Modulation of rosR Expression and Exopolysaccharide Production in Rhizobium leguminosarum bv. trifolii by Phosphate and Clover Root Exudates
Abstract
:1. Introduction
2. Results and Discussion
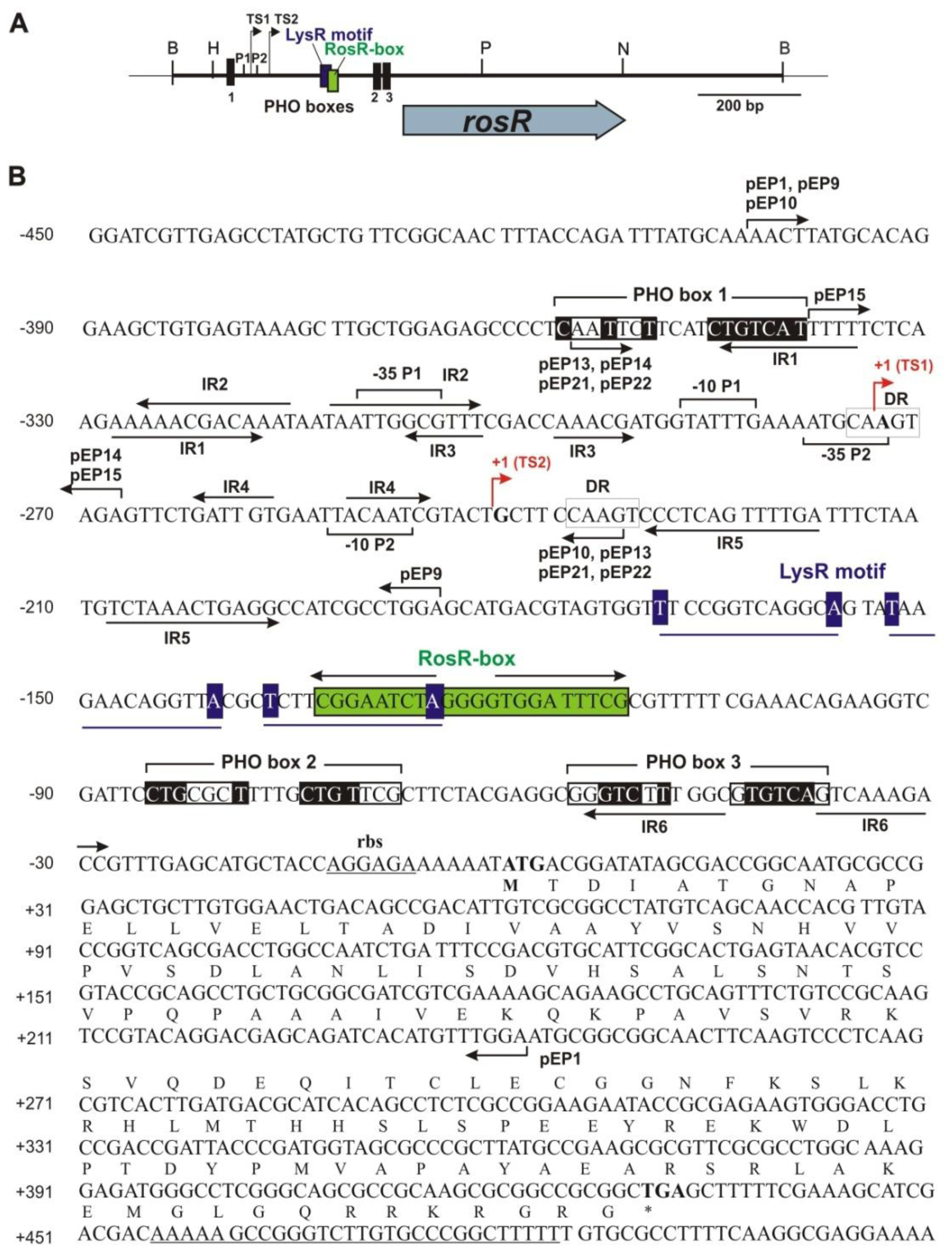
2.1. Identification of Putative Regulatory Motifs in the rosR Upstream Region
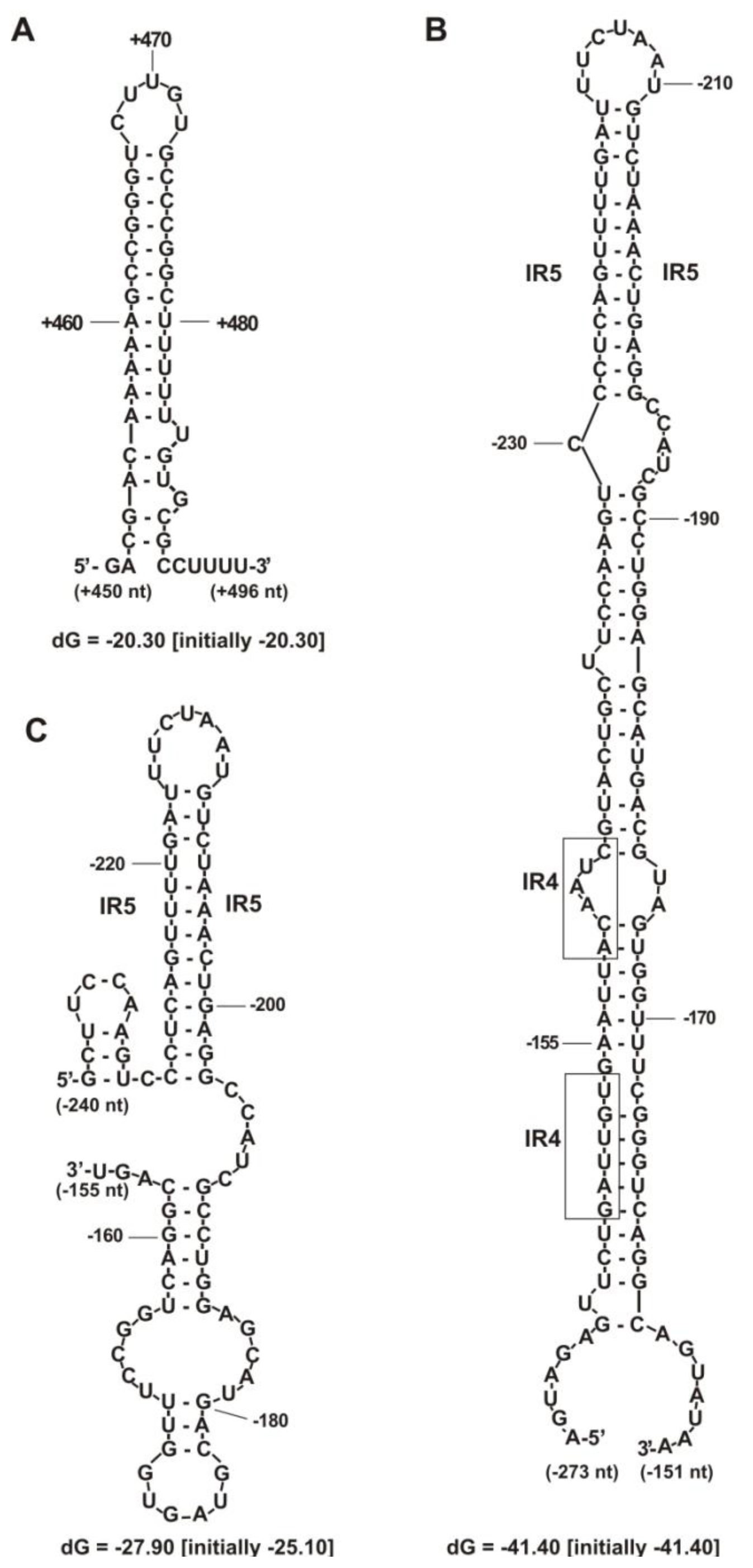
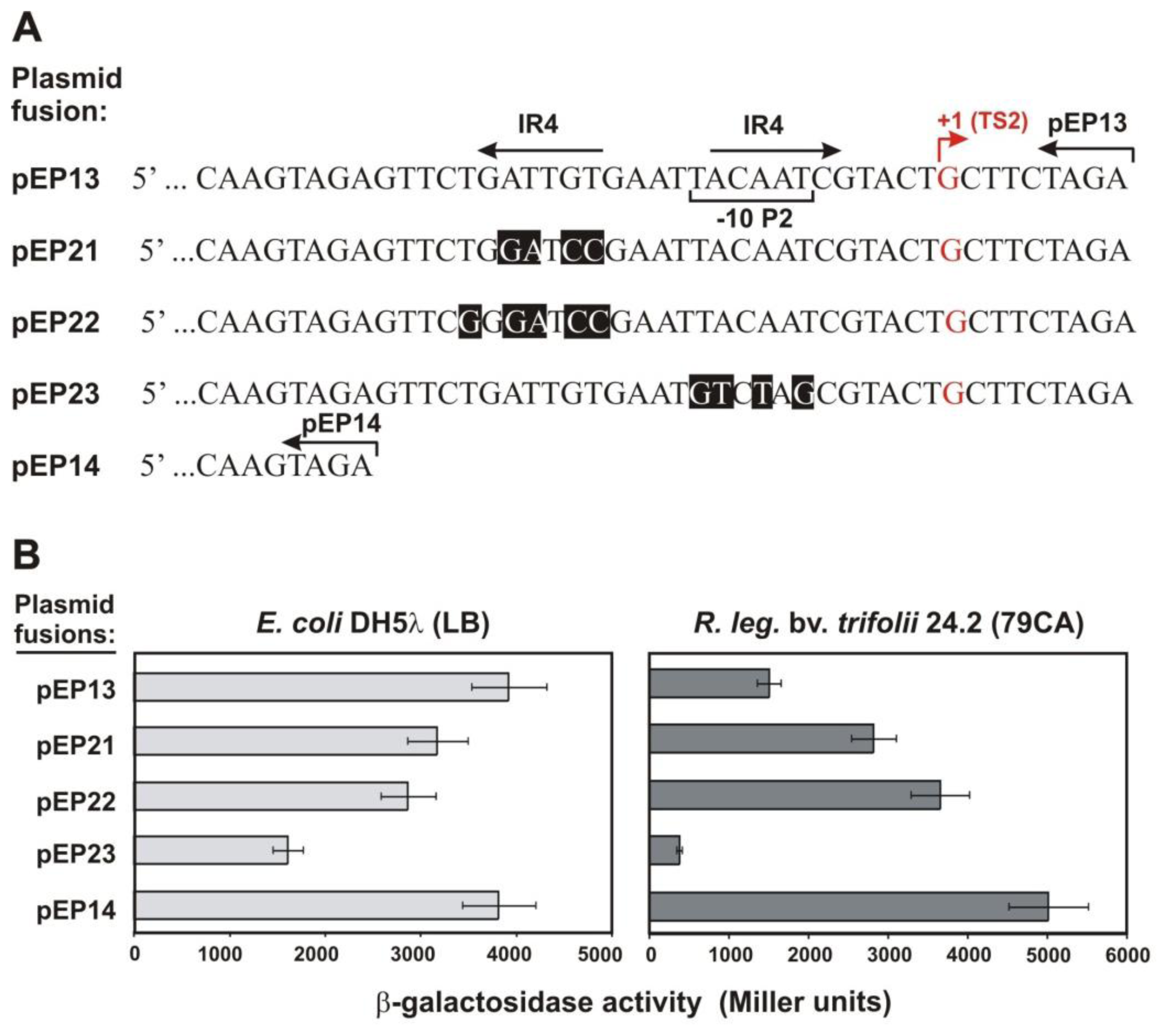
2.2. Functional Analysis of the Putative Regulatory Motif IR4 in the rosR Upstream Region
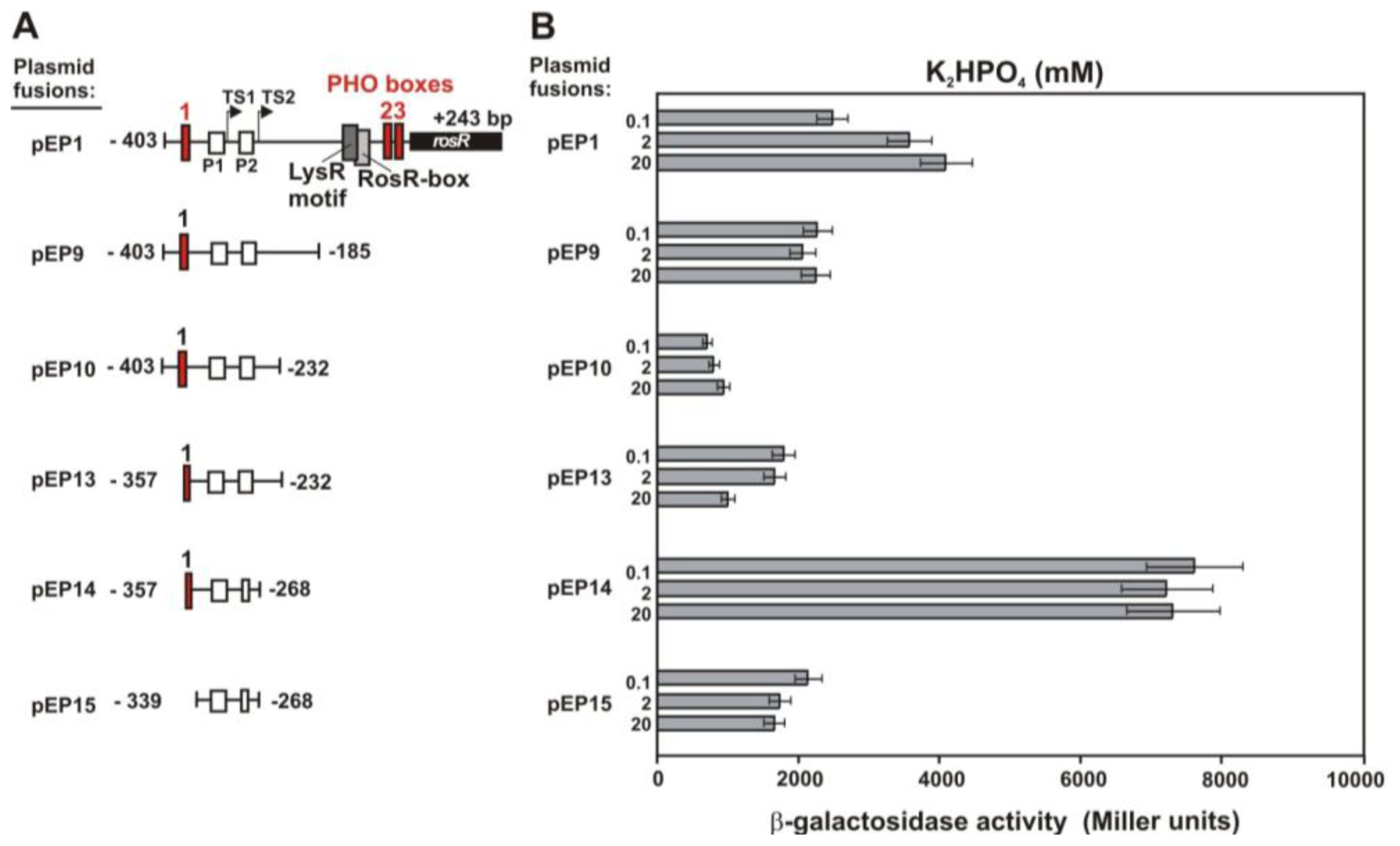
2.3. Functional Analysis of Other Putative Regulatory Motifs in the rosR Upstream Region
2.4. Functional Analysis of a Putative LysR Motif
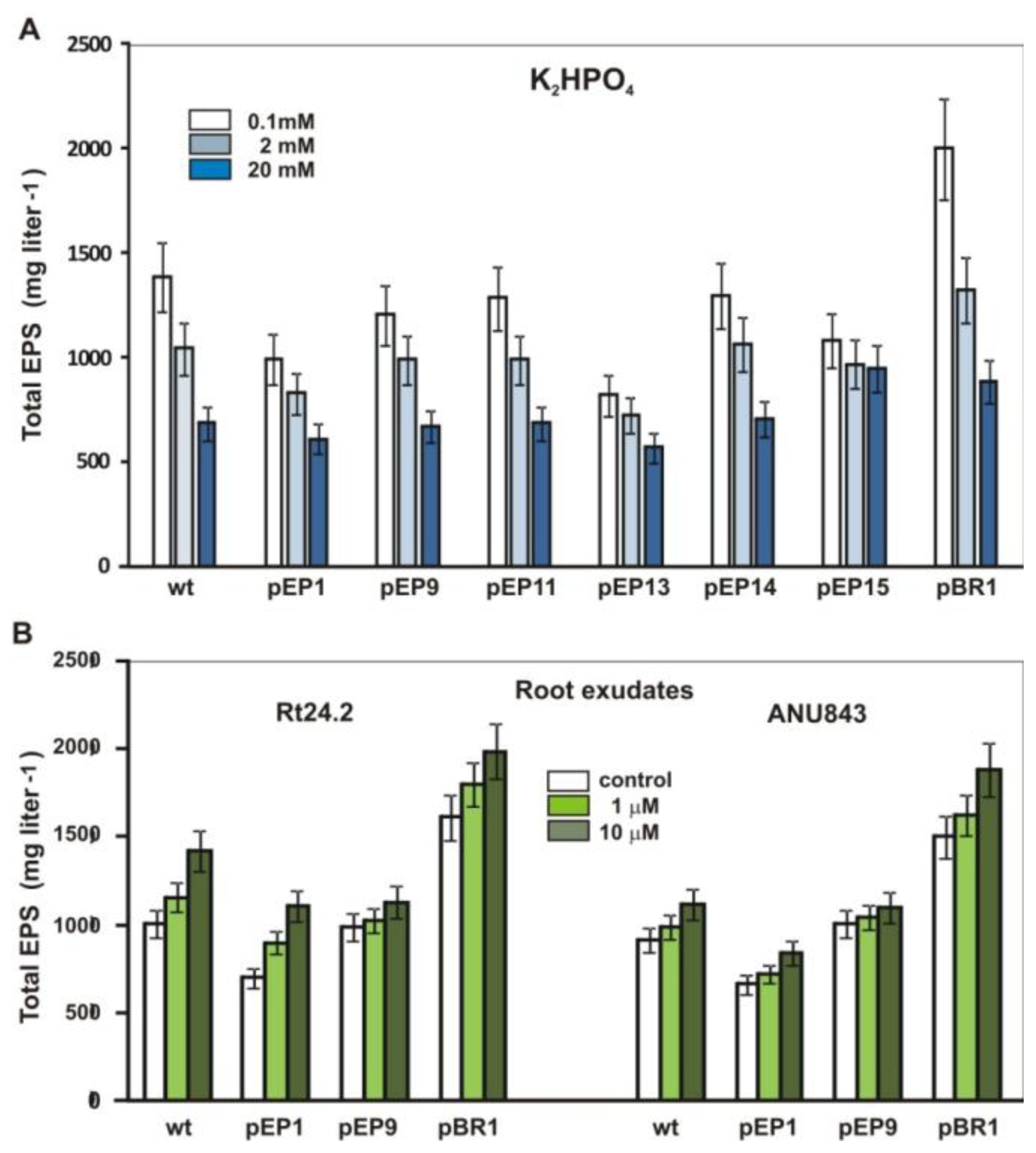
2.5. Effect of Phosphate and Clover Root Exudates on EPS Production
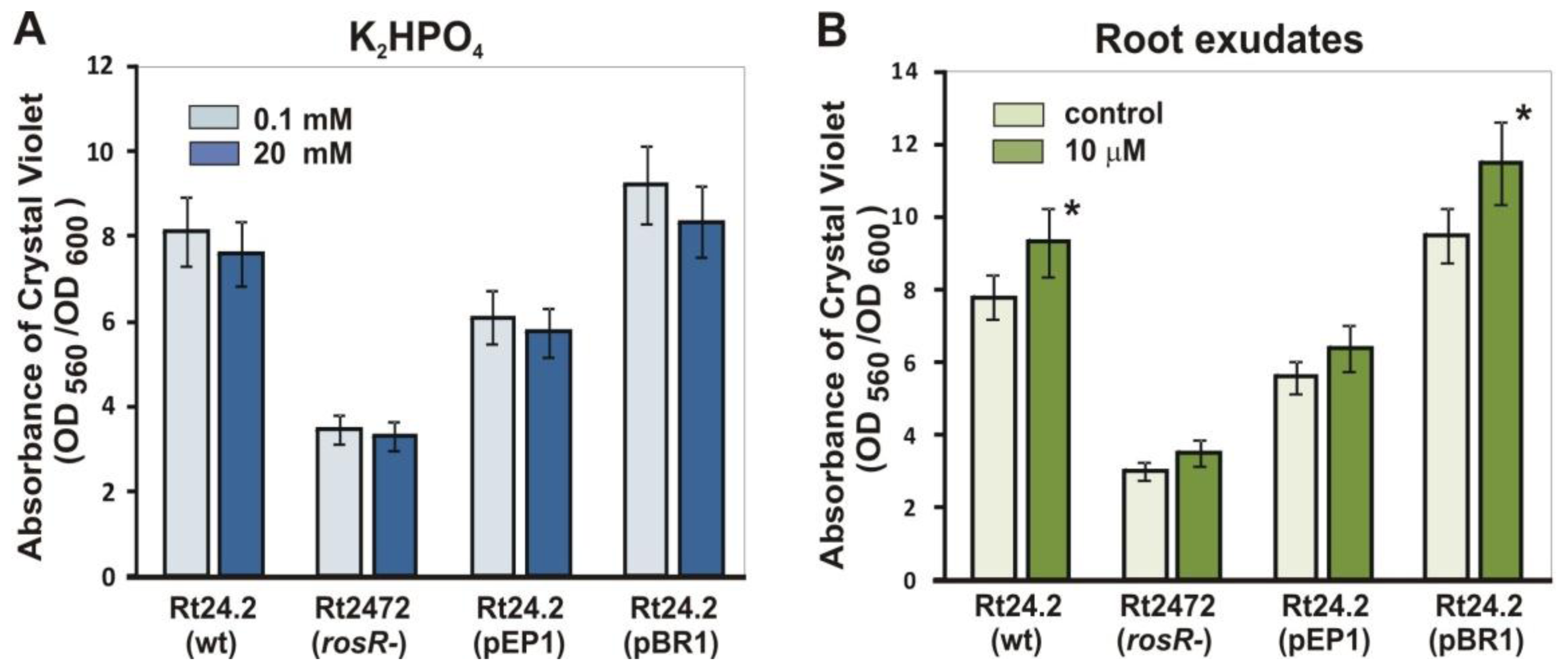
2.6. Biofilm Formation by R. leguminosarum bv. trifolii on Abiotic Surfaces in the Presence of Phosphate and Clover Root Exudates
2.7. Effect of Phosphate and Flavonoids on Symbiosis of Rt24.2 and Its Derivatives with Clover
3. Experimental Section
3.1. Bacterial Strains, Plasmids, and Growth Conditions
3.2. DNA Methods and Sequence Analysis
3.3. Construction of Transcriptional rosR-lacZ Fusions with Mutated IR4 Sites
3.4. β-Galactosidase Assay
3.5. EPS Isolation
3.6. Biofilm Formation Assay
3.7. Preparation of Exudates from Sprouted Seeds of Clover
3.8. Plant Tests
4. Conclusions
Acknowledgments
References
- Downie, JA. The roles of extracellular proteins, polysaccharides and signals in the interactions of rhizobia with legume roots. FEMS Microbiol. Rev 2010, 34, 150–170. [Google Scholar]
- Flemming, HC; Wingender, J. The biofilm matrix. Nat. Rev. Microbiol 2010, 8, 623–633. [Google Scholar]
- Becker, A; Pühler, A. Spaink, HP, Kondorosi, A, Hooykaas, PJJ, Eds.; Production of exopolysaccharides. In Rhizobiaceae Molecular Biology of Plant-Associated Bacteria; Kluwer Academic Press: Dordrecht, The Netherlands, 1998; pp. 97–118. [Google Scholar]
