Chin. J. Radiol. Med. Prot. 2012, 32, 249–255. [Google Scholar]
- Kulkarni, R.; Thomas, R.A.; Tucker, J.D. Expression of DNA repair and apoptosis genes in mitochondrial mutant and normal cells following exposure to ionizing radiation. Environ. Mol. Mutagen. 2011, 52, 229–237. [Google Scholar] [CrossRef] [PubMed]
- Veeraraghavan, J.; Aravindan, S.; Natarajan, M.; Awasthi, V.; Herman, T.S.; Aravindan, N. Neem leaf extract induces radiosensitization in human neuroblastoma xenograft through modulation of apoptotic pathway. Anticancer Res. 2011, 31, 161–170. [Google Scholar] [PubMed]
- Hneino, M.; Blirando, K.; Buard, V.; Tarlet, G.; Benderitter, M.; Hoodless, P.; François, A.; Milliat, F. The TG-interacting factor TGIF1 regulates stress-induced proinflammatory phenotype of endothelial cells. J. Biol. Chem. 2012, 287, 38913–38921. [Google Scholar] [CrossRef]
- Malik, I.A.; Moriconi, F.; Sheikh, N.; Naz, N.; Khan, S.; Dudas, J.; Mansuroglu, T.; Hess, C.F.; Rave-Fränk, M.; Christiansen, H.; et al. Single-dose gamma-irradiation induces upregulation of chemokine gene expression and recruitment of granulocytes into the portal area but not into other regions of rat hepatic tissue. Am. J. Pathol. 2010, 176, 1801–1815. [Google Scholar] [CrossRef] [PubMed]
- Han, K.H.; Ryu, J.W.; Lim, K.E.; Lee, S.H.; Kim, Y.; Hwang, C.S.; Choi, J.Y.; Han, K.O. Vascular expression of the chemokine CX3CL1 promotes osteoclast recruitment and exacerbates bone resorption in an irradiated murine model. Bone 2014, 61, 91–101. [Google Scholar] [CrossRef]
- Kyrkanides, S.; Moore, A.H.; Olschowka, J.A.; Daeschner, J.C.; Williams, J.P.; Hansen, J.T.; Kerry O’Banion, M. Cyclooxygenase-2 modulates brain inflammation-related gene expression in central nervous system radiation injury. Brain Res. Mol. Brain Res. 2002, 104, 159–169. [Google Scholar] [CrossRef] [PubMed]
- Bucci, B.; Misiti, S.; Cannizzaro, A.; Marchese, R.; Raza, G.H.; Miceli, R.; Stigliano, A.; Amendola, D.; Monti, O.; Biancolella, M.; et al. Fractionated ionizing radiation exposure induces apoptosis through caspase-3 activation and reactive oxygen species generation. Anticancer Res. 2006, 26, 4549–4557. [Google Scholar]
- Chang, J.T.; Chan, S.H.; Lin, C.Y.; Lin, T.Y.; Wang, H.M.; Liao, C.T.; Wang, T.H.; Lee, L.Y.; Cheng, A.J. Differentially expressed genes in radioresistant nasopharyngeal cancer cells: GP96 and GDF15. Mol. Cancer Ther. 2007, 6, 2271–2279. [Google Scholar] [CrossRef]
- Zhang, B.; Wang, Y.; Pang, X.; Su, Y.; Ai, G.; Wang, T. ER stress induced by ionising radiation in IEC-6 cells. Int. J. Radiat. Biol. 2010, 86, 429–435. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, V.N.; Ghandhi, S.A.; Zhou, H.; Huang, S.X.; Chai, Y.; Amundson, S.A.; Hei, T.K. Radiation response and regulation of apoptosis induced by a combination of TRAIL and CHX in cells lacking mitochondrial DNA: A role for NF-κB–STAT3-directed gene expression. Exp. Cell Res. 2011, 317, 1548–1566. [Google Scholar] [CrossRef] [PubMed]
