Evaluation of the Adenocarcinoma-Associated Gene AGR2 and the Intestinal Stem Cell Marker LGR5 as Biomarkers in Colorectal Cancer
Abstract
:1. Introduction
2. Results and Discussion
2.1. Results
2.1.1. Patients and Clinical Data
2.1.2. Expression of AGR2 and LGR5 mRNA Transcripts in Blood Samples
2.1.3. Clinic Pathological Characteristics and mRNA Markers in Blood
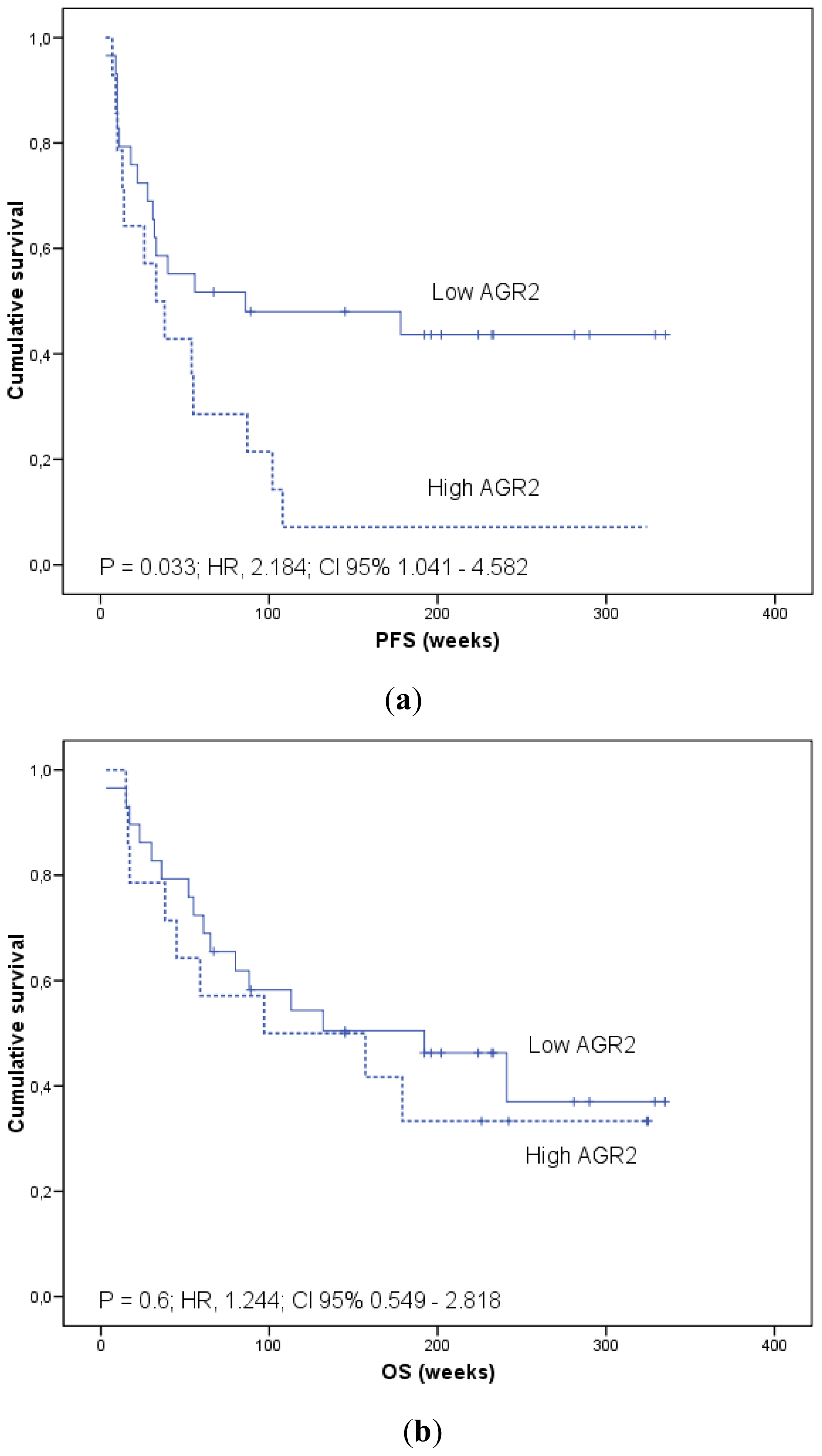
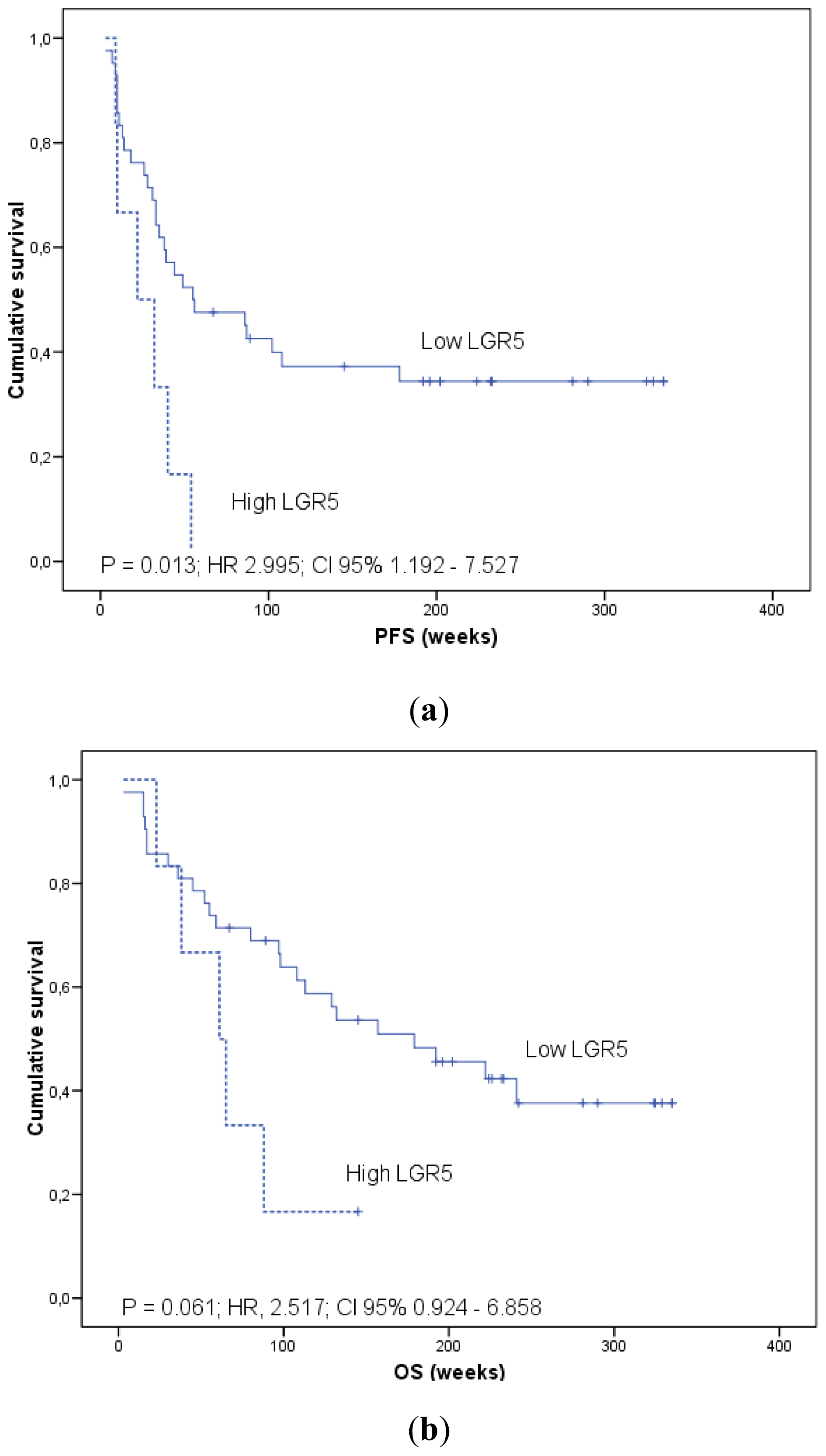
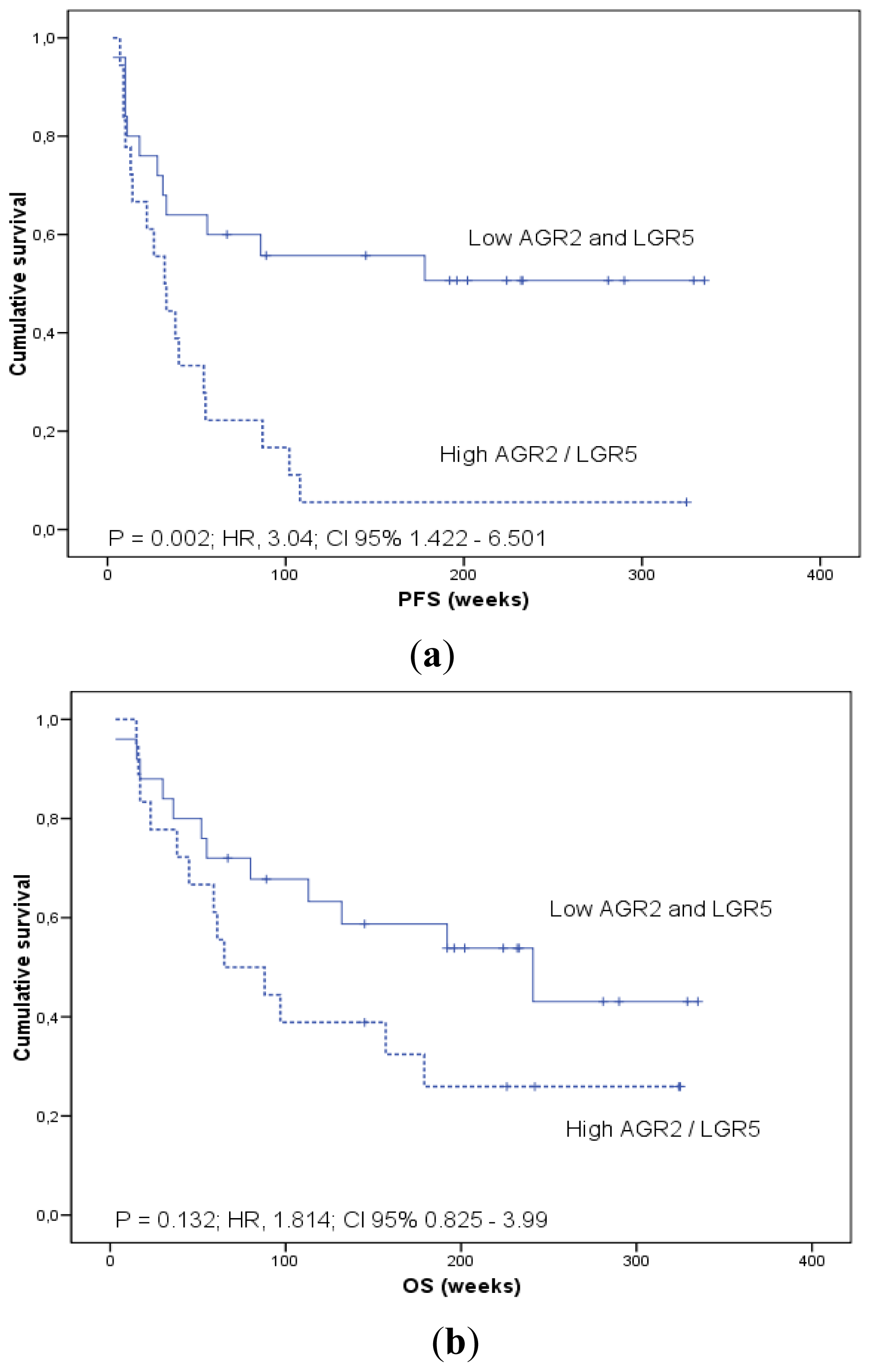
2.1.4. Prognostic Significance of AGR2 and LGR5 in Blood
2.2. Discussion
3. Experimental Section
3.1. Patients
3.2. Pathological Analysis
3.3. Processing of Blood Samples and mRNA Isolation
3.4. Reverse Transcription and Quantitative Real-Time PCR
3.5. Study Design and Statistical Analysis
4. Conclusions
Acknowledgments
References
- Malvezzi, M.; Arfé, A.; Bertuccio, P.; Levi, F.; La Vecchia, C.; Negri, E. European cancer mortality predictions for the year 2011. Ann. Oncol 2011, 22, 947–956. [Google Scholar]
- Bustin, S.A.; Mueller, R. Real-time reverse transcription PCR and the detection of occult disease in colorectal cancer. Mol. Asp. Med 2006, 27, 192–223. [Google Scholar]
- Sergeant, G.; Penninckx, F.; Topal, B. Quantitative RT-PCR detection of colorectal tumor cells in peripheral blood—A systematic review. J. Surg. Res 2008, 150, 144–152. [Google Scholar]
- Rahbari, N.N.; Aigner, M.; Thorlund, K.; Mollberg, N.; Motschall, E.; Jensen, K.; Diener, M.K.; Büchler, M.W.; Koch, M.; Weitz, J. Meta-analysis shows that detection of circulating tumor cells indicates poor prognosis in patients with colorectal cancer. Gastroenterology 2010, 138, 1714–1726. [Google Scholar]
- Tsouma, A.; Aggeli, C.; Lembessis, P.; Zografos, G.N.; Korkolis, D.P.; Pectasides, D.; Skondra, M.; Pissimissis, N.; Tzonou, A.; Koutsilieris, M. Multiplex RT-PCR-based detections of CEA, CK20 and EGFR in colorectal cancer patients. World J. Gastroenterol 2010, 16, 5965–5974. [Google Scholar]
- Thorsteinsson, M.; Jess, P. The clinical significance of circulating tumor cells in non-metastatic colorectal cancer. A review. Eur. J. Surg. Oncol 2011, 37, 459–465. [Google Scholar]