- Fraysse, N; Couderc, F; Poinsot, V. Surface polysaccharide involvement in establishing the rhizobium-legume symbiosis. Eur. J. Biochem 2003, 270, 1365–1380. [Google Scholar]
- Skorupska, A; Janczarek, M; Marczak, M; Mazur, A; Król, J. Rhizobial exopolysaccharides: Genetic control and symbiotic functions. Microb Cell Fact 2006, 5. [Google Scholar]
- Borthakur, D; Barker, CE; Lamb, JW; Daniels, MJ; Downie, JA; Johnston, AWB. A mutation that blocks exopolysaccharide synthesis prevents nodulation of peas by Rhizobium leguminosarum but not of beans by R. phaseolii and is corrected by cloned DNA from Rhizobium or the phytopathogen Xanthomonas. Mol. Gen. Genet 1986, 203, 320–323. [Google Scholar]
- Niehaus, K; Kapp, D; Pühler, A. Plant defence and delayed infection of alfalfa pseudonodules induced by an exopolysaccharide (EPSI)-deficient Rhizobium meliloti mutant. Planta 1993, 190, 415–425. [Google Scholar]
- Rolfe, BG; Carlson, RW; Ridge, RW; Dazzo, RW; Mateos, FB; Pankhurst, CE. Defective infection and nodulation of clovers by exopolysaccharide mutants of Rhizobium leguminosarum bv. trifolii. Aust. J. Plant Physiol 1996, 23, 285–303. [Google Scholar]
- Cheng, HP; Walker, GC. Succinoglycan is required for initiation and elongation of infection threads during nodulation of alfalfa by Rhizobium meliloti. J. Bacteriol 1998, 180, 5183–5191. [Google Scholar]
- Canter-Cremers, HCJ; Stevens, K; Lugtenberg, BJJ; Wijffelman, CA; Batley, M; Redmond, JW; Breedveld, M; Zevenhuizen, LPTM. Unusual structure of the exopolysaccharide of Rhizobium leguminosarum biovar viciae strain 248. Carbohydr. Res 1991, 218, 185–200. [Google Scholar]
- O’Neill, MA; Darvill, AG; Albersheim, P. The degree of esterification and points of substitution by O-acetyl and O-(3-hydroxybutanoyl) groups in the acidic extracellular polysaccharides secreted by Rhizobium leguminosarum biovars viciae, trifolii, and phaseoli are not related to host range. J. Biol. Chem 1991, 266, 9549–9555. [Google Scholar]