- El-Saghire, H.; Vandevoorde, C.; Ost, P.; Monsieurs, P.; Michaux, A.; de Meerleer, G.; Baatout, S.; Thierens, H. Intensity modulated radiotherapy induces pro-inflammatory and pro-survival responses in prostate cancer patients. Int. J. Oncol. 2014, 44, 1073–1083. [Google Scholar]
- Hwang, S.Y.; Jung, J.S.; Kim, T.H.; Lim, S.J.; Oh, E.S.; Kim, J.Y.; Ji, K.A.; Joe, E.H.; Cho, K.H.; Han, I.O. Ionizing radiation induces astrocyte gliosis through microglia activation. Neurobiol. Dis. 2006, 21, 457–467. [Google Scholar] [CrossRef] [PubMed]
- Han, S.K.; Song, J.Y.; Yun, Y.S.; Yi, S.Y. Gamma irradiation-reduced IFN-γ expression, STAT1 signals, and cell-mediated immunity. J. Biochem. Mol. Biol. 2002, 35, 583–589. [Google Scholar] [CrossRef]
- Datta, K.; Hyduke, D.R.; Suman, S.; Moon, B.H.; Johnson, M.D.; Fornace, A.J. Exposure to ionizing radiation induced persistent gene expression changes in mouse mammary gland. Radiat. Oncol. 2012, 7, 205. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, K.; Sakakura, C.; Miyagawa, K.; Kuriu, Y.; Kin, S.; Nakase, Y.; Hagiwara, A.; Mitsufuji, S.; Okazaki, Y.; Hayashizaki, Y.; et al. Differential gene expression profiles of radioresistant oesophageal cancer cell lines established by continuous fractionated irradiation. Br. J. Cancer 2004, 91, 1543–1550. [Google Scholar] [CrossRef] [PubMed]
- El-Saghire, H.; Michaux, A.; Thierens, H.; Baatout, S. Low doses of ionizing radiation induce immune-stimulatory responses in isolated human primary monocytes. Int. J. Mol. Med. 2013, 32, 1407–1414. [Google Scholar] [PubMed]
- Sawajiri, M.; Nomura, Y.; Banik, S.; Nishikiori, R.; Sonoda, Y.; Tanimoto, K. Irradiation effect on osteoclastogenesis stimulated by breast cancer cells. Health Phys. 2011, 101, 259–264. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Tang, Q.; Post, J.; Zhou, H.; Huang, X.B.; Zhang, X.D.; Wang, Q.; Sun, Y.M.; Fan, F.Y. Effect of radiation on the Notch signaling pathway in osteoblasts. Int. J. Mol. Med. 2013, 31, 698–706. [Google Scholar] [PubMed]
- Sung, H.Y.; Wu, H.G.; Ahn, J.H.; Park, W.Y. Dcr3 inhibit p53-dependent apoptosis in γ-irradiated lung cancer cells. Int. J. Radiat. Biol. 2010, 86, 780–790. [Google Scholar] [CrossRef]
- Aravindan, N.; Thomas, C.R.; Aravindan, S.; Mohan, A.S.; Veeraraghavan, J.; Natarajan, M. Irreversible EGFR inhibitor EKB-569 targets low-LET γ-radiation-triggered rel orchestration and potentiates cell death in squamous cell carcinoma. PLoS One 2011, 6, e29705. [Google Scholar] [CrossRef] [PubMed]
- Aravindan, S.; Natarajan, M.; Herman, T.S.; Awasthi, V.; Aravindan, N. Molecular basis of “hypoxic” breast cancer cell radio-sensitization: Phytochemicals converge on radiation induced Rel signaling. Radiat. Oncol. 2013, 8, 46. [Google Scholar] [CrossRef] [PubMed]
- Aravindan, S.; Natarajan, M.; Ramraj, S.K.; Pandian, V.; Khan, F.H.; Herman, T.S.; Aravindan, N. Abscopal effect of low-LET γ-radiation mediated through Rel protein signal transduction in a mouse model of nontargeted radiation response. Cancer Gene Ther. 2014, 21, 54–59. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Wang, Y.; Du, L.; Pandian, V.; Khan, F.H.; Herman, T.S.; Aravindan, N. Resveratrol inhibits ionising irradiation-induced inflammation in MSCs by activating SIRT1 and limiting NLRP-3 inflammasome activation. Int. J. Mol. Sci. 2013, 14, 14105–14118. [Google Scholar] [CrossRef]
