Expression of a Functional Recombinant Human Basic Fibroblast Growth Factor from Transgenic Rice Seeds
Abstract
:1. Introduction
2. Results
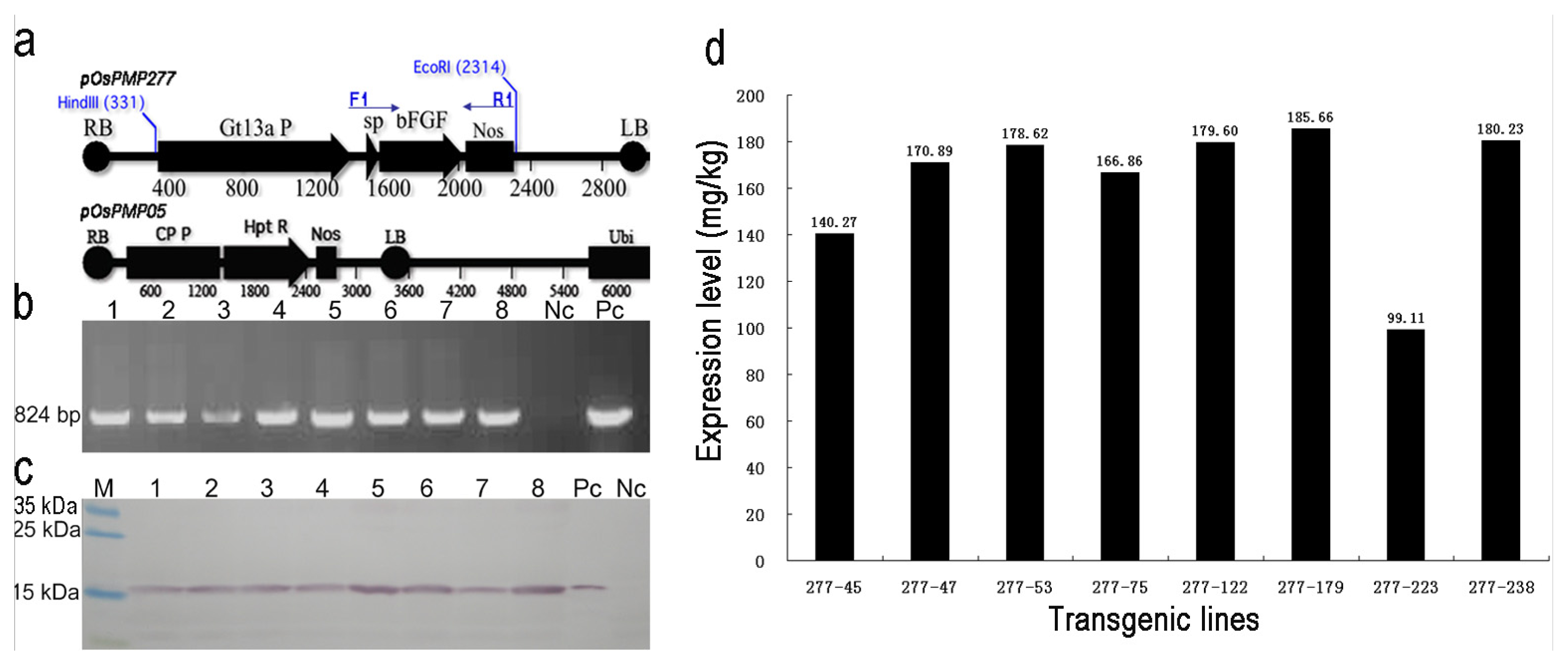
2.1. Generation and Selection of the Transgenic Lines for High Expression of OsrbFGF in Rice Seeds
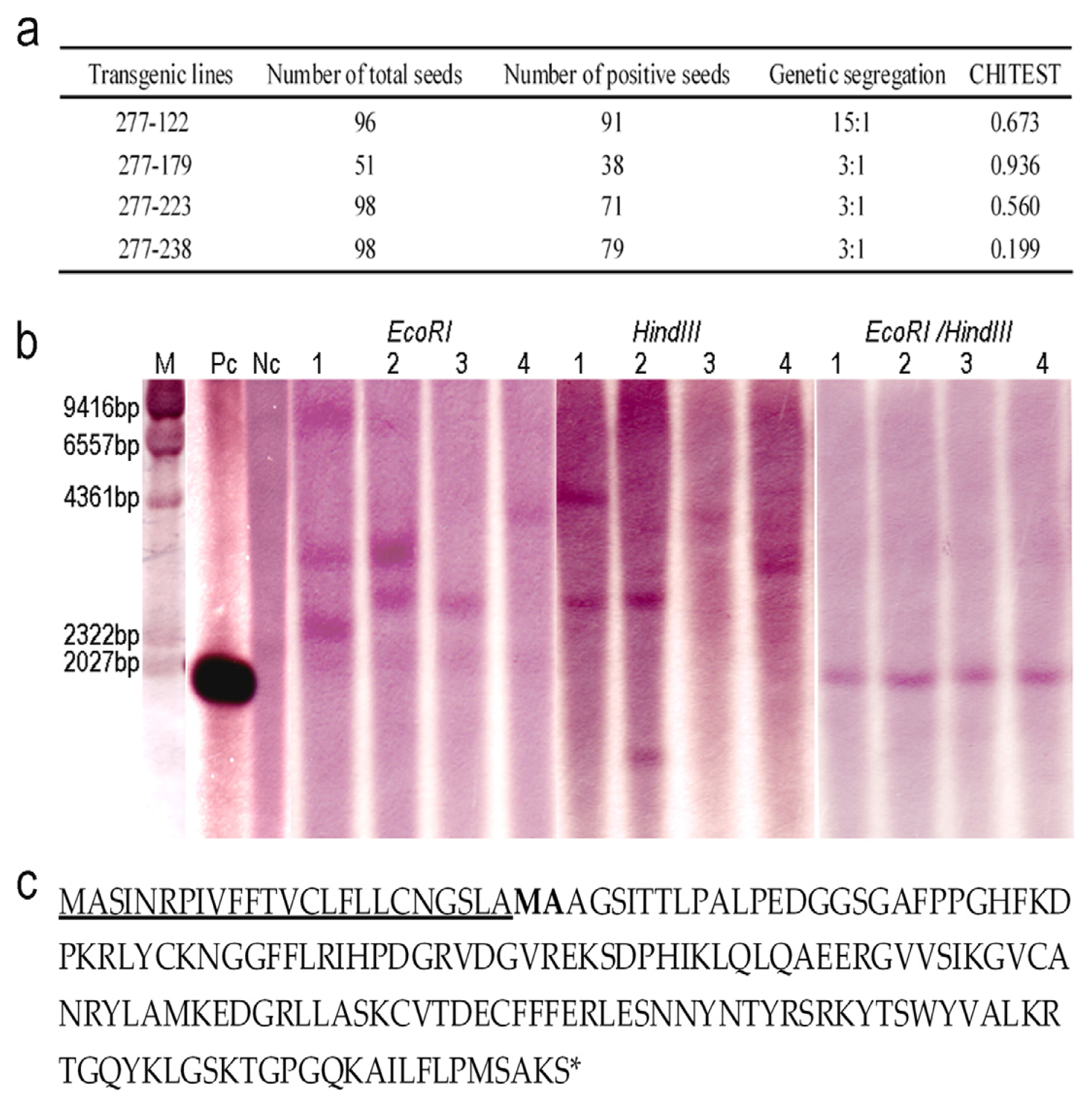
2.2. Genetic Analysis of the Transgenic Lines and Characterization of OsrbFGF
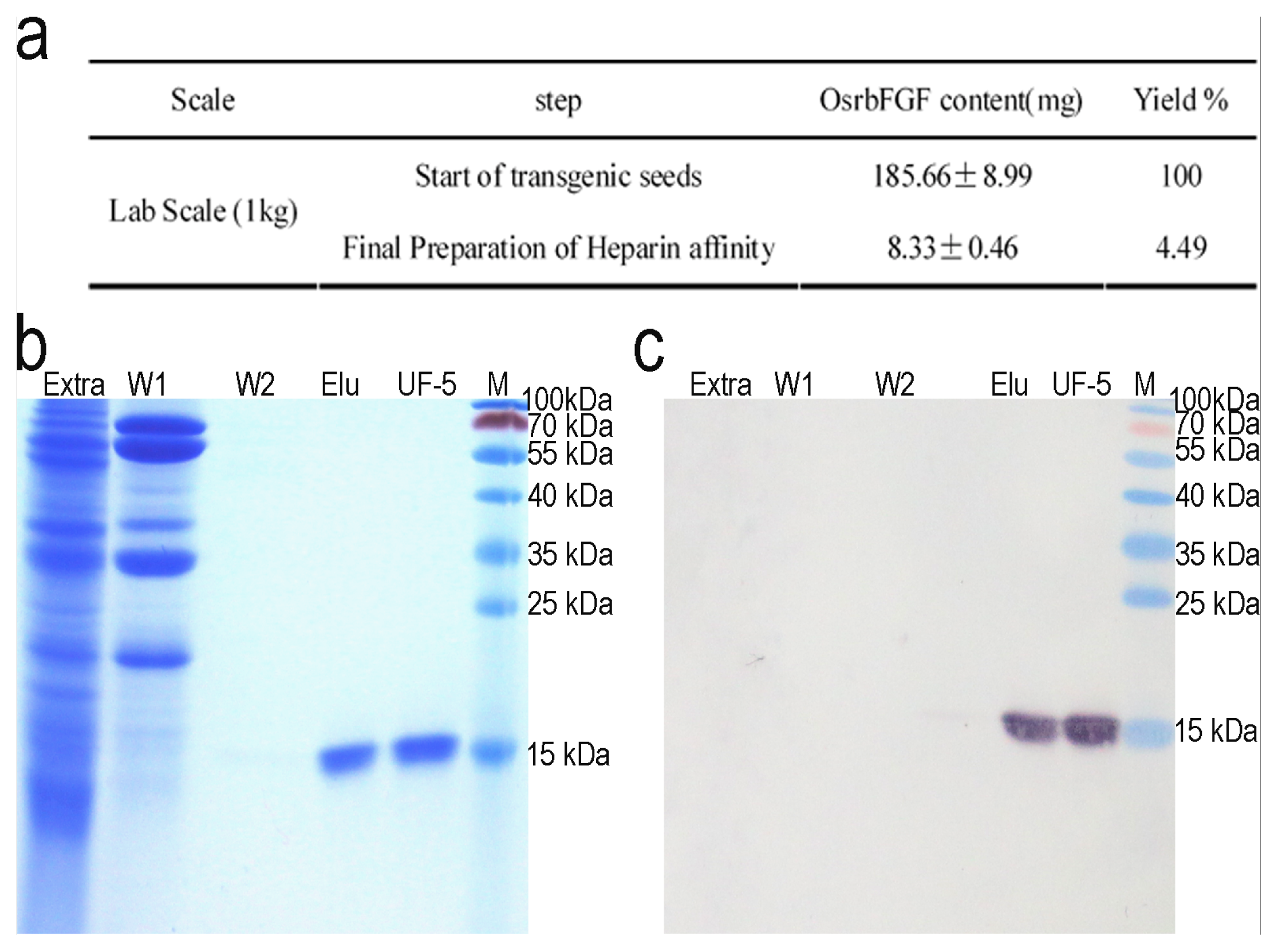
2.3. Purification of OsrbFGF from Transgenic Rice Seeds
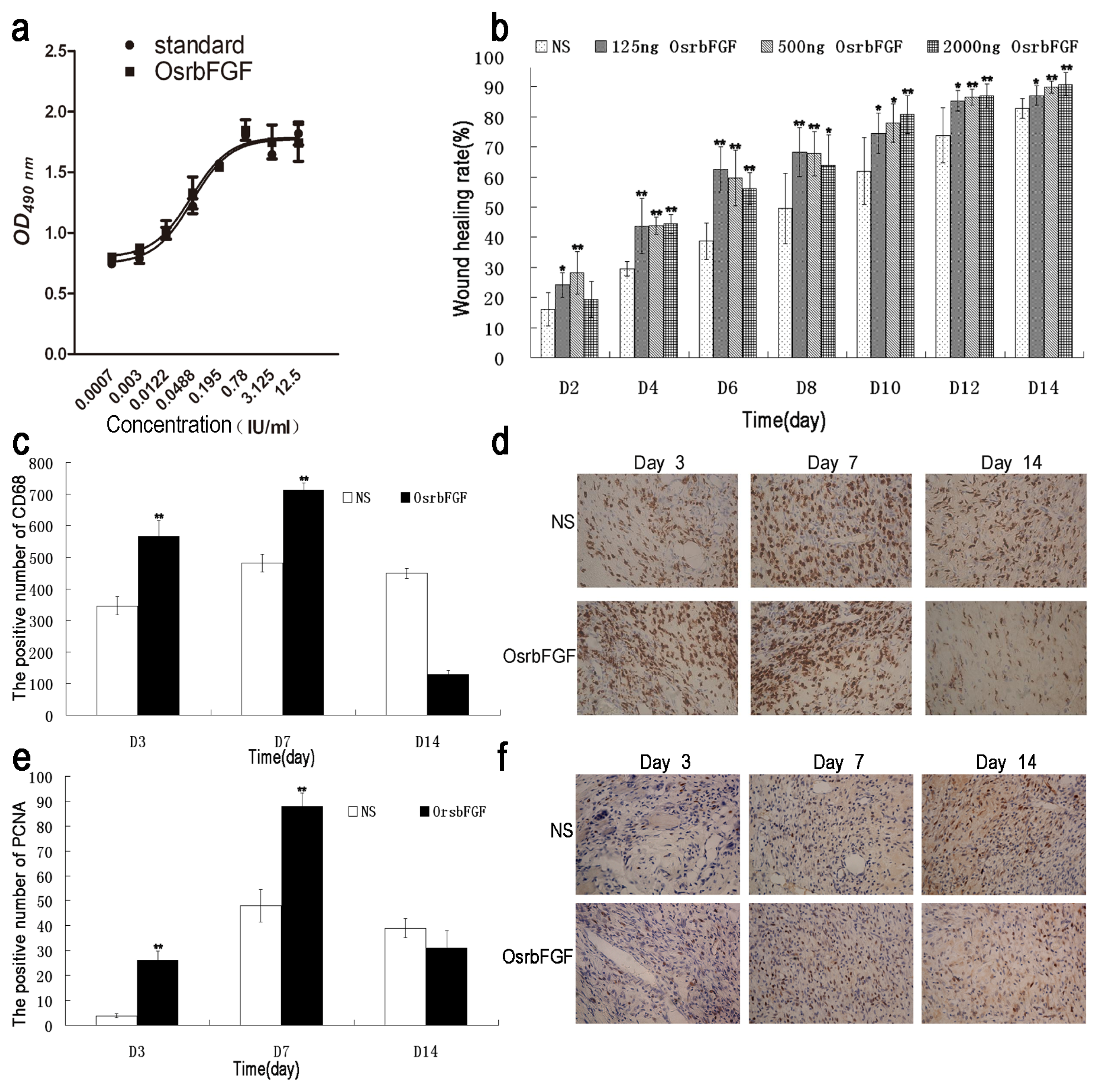
2.4. OsrbFGF Can Effectively Stimulate NIH/3T3 Cell Proliferation
2.5. OsrbFGF Has Equivalent Efficacy in Wound Healing
3. Discussion
4. Experimental Procedures
4.1. Plasmid Construction and Rice Transformation
4.2. Screening for High Expressing OsrbFGF Transgenic Lines and Determination of the Expression Level
4.3. Southern Blotting
4.4. OsrbFGF Purification
4.5. N-terminal Sequence Analysis
4.6. MTT Assay
4.7. Wound Healing Assay
4.8. Immunohistochemistry Assay
5. Conclusions
Acknowledgements
References
- Yamashita, T.; Yoshioka, M.; Itoh, N. Identification of a novel fibroblast growth factor, FGF-23, preferentially expressed in the ventrolateral thalamic nucleus of the brain. Biochem. Biophys. Res. Commun 2000, 277, 494–498. [Google Scholar]
- Gospodarowicz, D.; Bialecki, H.; Greenburg, G. Purification of the fibroblast growth factor activity from bovine brain. J. Biol. Chem 1978, 253, 3736–3743. [Google Scholar]