- Król, JE; Mazur, A; Marczak, M; Skorupska, A. Syntenic arrangements of the surface polysaccharide biosynthesis genes in Rhizobium leguminosarum. Genomics 2007, 89, 237–247. [Google Scholar]
- Young, JPW; Crossman, LC; Johnston, AWB; Thomson, NR; Ghazoui, ZF; Hull, KH; Wexler, M; Curson, ARJ; Todd, JD; Poole, PS; et al. The genome of Rhizobium leguminosarum has recognizable core and accessory components. Genome Biol 2006, 7. [Google Scholar]
- Rinaudi, LV; Giordano, W. An integrated view of biofilm formation in rhizobia. FEMS Microiol. Lett 2010, 304, 1–11. [Google Scholar]
- Williams, A; Wilkinson, A; Krehenbrink, M; Russo, D; Zorreguieta, A; Downie, JA. Glucomannan-mediated attachment of Rhizobium leguminosarum to pea root hairs is required for competitive nodule infection. J. Bacteriol 2008, 190, 4706–4715. [Google Scholar]
- Russo, DM; Williams, A; Edwards, A; Posadas, DM; Finnie, C; Dankert, M; Downie, JA; Zorreguieta, A. Proteins exported via the PrsD-PrsE type I secretion system and the acidic exopolysaccharide are involved in biofilm formation by Rhizobium leguminosarum. J. Bacteriol 2006, 188, 4474–4486. [Google Scholar]
- Rinaudi, LV; González, JE. The low-molecular-weight fraction of the exopolysaccharide II from Sinorhizobium meliloti is a crucial determinant of biofilm formation. J. Bacteriol 2009, 191, 7216–7224. [Google Scholar]
- Becker, A; Küster, H; Niehaus, K; Pühler, A. Extension of the Rhizobium meliloti succinoglycan biosynthesis gene cluster: Identification of the exsA gene encoding an ABC transporter protein, and the exsB gene which probably codes for a regulator of succinoglycan biosynthesis. Mol. Gen. Genet 1995, 249, 487–497. [Google Scholar]
- Yao, SY; Luo, L; Har, KJ; Becker, A; Rüberg, S; Yu, GQ; Zhu, JB; Cheng, HP. Sinorhizobium meliloti ExoR and ExoS proteins regulate both succinoglycan and flagellum production. J. Bacteriol 2004, 186, 6042–6049. [Google Scholar]
- Bahlawane, C; McIntosh, M; Krol, E; Becker, A. Sinorhizobium meliloti regulator MucR couples exopolysaccharide synthesis and motility. Mol. Plant Microbe Interact 2008, 21, 1498–1509. [Google Scholar]
