Characteristic of the Pepper CaRGA2 Gene in Defense Responses against Phytophthora capsici Leonian
Abstract
:1. Introduction
2. Results
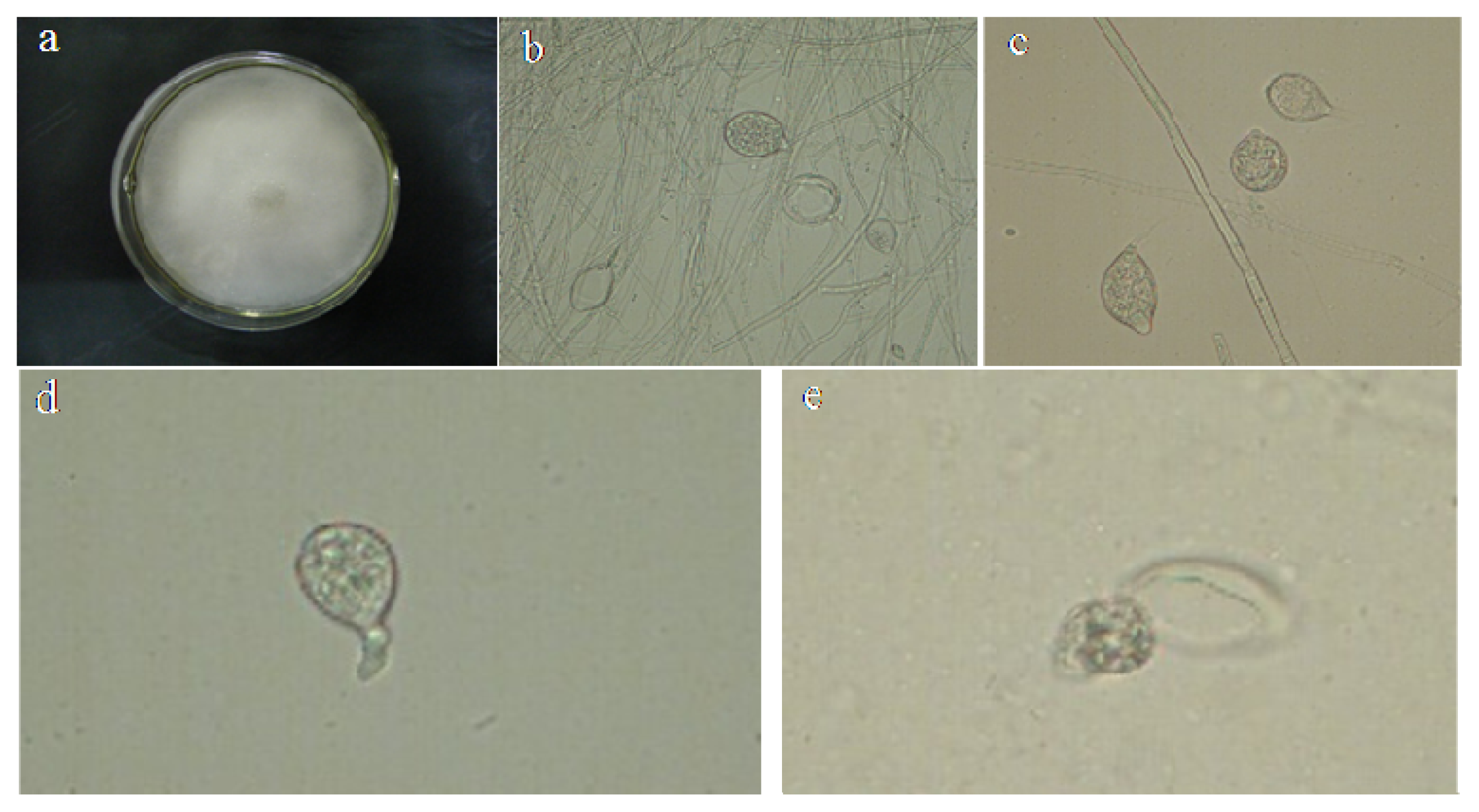
2.1. Identification of P. capsici
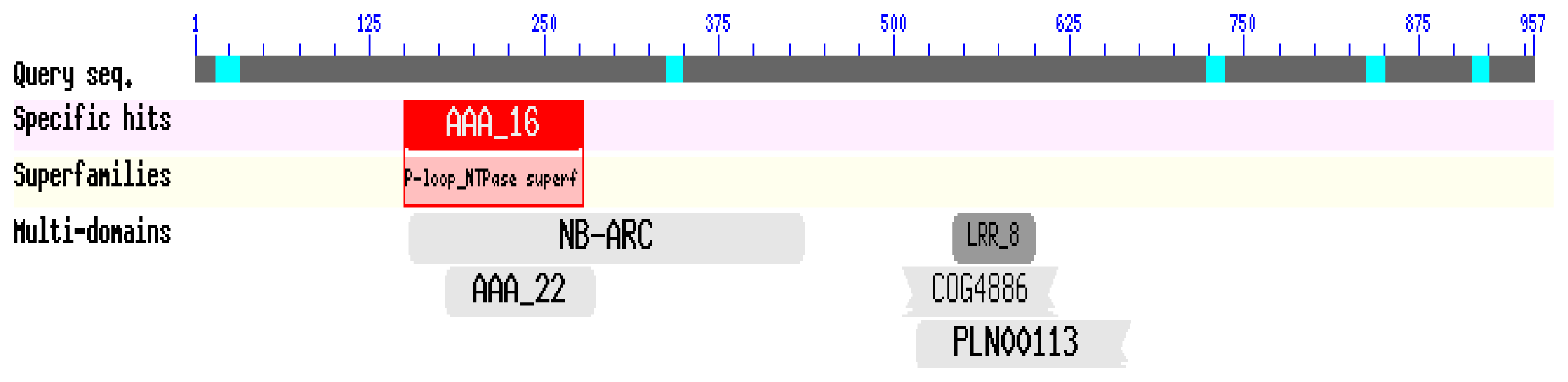
2.2. Isolation and Sequence Analysis of the CaRGA2 Gene
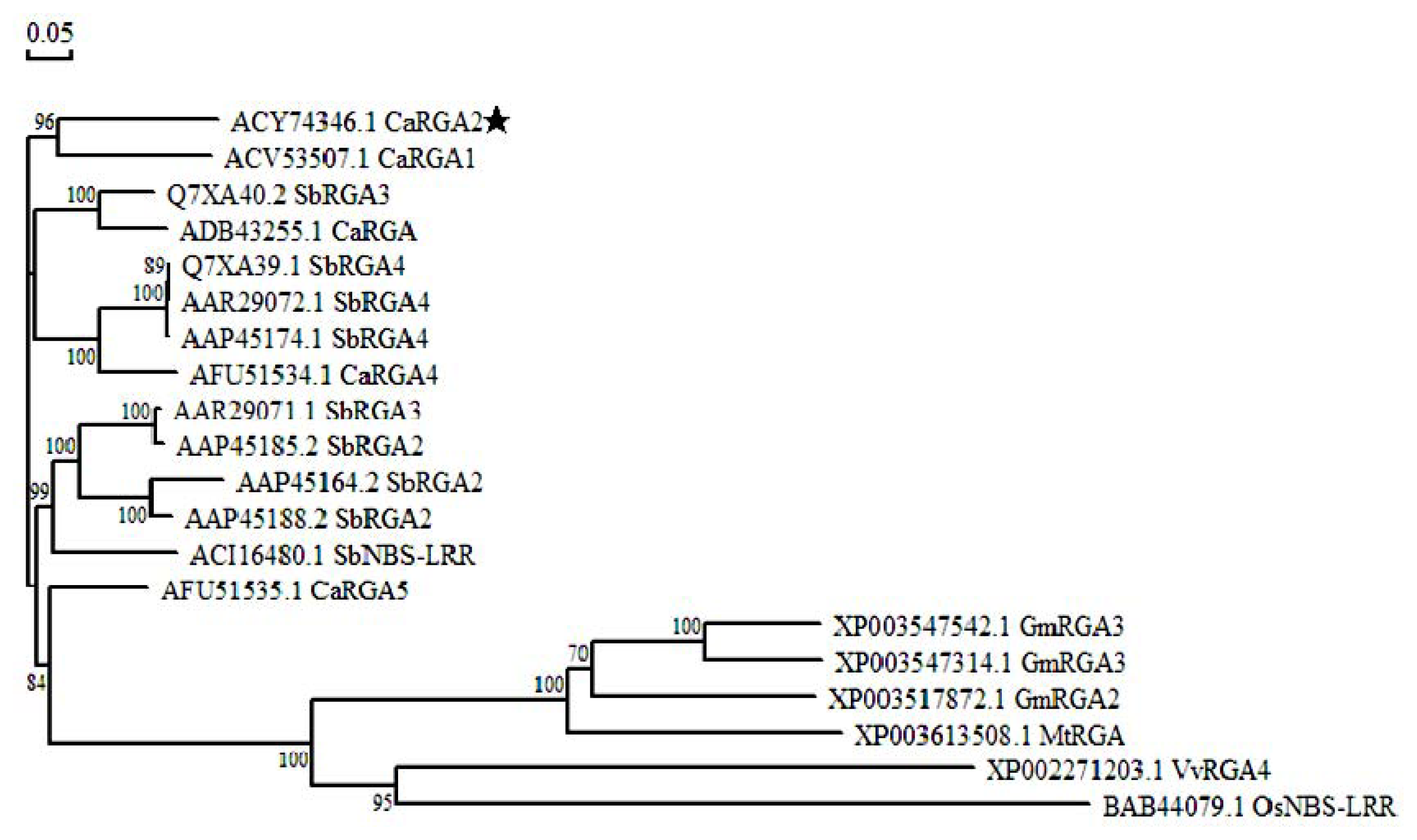
2.3. Protein Sequence Alignment and Phylogenetics Tree Analysis
2.4. Expression Analysis of CaRGA2 in Different Resistant Pepper Cultivars to P. capsici
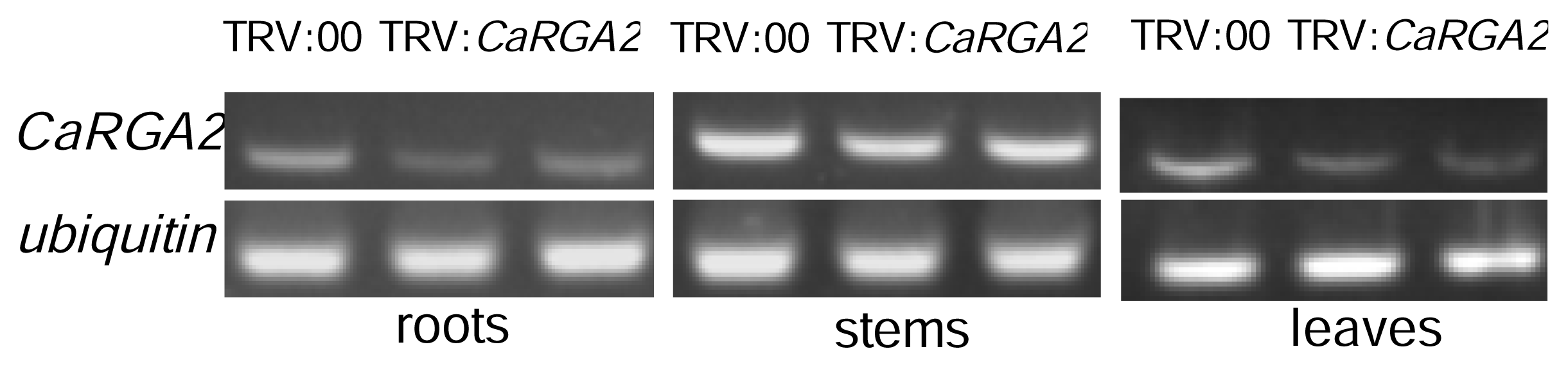
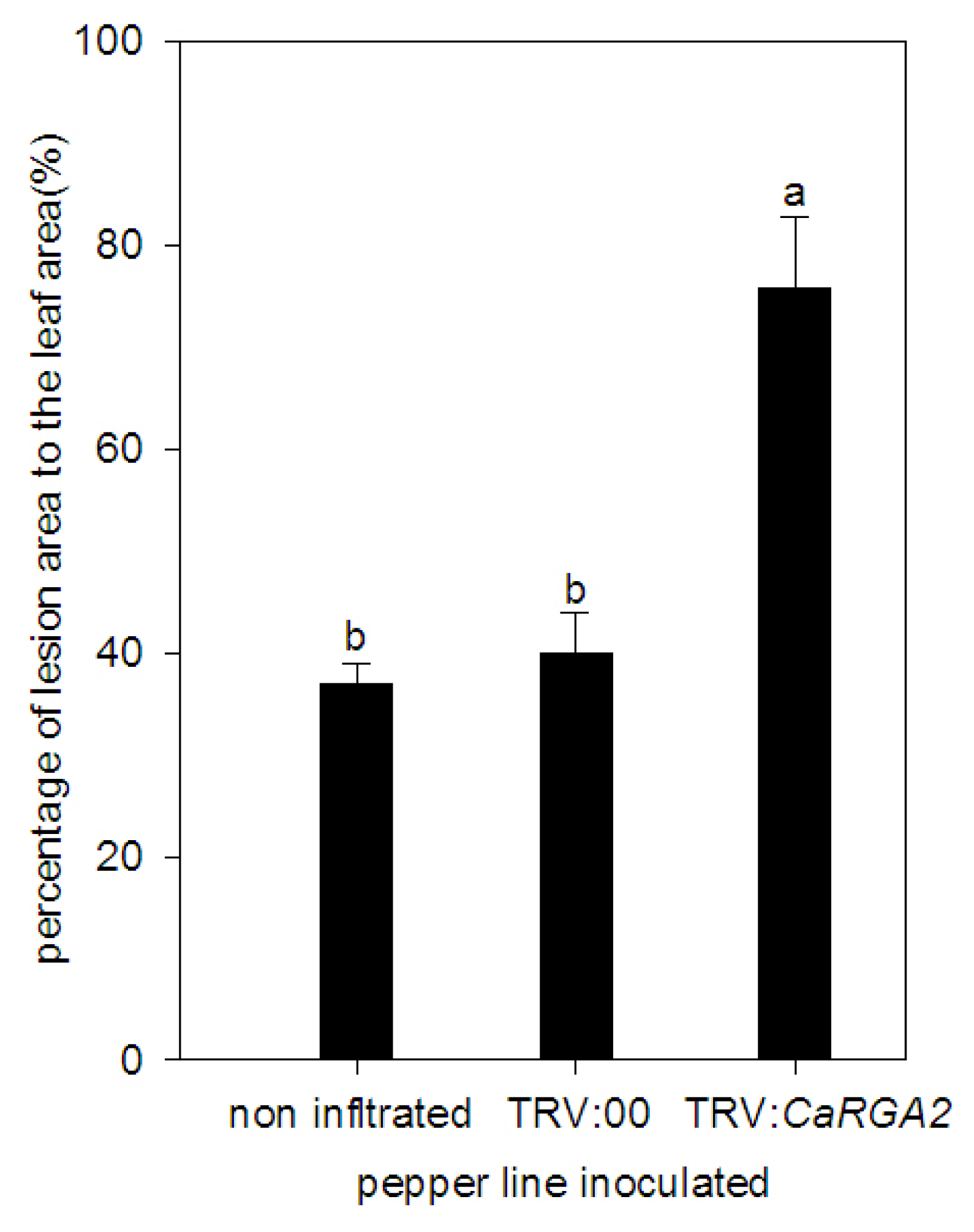
2.5. Enhanced Susceptibility to P. capsici Infection in CaRGA2-Silenced Plants
2.6. Semi-Quantitative RT-PCR Analysis
2.7. Detached Leaves Inoculation
3. Discussion
3.1. Identification of Pepper Phytophthora Blight Pathogen
3.2. Isolation and Bioinformatics Analysis of CaRGA2
3.3. Expression of CaRGA2 Gene in the Different Inoculation Methods
3.4. Virus-Induced Gene Silencing of CaRGA2 in Pepper
3.5. Semi-Quantitative RT-PCR Analysis
3.6. Functional Role of CaRGA2 in Disease Resistance
4. Experimental
4.1. Plant Materials, Pathogen and Inoculation
4.2. RNA Extraction and cDNA Synthesis
4.3. Cloning of CaRGA2
4.4. Sequence Analysis and Phylogenetic Analysis
4.5. Primer Design and Quantitative RT-PCR for Gene Expression Analysis
4.6. VIGS Plasmid Construction
4.7. Virus-Induced Gene Silencing (VIGS) of CaRGA2 in Pepper