- Bohlen, P.; Baird, A.; Esch, F.; Ling, N.; Gospodarowicz, D. Isolation and partial molecular characterization of pituitary fibroblast growth factor. Proc. Natl. Acad. Sci. USA 1984, 81, 5364–5368. [Google Scholar]
- Zou, H.; Nie, X.; Zhang, Y.; Hu, M.; Zhang, Y.A. Effect of basic fibroblast growth factor on the proliferation, migration and phenotypic modulation of airway smooth muscle cells. Chin. Med. J. Peking 2008, 121, 424–429. [Google Scholar]
- Lavandero, S.; Chappuzeau, A.; Sapag-Hagar, M.; Oka, T. In vivo and in vitro evidence of basic fibroblast growth factor action in mouse mammary gland development. FEBS Lett 1998, 439, 351–356. [Google Scholar]
- Akita, S.; Akino, K.; Imaizumi, T.; Hirano, A. A basic fibroblast growth factor improved the quality of skin grafting in burn patients. Burns 2005, 31, 855–858. [Google Scholar]
- Kwong, K.W.Y.; Ng, K.L.; Lam, C.C.; Wang, Y.Y.; Wong, W.K.R. Authentic human basic fibroblast growth factor produced by secretion in bacillus subtilis. Appl. Microbiol. Biotechnol. 2012. [Google Scholar] [CrossRef]
- Ke, Y.; Wilkinson, M.C.; Fernig, D.G.; Smith, J.A.; Rudland, P.S.; Barraclough, R. A rapid procedure for production of human basic fibroblast growth factor in Escherichia coli cells. BBA Gene Struct. Expr 1992, 1131, 307–310. [Google Scholar]