- Rüberg, S; Pühler, A; Becker, A. Biosynthesis of the exopolysaccharide galactoglucan in Sinorhizobium meliloti is subject to a complex control by the phosphate-dependent regulator PhoB and the proteins ExpG and MucR. Microbiology 1999, 145, 603–611. [Google Scholar]
- Bertram-Drogatz, PA; Quester, I; Becker, A; Pühler, A. The Sinorhizobium meliloti MucR protein, which is essential for the production of high-molecular-weight succinoglycan exopolysaccharide, binds to short DNA regions upstream of exoH and exoY. Mol. Gen. Genet 1998, 257, 433–441. [Google Scholar]
- Doherty, D; Leigh, JA; Glazebrook, J; Walker, GC. Rhizobium meliloti mutants that overproduce the R. meliloti acidic calcofluor-binding exopolysaccharide. J. Bacteriol 1988, 170, 4249–4256. [Google Scholar]
- Breedveld, MW; Zevenhuizen, LPTM; Zehnder, AJB. Osmotically induced oligo- and polysaccharide synthesis by Rhizobium meliloti SU-47. J. Gen. Microbiol 1990, 136, 2511–2519. [Google Scholar]
- Mendrygal, KE; González, JE. Environmental regulation of exopolysaccharide production in Sinorhizobium meliloti. J. Bacteriol 2000, 182, 599–606. [Google Scholar]
- Zhan, H; Lee, CC; Leigh, JA. Induction of the second exopolysaccharide (EPSb) in Rhizobium meliloti SU47 by low phosphate concentrations. J. Bacteriol 1991, 173, 7391–7394. [Google Scholar]
- Krol, E; Becker, A. Global transcriptional analysis of the phosphate starvation response in Sinorhizobium meliloti strains 1021 and 2011. Mol. Gen. Genomics 2004, 272, 1–17. [Google Scholar]
- Yuan, Z; Zaheer, R; Morton, R; Finan, TM. Genome prediction of PhoB regulated promoters in Sinorhizobium meliloti and twelve proteobacteria. Nucl. Acids Res 2006, 34, 2686–2697. [Google Scholar]
- Marketon, MM; Glenn, SA; Eberhard, A; González, JE. Quorum sensing controls exopolysaccharide production in Sinorhizobium meliloti. J. Bacteriol 2003, 185, 325–331. [Google Scholar]