4.8. Semi-Quantitative RT-PCR Analysis
4.9. Assay for Disease Resistance
4.10. Statistical Analysis
5. Conclusions
Acknowledgments
Conflict of Interest
References
- Hwang, B.K.; Kim, C.H. Phytophthora blight of bell pepper and its control in Korea. Plant Dis 1995, 79, 221–228. [Google Scholar]
- Lee, B.K.; Kim, B.S.; Chang, S.W.; Hwang, B.K. Aggressiveness to pumpkin cultivars of isolates of Phytophthora capsici from pumpkin and bell pepper. Plant Dis 2001, 85, 497–500. [Google Scholar]
- Ristaino, J.B.; Johnston, S.A. Ecologically based approaches to management of Phytophthora blight on bell pepper. Plant Dis 1999, 83, 1080–1089. [Google Scholar]
- Hausbeck, M.K.; Lamour, K.H. Phytophthora capsici on vegetable crops: Research progress and management challenges. Plant Dis 2004, 88, 1292–1303. [Google Scholar]
- Biles, C.L.; Brunton, B.D.; Wall, M.M.; Rivas, M. Phytophthora capsici zoospore infection of pepper fruit in various physical environments. Proc. Okla. Acad. Sci 1995, 75, 1–5. [Google Scholar]
- Oelke, L.M.; Bosland, P.W. Differentiation of race specific resistance to Phytophthora root rot and foliar blight in Capsicum annuum. J. Am. Soc. Hortisc. Sci 2003, 128, 213–218. [Google Scholar]
- Baysal, O.; Turgut, C.; Mao, G. Acibenzolar-S-methyl induced resistance to Phytophthora capsici in pepper leaves. Biol. Plant 2005, 49, 599–604. [Google Scholar]