- Iinuma, H.; Watanabe, T.; Mimori, K.; Adachi, M.; Hayashi, N.; Tamura, J.; Matsuda, K.; Fukushima, R.; Okinaga, K.; Sasako, M.; et al. Clinical significance of circulating tumor cells, including cancer stem-like cells, in peripheral blood for recurrence and prognosis in patients with Dukes’ stage B and C colorectal cancer. J. Clin. Oncol 2011, 29, 1547–1555. [Google Scholar]
- Valladares-Ayerbes, M.; Díaz-Prado, S.; Reboredo, M.; Medina, V.; Iglesias-Díaz, P.; Lorenzo-Patiño, M.J.; Campelo, R.G.; Haz, M.; Santamarina, I.; Antón-Aparicio, L.M. Bioinformatics approach to mRNA markers discovery for detection of circulating tumor cells in patients with gastrointestinal cancer. Cancer Detect. Prev 2008, 32, 236–250. [Google Scholar]
- Valladares-Ayerbes, M.; Díaz-Prado, S.; Reboredo, M.; Medina, V.; Lorenzo-Patiño, M.J.; Iglesias-Díaz, P.; Haz, M.; Pértega, S.; Santamarina, I.; Blanco, M.; et al. Evaluation of plakophilin-3 mRNA as a biomarker for detection of circulating tumor cells in gastrointestinal cancer patients. Cancer Epidemiol. Biomark. Prev 2010, 19, 1432–1440. [Google Scholar]
- Smirnov, D.A.; Zweitzig, D.R.; Foulk, B.W.; Miller, M.C.; Doyle, G.V.; Kenneth, J.P.; Neal, J.M.; Louis, M.W.; Steven, J.C.; Jose, G.M.; et al. Global gene expression profiling of circulating tumor cells. Cancer Res 2005, 65, 4993–4997. [Google Scholar]
- Sieuwerts, A.M.; Mostert, B.; Bolt-de Vries, J.; Peeters, D.; de Jongh, F.E.; Stouthard, J.M.L.; Dirix, L.Y.; van Dam, P.A.; Galen, A.V.; de Vanja, W.; et al. mRNA and microRNA expression profiles in circulating tumor cells and primary tumors of metastatic breast cancer patients. Clin. Cancer Res 2011, 17, 3600–3618. [Google Scholar]
- Thompson, D.A.; Weigel, R.J. hAG-2, the human homologue of the Xenopus laevis cement gland gene XAG-2, is coexpressed with estrogen receptor in breast cancer cell lines. Biochem. Biophys. Res. Commun 1998, 251, 111–116. [Google Scholar]
- Pohler, E.; Craig, A.L.; Cotton, J.; Lawrie, L.; Dillon, J.F.; Ross, P.; Kernohan, N.; Hupp, T.R. The Barrett’s antigen anterior gradient-2 silences the p53 transcriptional response to DNA damage. Mol. Cell Proteomics 2004, 3, 534–547. [Google Scholar]