- Barnett, M; Long, SR. Identification and characterization of a gene on Rhizobium meliloti pSymA, syrB, that negatively affects syrM expression. Mol. Plant Microbe Interact 1997, 10, 550–559. [Google Scholar]
- Dusha, I; Olah, B; Szegletes, Z; Erdei, L; Kondorosi, A. SyrM involved in the determination of the amount and ratio of the two forms of the acidic exopolysaccharide EPSI in Rhizobium meliloti. Mol. Plant Microbe Interact 1999, 12, 755–765. [Google Scholar]
- Borthakur, D; Johnston, AWB. Sequence of psi, a gene of the symbiotic plasmid of Rhizobium phaseoli which inhibits exopolysaccharide synthesis and nodulation and demonstration that its transcription is inhibited by psr, another gene on the symbiotic plasmid. Mol. Gen. Genet 1987, 207, 149–154. [Google Scholar]
- Latchford, JW; Borthakur, D; Johnston, AWB. The products of Rhizobium genes, psi and pss, which affect exopolysaccharide production, are associated with the bacterial cell surface. Mol. Microbiol 1991, 5, 2107–2114. [Google Scholar]
- Reeve, WG; Dilworth, MJ; Tiwari, RP; Glenn, AR. Regulation of exopolysaccharide production in Rhizobium leguminosarum biovar viciae WSM710 involves exoR. Microbiology 1997, 143, 1951–1958. [Google Scholar]
- Janczarek, M; Skorupska, A. The Rhizobium leguminosarum bv. trifolii pssB gene product is an inositol monophosphatase that influences exopolysaccharide synthesis. Arch. Microbiol 2001, 175, 143–151. [Google Scholar]
- Janczarek, M; Skorupska, A. The Rhizobium leguminosarum bv. trifolii RosR: Transcriptional regulator involved in exopolysaccharide production. Mol. Plant Microbe Interact 2007, 20, 867–881. [Google Scholar]
- Keller, M; Roxlau, A; Wenig, WM; Schmidt, M; Quandt, J; Niehaus, K; Jording, D; Arnold, W; Pühler, A. Molecular analysis of the Rhizobium meliloti mucR gene regulating the biosynthesis of the exopolysaccharides succinoglycan and galactoglucan. Mol. Plant Microbe Interact 1995, 8, 267–277. [Google Scholar]
- Bittinger, MA; Milner, JL; Saville, BJ; Handelsman, J. RosR, a determinant of nodulation competitiveness in Rhizobium etli. Mol. Plant Microbe Interact 1997, 10, 180–186. [Google Scholar]