- Wu, X.; Kamei, K.; Sato, H.; Sato, S.; Takano, R.; Ichida, M.; Mori, H.; Hara, S. High-level expression of human acidic fibroblast growth factor and basic fibroblast growth factor in silkworm (Bombyx mori L.) using recombinant baculovirus. Protein Expres. Purif 2001, 21, 192–200. [Google Scholar]
- Ding, S.H.; Huang, L.Y.; Wang, Y.D.; Sun, H.C.; Xiang, Z.H. High-level expression of basic fibroblast growth factor in transgenic soybean seeds and characterization of its biological activity. Biotechnol. Lett 2006, 28, 869–875. [Google Scholar]
- Mu, X.; Kong, N.; Chen, W.; Zhang, T.; Shen, M.; Yan, W. High-level expression, purification, and characterization of recombinant human basic fibroblast growth factor in pichia pastoris. Protein Expres. Purif 2008, 59, 282–288. [Google Scholar]
- Suzuki, Y.A.; Kelleher, S.L.; Yalda, D.; Wu, L.; Huang, J.; Huang, N.; Lönnerdal, B. Expression, characterization, and biologic activity of recombinant human lactoferrin in rice. J. Pediatr. Gastr. Nutr 2003, 36, 190–199. [Google Scholar]
- Yang, D.; Guo, F.; Liu, B.; Huang, N.; Watkins, S.C. Expression and localization of human lysozyme in the endosperm of transgenic rice. Planta 2003, 216, 597–603. [Google Scholar]
- Ning, T.; Xie, T.; Qiu, Q.; Yang, W.; Zhou, S.; Zhou, L.; Zheng, C.; Zhu, Y.; Yang, D. Oral administration of recombinant human granulocyte-macrophage colony stimulating factor expressed in rice endosperm can increase leukocytes in mice. Biotechnol. Lett 2008, 30, 1679–1686. [Google Scholar]