- Wang, Z.; Hao, Y.; Lowe, A.W. The adenocarcinoma-associated antigen, AGR2, promotes tumor growth, cell migration, and cellular transformation. Cancer Res 2008, 68, 492–497. [Google Scholar]
- Park, S.W.; Zhen, G.; Verhaeghe, C.; Nakagami, Y.; Nguyenvu, L.T.; Barczak, A.J.; Killeen, N.; Erle, D.J. The protein disulfide isomerase AGR2 is essential for production of intestinal mucus. Proc. Natl. Acad. Sci. USA 2009, 106, 6950–6955. [Google Scholar]
- Balic, M.; Lin, H.; Young, L.; Hawes, D.; Giuliano, A; McNamara, G.; Datar, R.H.; Cote, R.J. Most early disseminated cancer cells detected in bone marrow of breast cancer patients have a putative breast cancer stem cell phenotype. Clin. Cancer Res. 2006, 2, 5615–5621. [Google Scholar]
- Aktas, B.; Tewes, M.; Fehm, T.; Hauch, S.; Kimmig, R.; Kasimir-Bauer, S. Stem cell and epithelial-mesenchymal transition markers are frequently overexpressed in circulating tumor cells of metastatic breast cancer patients. Breast Cancer Res 2009, 11. [Google Scholar] [CrossRef]
- Visvader, J.E.; Lindeman, G.J. Cancer stem cells in solid tumors, accumulating evidence and unresolved questions. Nat. Rev. Cancer 2008, 8, 755–758. [Google Scholar]
- Barker, N.; van Es, J.H.; Kuipers, J.; Kujala, P.; van den Born, M.; Cozijnsen, M.; Haegebarth, A.; Korving, J.; Begthel, H.; Peters, P.J.; et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 2007, 449, 1003–1007. [Google Scholar]
- McClanahan, T.; Koseoglu, S.; Smith, K.; Grein, J.; Gustafson, E.; Black, S.; Kirschmeier, P.; Samatar, A. Identification of overexpression of orphan G protein-coupled receptor GPR49 in human colon and ovarian primary tumors. Cancer Biol. Ther 2006, 5, 419–426. [Google Scholar]
- Uchida, H.; Yamazaki, K.; Fukuma, M.; Yamada, T.; Hayashida, T.; Hasegawa, H.; Kitajima, M.; Kitagawa, Y.; Sakamoto, M. Overexpression of leucine-rich repeat-containing G protein-coupled receptor 5 in colorectal cancer. Cancer Sci 2010, 101, 1731–1737. [Google Scholar]