- Hussain, H; Johnston, AW. Iron-dependent transcription of the regulatory gene ros of Agrobacterium radiobacter. Mol. Plant Microbe Interact 1997, 10, 1087–1093. [Google Scholar]
- Chou, AY; Archdeacon, J; Kado, CI. Agrobacterium transcriptional regulator Ros is a prokaryotic zinc finger protein that regulates the plant oncogene ipt. Proc. Natl. Acad. Sci. USA 1998, 95, 5293–5298. [Google Scholar]
- Janczarek, M; Jaroszuk-Œciseł, J; Skorupska, A. Multiple copies of rosR and pssA genes enhance exopolysaccharide production, symbiotic competitiveness and clover nodulation in Rhizobium leguminosarum bv. trifolii. Antonie van Leeuwenhoek 2009, 96, 471–486. [Google Scholar]
- Janczarek, M; Skorupska, A. Rhizobium leguminosarum bv. trifolii rosR gene expression is regulated by catabolic repression. FEMS Microbiol. Lett 2009, 291, 112–119. [Google Scholar]
- Janczarek, M; Kutkowska, J; Piersiak, T; Skorupska, A. Rhizobium leguminosarum bv. trifolii rosR is required for interaction with clover, biofilm formation and adaptation to the environment. BMC Microbiol 2010, 10. [Google Scholar]
- Quester, I; Becker, A. Four promoters subject to regulation by ExoR and PhoB direct transcription of the Sinorhizobium meliloti exoYFQ operon involved in the biosynthesis of succinoglycan. J. Mol. Microbiol. Biotech 2004, 7, 115–132. [Google Scholar]
- Spinelli, SV; Pontel, LB; Vescovi, EG; Soncini, FC. Regulation of magnesium homeostasis in Salmonella: Mg2+ targets the mgtA transcript for degradation by RNase E. FEMS Microbiol. Lett 2008, 280, 226–234. [Google Scholar]
- Summers, ML; Elkins, JG; Elliot, BA; McDermott, TR. Expression and regulation of phosphate stress inducible genes in Sinorhizobium meliloti. Mol. Plant Microbe Interact 1999, 11, 1094–1101. [Google Scholar]
- Bieleski, RL. Phosphate pools, phosphate transport and phosphate availability. Annu. Rev. Plant Physiol 1973, 24, 225–252. [Google Scholar]