- Candole, B.L.; Conner, P.J.; McGregor, C.; Waters, V. The disease reactions of heirloom bell pepper “California Wonder” to Phytophthora capsici. Agric. Sci 2012, 3, 417–424. [Google Scholar]
- Kousik, C.S.; Donahoo, R.S.; Hassell, R. Resistance in watermelon rootstocks to crown rot caused by Phytophthora capsici. Crop Prot 2012, 39, 18–25. [Google Scholar]
- Martin, G.B.; Bogdanove, A.J.; Sessa, G. Understanding the functions of plant disease resistance proteins. Annu. Rev. Plant Biol 2003, 54, 23–61. [Google Scholar]
- Dangl, J.L.; Jones, J.D. Plant pathogens and integrated defence responses to infection. Nature 2001, 411, 826–833. [Google Scholar]
- Bai, J.; Pennill, L.A.; Ning, J.; Lee, S.W.; Ramalingam, J.; Webb, C.A.; Zhao, B.; Sun, Q.; Nelson, J.C.; Leach, J.E.; et al. Diversity in nucleotide binding site-leucine-rich repeat genes in cereals. Genome Res 2002, 12, 1871–1884. [Google Scholar]
- Cannon, S.B.; Zhu, H.; Baumgarten, A.M.; Spangler, R.; May, G.; Cook, D.R.; Young, N.D. Diversity, distribution, and ancient taxonomic relationships within the TIR and non-TIR NBS-LRR resistance gene subfamilies. J. Mol. Evol 2002, 54, 548–562. [Google Scholar]
- McDowell, J.M.; Simon, S.A. Recent insights into R gene evolution. Mol. Plant Pathol 2006, 7, 437–448. [Google Scholar]
- Leng, X.D.; Xiao, B.G.; Wang, S.; Gui, X.J.; Wang, Y.; Lu, X.P.; Xie, J.H.; Li, Y.P.; Fan, L.G. Identification of NBS-type resistance gene homologs in tobacco genome. Plant Mol. Biol. Rep 2010, 28, 152–161. [Google Scholar]