- Takahashi, H.; Ishii, H.; Nishida, N.; Takemasa, I.; Mizushima, T.; Ikeda, M.; Yokobori, T.; Mimori, K.; Yamamoto, H.; Sekimoto, M.; et al. Significance of Lgr5+ve Cancer Stem Cells in the Colon and Rectum. Ann. Surg. Oncol 2011, 18, 1166–1174. [Google Scholar]
- Köhne, C.H.; Cunningham, D.; di Costanzo, F.; Glimelius, B.; Blijham, G.; Aranda, E.; Scheithauer, W.; Rougier, P.; Palmer, M.; Wils, J.; et al. Clinical determinants of survival in patients with 5-fluorouracil-based treatment for metastatic colorectal cancer, results of a multivariate analysis of 3825 patients. Ann. Oncol 2002, 13, 308–317. [Google Scholar]
- DeLong, E.R.; DeLong, D.M.; Clarke-Pearson, D.L. Comparing the areas under two or more correlated receiver operating curves, a nonparametric approach. Biometrics 1988, 44, 837–845. [Google Scholar]
- Taylor, J.M.G.; Ankerst, D.P.; Andridge, R.R. Validation of biomarker based risk prediction models. Clin. Cancer Res 2008, 14, 5977–5983. [Google Scholar]
- Bustin, S.A.; Gyselman, V.G.; Siddiqi, S.; Dorudi, S. Cytokeratin 20 is not a tissue-specific marker for the detection of malignant epithelial cells in the blood of colorectal cancer patients. Int. J. Surg. Invest 2000, 2, 49–57. [Google Scholar]
- Hundt, S.; Ulrike, H.; Brenner, H. Blood markers for early detection of colorectal cancer, a systematic review. Cancer Epidemiol. Biomark. Prev 2007, 16, 1935–1953. [Google Scholar]
- Liu, D.; Rudland, P.S.; Sibson, D.R.; Platt-Higgins, A.; Barraclough, R. Human homologue of cement gland protein, a novel metastasis inducer associated with breast carcinomas. Cancer Res 2005, 65, 3796–3805. [Google Scholar]
- Barraclough, D.L.; Platt-Higgins, A.; de Silva Rudland, S.; Barraclough, R.; Winstanley, J.; West, C.R.; Rudland, P.S. The metastasis-associated anterior gradient 2 protein is correlated with poor survival of breast cancer patients. Am. J. Pathol 2009, 175, 1848–1857. [Google Scholar]
- Zhang, Y.; Forootan, S.S.; Liu, D.; Barraclough, R.; Foster, C.S.; Rudland, P.S.; Ke, Y. Increased expression of anterior gradient-2 is significantly associated with poor survival of prostate cancer patients. Prostate Cancer Prostatic Dis 2007, 10, 293–300. [Google Scholar]