- He, Y.; Ning, T.; Xie, T.; Qiu, Q.; Zhang, L.; Sun, Y.; Jiang, D.; Fu, K.; Yin, F.; Zhang, W. Large-scale production of functional human serum albumin from transgenic rice seeds. Proc. Natl. Acad. Sci. USA 2011, 108, 19078–19083. [Google Scholar]
- Ramessar, K.; Capell, T.; Christou, P. Molecular pharming in cereal crops. Phytochem. Rev 2008, 7, 579–592. [Google Scholar]
- Xie, T.; Qiu, Q.; Zhang, W.; Ning, T.; Yang, W.; Zheng, C.; Wang, C.; Zhu, Y.; Yang, D. A biologically active rhIGF-1 fusion accumulated in transgenic rice seeds can reduce blood glucose in diabetic mice via oral delivery. Peptides 2008, 29, 1862–1870. [Google Scholar]
- Daley, M.; Knauf, V.C.; Summerfelt, K.R.; Turner, J.C. Co-transformation with one agrobacterium tumefaciens strain containing two binary plasmids as a method for producing marker-free transgenic plants. Plant Cell Rep 1998, 17, 489–496. [Google Scholar]
- Gospodarowicz, D. Localisation of a fibroblast growth factor and its effect alone and with hydrocortisone on 3T3 cell growth. Nature 1974, 249, 123–127. [Google Scholar]
- McGee, G.S.; Davidson, J.M.; Buckley, A.; Sommer, A.; Woodward, S.C.; Aquino, A.M.; Barbour, R.; Demetriou, A.A. Recombinant basic fibroblast growth factor accelerates wound healing. J. Surg. Res 1988, 45, 145–153. [Google Scholar]
- Eming, S.A.; Hammerschmidt, M.; Krieg, T.; Roers, A. In Interrelation of immunity and tissue repair or regeneration. Semin. Cell Dev. Biol 2009, 20, 517–527. [Google Scholar]