- He, C.Y.; Tian, A.G.; Zhang, J.S.; Zhang, Z.Y.; Gai, J.Y.; Chen, S.Y. Isolation and characterization of a full-length resistance gene homolog from soybean. Theor. Appl. Genet 2003, 106, 786–793. [Google Scholar]
- Radwan, O.; Bouzidi, M.F.; Vear, F.; Philippon, J.; Tourvieille Labrouche, D.; Nicolas, P.; Mouzeyar, S. Identification of non TIR-NBS-LRR markers linked to the PI5/PI8 locus for resistance to downy mildew in sunflower. Theor. Appl. Genet 2003, 106, 1438–1446. [Google Scholar]
- Yan, G.; Chen, X.; Line, R.; Wellings, C. Resistance geneanalog polymorphism markers co-segregating with the YR5 gene for resistance to wheat stripe rust. Theor. Appl. Genet 2003, 106, 636–643. [Google Scholar]
- Gutierrez, N.; Giménez, M.J.; Torres, A.M.; Atienza, S.G.; Avila, C.M.; Palomino, C. Up-regulation of resistance gene analogs (RGA) in chickpea in the early response to Fusarium wilt. Euphytica 2012, 186, 793–804. [Google Scholar]
- Holzberg, S.; Brosio, P.; Gross, C.; Pogue, G. Barley stripe mosaic virus induced gene silencing in a monocot plant. Plant J 2002, 30, 315–327. [Google Scholar]
- Cai, X.Z.; Xu, Q.F.; Wang, C.C.; Zheng, Z. Development of a virus-induced gene-silencing system for functional analysis of the RPS2-dependent resistance signalling pathways in Arabidopsis. Plant Mol. Biol 2006, 62, 223–232. [Google Scholar]