- Edgell, T.A.; Barraclough, D.L.; Rajic, A.; Dhulia, J.; Lewis, K.J.; Armes, J.E.; Barraclough, R.; Rudland, P.S.; Rice, G.E.; Autelitano, D.J. Increased plasma concentrations of anterior gradient 2 protein are positively associated with ovarian cancer. Clin. Sci. (Lond.) 2010, 118, 717–725. [Google Scholar]
- Dong, A.; Gupta, A.; Pai, R.K.; Tun, M.; Lowe, A.W. The human adenocarcinoma-associated gene, AGR2, induces expression of amphiregulin through HIPPO pathway co-activator YAP1 activation. J. Biol. Chem 2011, 286, 18301–18310. [Google Scholar]
- Merlos-Suárez, A.; Barriga, F.M.; Jung, P.; Iglesias, M.; Céspedes, M.V.; Rossell, D.; Sevillano, M.; Hernando-Momblona, X.; Silva-Diaz, V.; Muñoz, P.; et al. The intestinal stem cell signature identifies colorectal cancer stem cells and predicts disease relapse. Cell Stem Cell 2011, 8, 511–524. [Google Scholar]
- de Sousa, E.; Melo, F.; Colak, S.; Buikhuisen, J.; Koster, J.; Cameron, K.; de Jong, J.H.; Tuynman, J.B.; Prasetyanti, P.R.; Fessler, E.; et al. Methylation of cancer-stem-cell-associated WNT target genes predicts poor prognosis in colorectal cancer patients. Cell Stem Cell 2011, 9, 476–485. [Google Scholar]
- Pilati, P.; Mocellin, S.; Bertazza, L.; Galdi, F.; Briarava, M.; Mammano, E.; Tessari, E.; Zavagno, G.; Nitti, D. Prognostic value of putative circulating cancer stem cells in patients undergoing hepatic resection for colorectal liver metastasis. Ann. Surg. Oncol 2012, 19, 402–408. [Google Scholar]
- Weitz, J.; Kienle, P.; Lacroix, J.; Willeke, F.; Benner, A.; Lehnert, T.; Herfarth, C.; von Knebel Doeberitz, M. Dissemination of tumor cells in patients undergoing surgery for colorectal cancer. Clin. Cancer Res 1998, 4, 343–348. [Google Scholar]
- Peach, G.; Kim, C.; Zacharakis, E.; Purkayastha, S.; Ziprin, P. Prognostic significance of circulating tumor cells following surgical resection of colorectal cancers, a systematic review. Br. J. Cancer 2010, 102, 1327–1334. [Google Scholar]
- Cohen, S.J.; Punt, C.J.; Iannotti, N.; Saidman, B.H.; Sabbath, K.D.; Gabrail, N.Y.; Picus, J.; Morse, M.; Mitchell, E.; Miller, M.C.; et al. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J. Clin. Oncol 2008, 26, 3213–3221. [Google Scholar]