- Bardin, S; Dan, S; Osteras, M; Finan, TM. A phosphate transport system is required for symbiotic nitrogen fixation by Rhizobium meliloti. J. Bacteriol 1996, 178, 4540–4547. [Google Scholar]
- Riley, M; Abe, T; Arnaud, MB; Berlyn, MK; Blattner, FR; Chaudhuri, RR; Glasner, JD; Horiuchi, T; Keseler, IM; Kosuge, T; et al. Escherichia coli K-12: A cooperatively developed annotation snapshot-2005. Nucl Acids Res 2006, 34, 1–9. [Google Scholar]
- Bardin, SD; Voegele, RT; Finan, TM. Phosphate assimilation in Rhizobium (Sinorhizobium) meliloti: Identification of a pit-like gene. J. Bacteriol 1998, 180, 4219–4226. [Google Scholar]
- Rosenberg, H; Gerdes, RG; Chegwidden, K. Two systems for the uptake of phosphate in Escherichia coli. J. Bacteriol 1977, 131, 505–511. [Google Scholar]
- Long, SR. Genes and signals in the Rhizobium—Legume symbiosis. Plant Physiol 2001, 125, 69–72. [Google Scholar]
- Fujishige, NA; Kapadia, NN; Hirsch, AM. A feeling for the micro-organism: Structure on a small scale. Biofilms on plant roots. Bot. J. Linn. Soc 2006, 150, 79–88. [Google Scholar]
- Fujishige, NA; Lum, MR; De Hoff, PL; Whitelegge, JP; Faull, KF; Hirsch, AM. Rhizobium common nod genes are required for biofilm formation. Mol. Microbiol 2008, 67, 504–515. [Google Scholar]
- Rinaudi, LV; Fujishige, NA; Hirsch, AM; Banchio, E; Zorreguieta, A; Giordano, W. Effects of nutritional and environmental conditions on Sinorhizobium meliloti biofilm formation. Res. Microbiol 2006, 157, 867–875. [Google Scholar]
- Danhorn, T; Hentzer, M; Givskov, M; Parsek, MR; Fuqua, C. Phosphorus limitation enhances biofilm formation of the plant pathogen Agrobacterium tumefaciens through the PhoR-PhoB regulatory system. J. Bacteriol 2004, 186, 4492–4501. [Google Scholar]
- Djordjevic, MA; Żurkowski, W; Shine, J; Rolfe, BG. Sym-plasmid transfer to various symbiotic mutants of Rhizobium trifolii, R. leguminosarum and R. meliloti. J. Bacteriol 1983, 156, 1035–1045. [Google Scholar]