- Baulcombe, D.C. Fast forward genetics based on virus-induced gene silencing. Curr. Opin. Plant Biol 1999, 2, 109–113. [Google Scholar]
- Dinesh-Kumar, S.P.; Anandalakshmi, R.; Marathe, R.; Schiff, M.; Liu, Y. Virus-induced gene silencing. Methods Mol. Biol 2003, 236, 287–294. [Google Scholar]
- Kumar, M.S.; Govind, G.; Kang, L.; Mysore, K.S.; Udayakumar, M. Functional characterization of Nicotiana benthamiana homologs of peanut water deficit-induced genes by virus-induced gene silencing. Planta 2007, 225, 523–539. [Google Scholar]
- Liu, Y.; Schiff, M.; Marathe, R.; Dinesh-Kumar, S.P. Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J 2002, 30, 415–429. [Google Scholar]
- Ratcliff, F.; Martin-Hernandez, A.M.; Baulcombe, D.C. Technical advance: Tobacco rattle virus as a vector for analysis of gene function by silencing. Plant J 2001, 25, 237–245. [Google Scholar]
- Chung, E.; Seong, E.; Kim, Y.C.; Chung, E.J.; Oh, S.K.; Lee, S.; Park, J.M.; Joung, Y.H.; Choi, D. A method of high frequency virus-induced gene silencing in chili pepper (Capsicum annuum L. cv. Bukang). Mol. Cells 2004, 17, 377–380. [Google Scholar]
- Brigneti, G.; Martin-Hernandez, A.M.; Jin, H.; Chen, J.; Baulcombe, D.C.; Baker, B.; Jones, J.D. Virus-induced gene silencing in Solanum species. Plant J 2004, 39, 264–272. [Google Scholar]