- Tol, J.; Koopman, M.; Miller, M.C.; Tibbe, A.; Cats, A.; Creemers, G.J.; Vos, A.H.; Nagtegaal, I.D.; Terstappen, L.W.; Punt, C.J. Circulating tumor cells early predict progression-free and overall survival in advanced colorectal cancer patients treated with chemotherapy and targeted agents. Ann. Oncol 2010, 21, 1006–1012. [Google Scholar]
- Wicha, M.S.; Hayes, D.F. Circulating tumor cells, not all detected cells are bad and not all bad cells are detected. J. Clin. Oncol 2011, 29, 1508–1511. [Google Scholar]
- Wittekind, C.; Greene, F.L.; Hutter, R.V.P.; Klimpfinger, M.; Sobin, L.H. TNM Atlas, 5th ed; Springer-Verlag: Berlin, Germany, 2005. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar]
- Pepe, M.S.; Etzioni, R.; Feng, Z.; Potter, J.D.; Thompson, M.L.; Thornquist, M.; Winget, M.; Yasui, Y. Phases of biomarker development for early detection of cancer. J. Natl. Cancer Inst 2001, 93, 1054–1061. [Google Scholar]
- McShane, L.M.; Altman, D.G.; Sauerbrei, W.; Taube, S.E.; Gion, M.; Clark, G.M. Statistics subcommittee of the NCI-EORTC working group on cancer diagnostics. Reporting recommendations for tumor marker prognostic studies (REMARK). J. Natl. Cancer Inst 2005, 97, 1180–1184. [Google Scholar]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines, minimum information for publication of quantitative real-time PCR experiments. Clin. Chem 2009, 55, 611–622. [Google Scholar]

| Mean (SD) | Range | |
|---|---|---|
| Age (years) | 62.7 (9.6) | 31–80 |
| N | % | |
| <60 years | 20 | 37.0 |
| ≥60 years | 34 | 63.0 |
| Gender | ||
| Female | 21 | 38.9 |
| Male | 33 | 61.1 |
| Stage | ||
| I–II | 6 | 11.1 |
| III | 12 | 22.2 |
| IV | 36 | 66.7 |
| pT | ||
| pT1–pT2 | 6 | 14.1 |
| pT3 | 36 | 66.7 |
| pT4 | 8 | 14.8 |
| pTx | 4 | 7.4 |
| pN | ||
| pN0 | 13 | 24.1 |
| pN1 | 26 | 48.1 |
| pN2 | 11 | 20.4 |
| pNx | 4 | 7.4 |
| M | ||
| M0 | 18 | 33.3 |
| M1 | 36 | 66.7 |
| Residual disease status | ||
| R0–R1 | 16 | 29.6 |
| R2 | 38 | 70.4 |
| Number of Metastatic Sites | ||