- Yamei, W.; Licui, S.; Yahui, Q.; Yang, S.; Yudong, Y.; Hong, Y.; Jingyi, Z. Expression of human basic fibroblast growth factor gene in sf9 insect cell. J. Cap. Univ. Med. Sci 2004, 25, 431–434. [Google Scholar]
- Barr, P.J.; Cousens, L.S.; Lee-Ng, C.T.; Medina-Selby, A.; Masiarz, F.R.; Hallewell, R.A.; Chamberlain, S.H.; Bradley, J.D.; Lee, D.; Steimer, K.S. Expression and processing of biologically active fibroblast growth factors in the yeast Saccharomyces cerevisiae. J. Biol. Chem 1988, 263, 16471–16478. [Google Scholar]
- Luo, J.; Ning, T.; Sun, Y.; Zhu, J.; Zhu, Y.; Lin, Q.; Yang, D. Proteomic analysis of rice endosperm cells in response to expression of hGM-CSF. J. Proteome Res 2008, 8, 829–837. [Google Scholar]
- Hennegan, K.; Yang, D.; Nguyen, D.; Wu, L.; Goding, J.; Huang, J.; Guo, F.; Huang, N.; Watkins, S.C. Improvement of human lysozyme expression in transgenic rice grain by combining wheat (Triticum aestivum) puroindoline b and rice (Oryza sativa) Gt1 promoters and signal peptides. Transgenic Res 2005, 14, 583–592. [Google Scholar]
- Yang, D.; Wu, L.; Hwang, Y.S.; Chen, L.; Huang, N. Expression of the reb transcriptional activator in rice grains improves the yield of recombinant proteins whose genes are controlled by a reb-responsive promoter. Proc. Natl. Acad. Sci. USA 2001, 98, 11438–11443. [Google Scholar]
- Galiano, R.D.; Michaels, V.; Dobryansky, M.; Levine, J.P.; Gurtner, G.C. Quantitative and reproducible murine model of excisional wound healing. Wound Repair Regen 2004, 12, 485–492. [Google Scholar]