- Hileman, L.C.; Drea, S.; Martino, G.; Litt, A.; Irish, V.F. Virus-induced gene silencing is an effective tool for assaying gene function in the basal eudicot species Papaver somniferum (opium poppy). Plant J 2005, 44, 334–341. [Google Scholar]
- Gould, B.; Kramer, E.M. Virus-induced gene silencing as a tool for functional analyses in the emerging model plant Aquilegia(columbine, Ranunculaceae). Plant Methods 2007. [Google Scholar] [CrossRef]
- Wege, S.; Scholz, A.; Gleissberg, S.; Becker, A. Highly efficient virus induced gene silencing (VIGS) in California poppy (Eschscholzia californica): An evaluation of VIGS as a strategy to obtain functional data from non-model plants. Ann. Bot. (Lond.) 2007, 100, 641–649. [Google Scholar]
- Burch-Smith, T.M.; Schiff, M.; Liu, Y.; Dinesh-Kumar, S.P. Efficient virus induced gene silencing in Arabidopsis. Plant Physiol 2006, 142, 21–27. [Google Scholar]
- Liu, E.W.; Page, J.E. Optimized cDNA libraries for virus-induced gene silencing (VIGS) using tobacco rattle virus. Plant Methods 2008. [Google Scholar] [CrossRef]
- Liu, Y.; Schiff, M.; Dinesh-Kumar, S.P. Virus-induced gene silencing in tomato. Plant J 2002, 31, 777–786. [Google Scholar]
- Kumagai, M.H.; Donson, J.; Della-Cioppa, G.; Harvey, D.; Hanley, K.; Grill, L.K. Cytoplasmic inhibition of carotenoid biosynthesis with virus-derived RNA. Proc. Natl. Acad. Sci. USA 1995, 92, 1679–1683. [Google Scholar]
- Bioinformatics Tool. Available online: http://www.ncbi.nlm.nih.gov/BLAST/ (accessed on 23 December 2012).