| 0 | 18 | 33.3 |
| 1 | 25 | 46.3 |
| ≥2 | 11 | 20.4 |
| Location of Metastasis | ||
| None | 18 | 33.3 |
| Liver Only | 23 | 42.6 |
| Liver and Other | 11 | 20.4 |
| Non-liver Metastasis | 2 | 3.7 |
| Grade | ||
| Low Grade | 10 | 18.5 |
| High Grade | 44 | 81.5 |
| Vascular/Perineural Invasion | ||
| Unknown | 6 | 11.1 |
| No | 24 | 44.4 |
| Yes | 24 | 44.4 |
| Parameter | AGR2 | LGR5 | ||||
|---|---|---|---|---|---|---|
| Mean | SEM | p | Mean | SEM | p | |
| Age (y) | 0.459 | 0.128 | ||||
| <60 | 497.0 | 142.2 | 22.5 | 10.0 | ||
| ≥60 | 367.3 | 105.1 | 5.0 | 5.0 | ||
| Gender | 0.075 | 0.203 | ||||
| Male | 291.1 | 87.9 | 5.8 | 3.9 | ||
| Female | 633.7 | 161.5 | 22.1 | 11.7 | ||
| Stage | 0.137* | 0.204* | ||||
| I–II | 1.1 | 0.2 | 0.3 | 0.05 | ||
| III | 471.3 | 171.2 | 0.1 | 0.08 | ||
| IV | 492.6 | 113.8 | 18.4 | 7.7 | ||
| pT | 0.002a | 0.915 | ||||
| pT1–T2 | 82.1 | 57.5 | 10.7 | 10.5 | ||
| pT3–T4 | 453.7 | 92.4 | 12.5 | 5.8 | ||
| pN | 0.063* | 0.309* | ||||
| Node Negative | 306.9 | 162.3 | 0.26 | 0.05 | ||
| pN1 | 311.7 | 80.4 | 13.1 | 7.01 | ||
| pN2 | 795.2 | 266.3 | 23.9 | 16.4 | ||
| M | 0.283 | 0.024a | ||||
| M0 | 305.4 | 122.5 | 0.18 | 0.06 | ||
| M1 | 492.6 | 113.8 | 18.4 | 7.7 | ||
| R Status | 0.671 | 0.024a | ||||
| R0–R1 | 363.2 | 156.1 | 0.13 | 0.03 | ||
| R2 | 442.6 | 101.7 | 40.3 | 7.01 | ||
| Number of Metastatic sites | 0.373* | 0.159* | ||||
| 0 | 305.4 | 122.5 | 0.18 | 0.06 | ||
| 1 | 407.7 | 145.0 | 21.5 | 10.6 | ||
| ≥2 | 628.4 | 184.7 | 12.3 | 9.3 | ||
| Grade | 0.023a | 0.024a | ||||
| Low grade | 183.0 | 71.9 | 0.1 | 0.04 | ||
| High grade | 480.9 | 102.8 | 14.6 | 6.1 | ||
| Vascular/Perineural Invasion | 0.751 | 0.269 | ||||
| No | 385.8 | 100.6 | 6.6 | 6.5 | ||
| Yes | 441.2 | 146.1 | 18.6 | 8.5 | ||
| Wald | Hazard Ratio | 95% CI | p | |||
|---|---|---|---|---|---|---|
| Progression free survival | ||||||
| Depth of invasion | pT1–2/pT3/pT4 | 1.042 | 1.430 | 0.720 | 2.841 | 0.307 |
| Lymph Nodes | Negative/Positive | 0.834 | 1.714 | 0.539 | 5.445 | 0.361 |
| Residual disease | R0–1/R2 | 8.047 | 5.824 | 1.724 | 19.68 | 0.005 |