- Gridley, D.S.; Pecaut, M.; Rizvi, A.; Coutrakon, G.B.; Luo-Owen, X.; Makinde, A.Y.; Slater, J.M. Low-dose, low-dose-rate proton radiation modulates CD4+ T cell gene expression. Int. J. Radiat. Biol. 2009, 85, 250–261. [Google Scholar] [CrossRef]
- Lindgren, T.; Stigbrand, T.; Riklund, K.; Johansson, L.; Eriksson, D. Gene expression profiling in MOLT-4 cells during gamma-radiation-induced apoptosis. Tumour. Biol. 2012, 33, 689–700. [Google Scholar] [CrossRef] [PubMed]
- Gridley, D.S.; Slater, J.M.; Luo-Owen, X.; Rizvi, A.; Chapes, S.K.; Stodieck, L.S.; Ferguson, V.L.; Pecaut, M.J. Spaceflight effects on Tlymphocyte distribution, function and gene expression. J. Appl. Physiol. 2009, 106, 194–202. [Google Scholar] [CrossRef]
- Bo, H.; Ghazizadeh, M.; Shimizu, H.; Kurihara, Y.; Egawa, S.; Moriyama, Y.; Tajiri, T.; Kawanami, O. Effect of ionizing irradiation on human esophageal cancer cell lines by cDNA microarray gene expression analysis. J. Nippon Med. Sch. 2004, 71, 172–180. [Google Scholar] [CrossRef] [PubMed]
- Sanzari, J.K.; Wambi, C.; Lewis-Wambi, J.S.; Kennedy, A.R. Antioxidant dietary supplementation in mice exposed to proton radiation attenuates expression of programmed cell death-associated genes. Radiat. Res. 2011, 175, 650–656. [Google Scholar] [CrossRef] [PubMed]
- Veeraraghavan, J.; Natarajan, M.; Aravindan, S.; Herman, T.S.; Aravindan, N. Radiation-triggered tumor necrosis factor (TNF) α-NFκB cross-signaling favors survival advantage in human neuroblastoma cells. J. Biol. Chem. 2011, 286, 21588–21600. [Google Scholar] [CrossRef] [PubMed]
- Sugihara, T.; Murano, H.; Nakamura, M.; Ichinohe, K.; Tanaka, K. Activation of interferon-stimulated genes by γ-ray irradiation independently of the ataxia telangiectasia mutated-p53 pathway. Mol. Cancer Res. 2011, 9, 476–484. [Google Scholar] [CrossRef] [PubMed]
- Weston, V.J.; Austen, B.; Wei, W.; Marston, E.; Alvi, A.; Lawson, S.; Darbyshire, P.J.; Griffiths, M.; Hill, F.; Mann, J.R.; et al. Apoptotic resistance to ionizing radiation in pediatric B-precursor acute lymphoblastic leukemia frequently involves increased NF-κB survival pathway signaling. Blood 2004, 104, 1465–1473. [Google Scholar] [CrossRef] [PubMed]
- Paul, S.; Amundson, S.A. Development of gene expression signatures for practical radiation biodosimetry. Int. J. Radiat. Oncol. Biol. Phys. 2008, 71, 1236–1244. [Google Scholar] [CrossRef]
- Dressman, H.K.; Muramoto, G.G.; Chao, N.J.; Meadows, S.; Marshall, D.; Ginsburg, G.S.; Nevins, J.R.; Chute, J.P. Gene expression signatures that predict radiation exposure in mice and humans. PLoS Med. 2007, 4, e106. [Google Scholar] [CrossRef]
- Pogosova-Agadjanyan, E.L.; Fan, W.; Georges, G.E.; Schwartz, J.L.; Kepler, C.M.; Lee, H.; Suchanek, A.L.; Cronk, M.R.; Brumbaugh, A.; Engel, J.H.; et al. Identification of radiation-induced expression changes in nonimmortalized human T cells. Radiat. Res. 2011, 175, 172–184. [Google Scholar] [CrossRef] [PubMed]