- Wu, Y.; Chen, L.; Scott, P.G.; Tredget, E.E. Mesenchymal stem cells enhance wound healing through differentiation and angiogenesis. Stem Cells 2007, 25, 2648–2659. [Google Scholar]
© 2013 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
An, N.; Ou, J.; Jiang, D.; Zhang, L.; Liu, J.; Fu, K.; Dai, Y.; Yang, D. Expression of a Functional Recombinant Human Basic Fibroblast Growth Factor from Transgenic Rice Seeds. Int. J. Mol. Sci. 2013, 14, 3556-3567. https://doi.org/10.3390/ijms14023556
An N, Ou J, Jiang D, Zhang L, Liu J, Fu K, Dai Y, Yang D. Expression of a Functional Recombinant Human Basic Fibroblast Growth Factor from Transgenic Rice Seeds. International Journal of Molecular Sciences. 2013; 14(2):3556-3567. https://doi.org/10.3390/ijms14023556
Chicago/Turabian StyleAn, Na, Jiquan Ou, Daiming Jiang, Liping Zhang, Jingru Liu, Kai Fu, Ying Dai, and Daichang Yang. 2013. "Expression of a Functional Recombinant Human Basic Fibroblast Growth Factor from Transgenic Rice Seeds" International Journal of Molecular Sciences 14, no. 2: 3556-3567. https://doi.org/10.3390/ijms14023556
APA StyleAn, N., Ou, J., Jiang, D., Zhang, L., Liu, J., Fu, K., Dai, Y., & Yang, D. (2013). Expression of a Functional Recombinant Human Basic Fibroblast Growth Factor from Transgenic Rice Seeds. International Journal of Molecular Sciences, 14(2), 3556-3567. https://doi.org/10.3390/ijms14023556