| AGR2/LGR5 | Negative/Positive | 6.025 | 2.803 | 1.231 | 6.385 | 0.014 |
| Overall survival | ||||||
| Depth of invasion | pT1–2/pT3/pT4 | 0.741 | 1.443 | 0.626 | 3.322 | 0.389 |
| Lymph Nodes | Negative/Positive | 0.020 | 1.085 | 0.348 | 3.384 | 0.888 |
| Residual disease | R0–1/R2 | 7.041 | 7.338 | 1.683 | 31.99 | 0.008 |
| AGR2/LGR5 | Negative/Positive | 1.158 | 1.594 | 0.682 | 3.724 | 0.282 |
© 2012 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Valladares-Ayerbes, M.; Blanco-Calvo, M.; Reboredo, M.; Lorenzo-Patiño, M.J.; Iglesias-Díaz, P.; Haz, M.; Díaz-Prado, S.; Medina, V.; Santamarina, I.; Pértega, S.; et al. Evaluation of the Adenocarcinoma-Associated Gene AGR2 and the Intestinal Stem Cell Marker LGR5 as Biomarkers in Colorectal Cancer. Int. J. Mol. Sci. 2012, 13, 4367-4387. https://doi.org/10.3390/ijms13044367
Valladares-Ayerbes M, Blanco-Calvo M, Reboredo M, Lorenzo-Patiño MJ, Iglesias-Díaz P, Haz M, Díaz-Prado S, Medina V, Santamarina I, Pértega S, et al. Evaluation of the Adenocarcinoma-Associated Gene AGR2 and the Intestinal Stem Cell Marker LGR5 as Biomarkers in Colorectal Cancer. International Journal of Molecular Sciences. 2012; 13(4):4367-4387. https://doi.org/10.3390/ijms13044367
Chicago/Turabian StyleValladares-Ayerbes, Manuel, Moisés Blanco-Calvo, Margarita Reboredo, María J. Lorenzo-Patiño, Pilar Iglesias-Díaz, Mar Haz, Silvia Díaz-Prado, Vanessa Medina, Isabel Santamarina, Sonia Pértega, and et al. 2012. "Evaluation of the Adenocarcinoma-Associated Gene AGR2 and the Intestinal Stem Cell Marker LGR5 as Biomarkers in Colorectal Cancer" International Journal of Molecular Sciences 13, no. 4: 4367-4387. https://doi.org/10.3390/ijms13044367
APA StyleValladares-Ayerbes, M., Blanco-Calvo, M., Reboredo, M., Lorenzo-Patiño, M. J., Iglesias-Díaz, P., Haz, M., Díaz-Prado, S., Medina, V., Santamarina, I., Pértega, S., Figueroa, A., & Antón-Aparicio, L. M. (2012). Evaluation of the Adenocarcinoma-Associated Gene AGR2 and the Intestinal Stem Cell Marker LGR5 as Biomarkers in Colorectal Cancer. International Journal of Molecular Sciences, 13(4), 4367-4387. https://doi.org/10.3390/ijms13044367