- Zheng, X.B. Phytophthora and Its Research Techniques; Agricultural Press: Beijing, China, 1997; pp. 10–132. [Google Scholar]
- Silvar, C.; Díaz, J.; Merino, F. Real-time polymerase chain reaction quantification of Phytophthora capsici in different pepper genotypes. Phytopathology 2005, 95, 1423–1429. [Google Scholar]
- Pena-Cortés, H.; Albrecht, T.; Prat, S.; Weiler, E.W.; Willmitzer, L. Aspirin prevents wound-induced gene-expression in tomato leaves blocking jasmonic acid biosynthesis. Planta 1993, 191, 123–128. [Google Scholar]
- Yoshimura, S.; Yamanouchi, U.; Katayose, Y.; Toki, S. Expression of Xa1, a bacterial blight-resistance gene in rice, is induced by bacterial inoculation. Proc. Natl. Acad. Sci. USA 1998, 95, 1663–1668. [Google Scholar]
- Wan, H.J.; Zhao, Z.Z.; Malik, A.A.; Qian, C.; Chen, J.F. Identification and characterization of potential NBS-encoding resistance genes and induction kinetics of a putative candidate gene associated with downy mildew resistance in Cucumis. BMC Plant Biol 2010, 10, e186. [Google Scholar]
- Wang, Z.X.; Yamanouchi, U.; Katayose, Y.; Sasaki, T.; Yano, M. Expression of the Pib rice-blast-resistance gene family is up-regulated by environmental conditions favoring infection and by chemical signals that trigger secondary plant defences. Plant Mol. Biol 2001, 47, 653–661. [Google Scholar]
- Wang, Z.X.; Yano, M.; Yamanouchi, U.; Iwamoto, M.; Monna, L.; Hayasaka, H.; Katayose, Y.; Sasaki, T. The Pib gene for rice blast resistance belongs to the nucleotide binding and leucine-rich repeat class of plant disease resistance genes. Plant J 1999, 19, 55–64. [Google Scholar]
- Thurau, T.; Sirak, K.; Jung, C.; Cai, D. The promoter of the nematode resistance gene Hs1pro−1 activates a nematode-responsive and feeding site-specific gene expression in sugar beet (Beta vulgaris L.) and Arabidopsis thaliana. Plant Mol. Biol 2003, 52, 643–660. [Google Scholar]
- Silvar, C.; Merino, F.; Díaz, J. Differential activation of defence-related genes in different pepper cultivars infected with Phytophthora capsici. J. Plant Physiol 2008, 165, 1120–1124. [Google Scholar]
- Zhang, Y.; Wang, X.F.; Yang, S.; Chi, J.; Zhang, G.Y.; Ma, Z.Y. Cloning and characterization of a Verticillium wilt resistance gene from Gossypium barbadense and functional analysis in Arabidopsis thaliana. Plant Cell Rep 2011, 30, 2085–2096. [Google Scholar]
- Lu, R.; Malcuit, I.; Moffett, P.; Ruiz, M.T.; Peart, J.; Wu, A.J.; Rathjen, J.P.; Bendahmane, A.; Day, L.; Baulcombe, D.C. High throughput virus-induced gene silencing implicates heat shock protein 90 in plant disease resistance. EMBO J 2003, 22, 5690–5699. [Google Scholar]
- Burch-Smith, T.M.; Anderson, J.C.; Martin, G.B.; Dinesh-Kumar, S.P. Applications and advantages of virus induced gene silencing for gene function studies in plants. Plant J 2004, 39, 734–746. [Google Scholar]
- Fu, D.Q.; Zhu, B.Z.; Zhu, H.L.; Jiang, W.B.; Luo, Y.B. Virus induced gene silencing in tomato fruit. Plant J 2005, 43, 299–308. [Google Scholar]
- Lu, R.; Martın-Hernandez, A.M.; Peart, J.R.; Malcuit, I.; Baulcombe, D.C. Virus-induced gene silencing in plants. Methods 2003, 30, 296–303. [Google Scholar]
- Wang, J.E.; Liu, K.K.; Li, D.W.; Zhang, Y.L.; Zhao, Q.; He, Y.M.; Gong, Z.H. A novel peroxidase canPOD gene of pepper is involved in defense responses to Phytophtora capsici infection as well as abiotic stress tolerance. Int. J. Mol. Sci 2013, 14, 3158–3177. [Google Scholar]
- Hong, J.K.; Choi, D.S.; Kim, S.H.; Yi, S.H.; Kim, Y.J.; Hwang, B.K. Distinct roles of the pepper pathogen-induced membrane protein gene CaPIMP1 in bacterial disease resistance and oomycete disease susceptibility. Planta 2008, 228, 485–497. [Google Scholar]