- Li, S.E.; Guo, F.; Wang, P.; Han, L.; Wang, X.A.; Li, J.; Guo, Y.; LÜ, Y.M. X-Ray-induced expression changes of TNFSF4 gene in human peripheral blood. Biomed. Environ. Sci. 2014, 27, 729–732. [Google Scholar] [PubMed]
- Tao, L.L.; Shi, S.J.; Chen, L.B.; Huang, G.C. Expression of monocyte chemotactic protein-1/CCL2 in gastric cancer and its relationship with tumor hypoxia. World J. Gastroenterol. 2014, 20, 4421–4427. [Google Scholar] [CrossRef] [PubMed]
- Affò, S.; Sancho-Bru, P. CCL2: A link between hepatic inflammation, fibrosis and angiogenesis? Gut 2014. [Google Scholar] [CrossRef]
- Lee, W.H.; Sonntag, W.E.; Mitschelen, M.; Yan, H.; Lee, Y.W. Irradiation induces regionally specific alterations in proinflammatory environments in rat brain. Int. J. Radiat. Biol. 2010, 86, 132–144. [Google Scholar] [CrossRef] [PubMed]
- Kalm, M.; Roughton, K.; Blomgren, K. Lipopolysaccharide sensitized male and female juvenile brains to ionizing radiation. Cell Death Dis. 2013, 4, e962. [Google Scholar] [CrossRef]
- Nalla, A.K.; Gogineni, V. R.; Gupta, R.; Dinh, D.H.; Rao, J.S. Suppression of uPA and uPAR blocks radiation-induced MCP-1 mediated recruitment of endothelial cells in meningioma. Cell Signal. 2011, 23, 1299–1310. [Google Scholar] [CrossRef] [PubMed]
- Riquier, H.; Wera, A.C.; Heuskin, A.C.; Feron, O.; Lucas, S.; Michiels, C. Comparison of X-ray and α particle effects on a human cancer and endothelial cells: Survival curves and gene expression profiles. Radiother. Oncol. 2013, 106, 397–403. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Wang, P.; Han, L.; Wang, X.A.; Lv, Y.M. Changes of MN23-H1 gene expression in human peripheral blood cell after X-ray irradiation. Chin. J. Radiol. Med. Prot. 2010, 30, 161–164. [Google Scholar] [CrossRef]
- MITOMAP. Available online: http://www.mitomap.org (accessed on 23 October 2014).
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, P.; Guo, F.; Han, L.; Wang, X.; Li, J.; Guo, Y.; LÜ, Y. X-ray-Induced Changes in the Expression of Inflammation-Related Genes in Human Peripheral Blood. Int. J. Mol. Sci. 2014, 15, 19516-19534. https://doi.org/10.3390/ijms151119516
Wang P, Guo F, Han L, Wang X, Li J, Guo Y, LÜ Y. X-ray-Induced Changes in the Expression of Inflammation-Related Genes in Human Peripheral Blood. International Journal of Molecular Sciences. 2014; 15(11):19516-19534. https://doi.org/10.3390/ijms151119516
Chicago/Turabian StyleWang, Ping, Fei Guo, Lin Han, Xi'ai Wang, Jie Li, Yan Guo, and Yumin LÜ. 2014. "X-ray-Induced Changes in the Expression of Inflammation-Related Genes in Human Peripheral Blood" International Journal of Molecular Sciences 15, no. 11: 19516-19534. https://doi.org/10.3390/ijms151119516
APA StyleWang, P., Guo, F., Han, L., Wang, X., Li, J., Guo, Y., & LÜ, Y. (2014). X-ray-Induced Changes in the Expression of Inflammation-Related Genes in Human Peripheral Blood. International Journal of Molecular Sciences, 15(11), 19516-19534. https://doi.org/10.3390/ijms151119516