- Sarowar, S.; Kim, Y.J.; Kim, K.D.; Hwang, B.K.; Ok, S.H. Overexpression of lipid transfer protein (LTP) genes enhances resistance to plant pathogens and LTP functions in long-distance systemic signaling in tobacco. Plant Cell Rep 2009, 28, 419–427. [Google Scholar]
- Shan, X.C.; Goodwin, P.H. Silencing an ACC oxidase gene affects the susceptible host response of Nicotiana benthamiana to infection by Colletotrichum orbiculare. Plant Cell Rep 2006, 25, 241–247. [Google Scholar]
- Shi, X.L.; Tian, Z.D.; Liu, J.; Edwin, A.G.; Xie, C.H. A potato pathogenesis-related protein gene, StPRp27, contributes to race-nonspecific resistance against Phytophthora infestans. Mol. Biol. Rep 2012, 39, 1909–1916. [Google Scholar]
- Mihovilovich, E.; Munive, S.; Bonierbale, M. Influence of daylength and isolates of Phytophthora infestans on field resistance to late blight of potato. Theor. Appl. Genet 2010, 120, 1265–1278. [Google Scholar]
- Brodersen, P.; Petersen, M.; Bjørn-Nielsen, H.; Zhu, S.; Newman, M.A.; Shokat, K.M.; Rietz, S.; Parker, J.; Mundy, J. Arabidopsis MAPkinase 4 regulates salicylic acid- and jasmonic acid/ethylenedependent responses via EDS1 and PAD4. Plant J 2006, 47, 532–546. [Google Scholar]
- Yeom, S.I.; Baek, H.K.; Oh, S.K.; Kang, W.H. Use of a secretion trap screen in pepper following Phytophthora capsici infection reveals novel functions of secreted plant proteins in modulating cell death. Mol. Plant Microbe Interact 2011, 24, 671–684. [Google Scholar]
- Hong, J.K.; Hwang, B.K. Induction by pathogen, salt and drought of a basic class II chitinase mRNA and its in situ localization in pepper (Capsicum annuum). Physiol. Plant 2002, 114, 549–558. [Google Scholar]
- Wan, H.J.; Yuan, W.; Ruan, M.Y.; Ye, Q.J.; Wang, R.Q.; Li, Z.M.; Zhou, G.Z.; Yao, Z.P.; Zhao, J.; Liu, S.J.; et al. Identification of reference genes for reverse transcription quantitative real-time PCR normalization in pepper (Capsicum annuum L.). Biochem. Biophys. Res. Commun 2011, 416, 24–30. [Google Scholar]
- Wang, C.; Cai, X.; Wang, X.; Zheng, Z. Optimisation of tobacco rattle virus-induced gene silencing in Arabidopsis. Funct. Plant Biol 2006, 33, 347–355. [Google Scholar]
- An, S.H.; Choi, H.W.; Hwang, I.S.; Hong, J.K.; Hwang, B.K. A novel pepper membrane-ocated receptor-like protein gene CaMRP1 is required for disease susceptibility, methyl jasmonate insensitivity and salt tolerance. Plant Mol. Biol 2008, 67, 519–533. [Google Scholar]




© 2013 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Zhang, Y.-L.; Jia, Q.-L.; Li, D.-W.; Wang, J.-E.; Yin, Y.-X.; Gong, Z.-H. Characteristic of the Pepper CaRGA2 Gene in Defense Responses against Phytophthora capsici Leonian. Int. J. Mol. Sci. 2013, 14, 8985-9004. https://doi.org/10.3390/ijms14058985
Zhang Y-L, Jia Q-L, Li D-W, Wang J-E, Yin Y-X, Gong Z-H. Characteristic of the Pepper CaRGA2 Gene in Defense Responses against Phytophthora capsici Leonian. International Journal of Molecular Sciences. 2013; 14(5):8985-9004. https://doi.org/10.3390/ijms14058985
Chicago/Turabian StyleZhang, Ying-Li, Qing-Li Jia, Da-Wei Li, Jun-E Wang, Yan-Xu Yin, and Zhen-Hui Gong. 2013. "Characteristic of the Pepper CaRGA2 Gene in Defense Responses against Phytophthora capsici Leonian" International Journal of Molecular Sciences 14, no. 5: 8985-9004. https://doi.org/10.3390/ijms14058985
APA StyleZhang, Y.-L., Jia, Q.-L., Li, D.-W., Wang, J.-E., Yin, Y.-X., & Gong, Z.-H. (2013). Characteristic of the Pepper CaRGA2 Gene in Defense Responses against Phytophthora capsici Leonian. International Journal of Molecular Sciences, 14(5), 8985-9004. https://doi.org/10.3390/ijms14058985