- Gaal, T; Bartlett, MS; Ross, W; Turnbough, JCL; Gourse, RL. Transcription regulation by initiating NTP concentration: rRNA synthesis in bacteria. Science 1997, 278, 2092–2097. [Google Scholar]
- Spira, B. Departamento de Microbiologia, Instituto de Ciěncias Biomédicas, Universidade de Săo Paulo, Săo Paulo-SP: Brazil; Personal collection; 2009. [Google Scholar]
- Sambrook, J; Fitsch, EF; Maniatis, T. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1989. [Google Scholar]
- Spaink, HP; Okker, RJH; Wijffelman, CA; Pees, E; Lugtenberg, BJJ. Promoters in the nodulation region of the Rhizobium leguminosarum Sym plasmid pRL1JI. Plant Mol. Biol 1987, 9, 27–39. [Google Scholar]
- Vincent, JM. A manual for the practical study of root nodule bacteria. In International Biological Program Handbook No 15; Blackwell Scientific Publications Ltd: Oxford, UK, 1970. [Google Scholar]
- Brown, CM; Dilworth, MJ. Ammonia assimilation by Rhizobium cultures and bacteroids. J. Gen. Microbiol 1975, 86, 39–48. [Google Scholar]
- GeneBee, AN. Belozersky Institute of Physico-Chemical Biology, Moscow State University: Moscow, Russia, 1965. Available online: http://www.genebee.msu.su/ accessed on 16 June 2011.
- Rice, P; Longden, I; Bleasby, A. EMBOSS: The European Molecular Biology Open Software Suite. Trends in Genetics. 2000, pp. 276–277. Available online: http://emboss.ch.embnet.org/Pise accessed on 16 June 2011.
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucl Acids Res. 2003, 31, pp. 3406–3415. Available online: http://mfold.rna.albany.edu accessed on 16 June 2011.
- Loewus, FA. Improvement in the anthrone method for determination of carbohydrates. Anal. Chem 1952, 24, 219. [Google Scholar]
- Newbury, SF; Smith, NH; Higgins, CF. Differential mRNA stability controls relative gene expression within a polycistronic operon. Cell 1987, 51, 1131–1143. [Google Scholar]
- Masse, E; Escorcia, FE; Gottesman, S. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev 2003, 17, 2374–2383. [Google Scholar]
- Narberhaus, F; Waldminghaus, T; Chowdhury, S. RNA thermometers. FEMS Microbiol. Rev 2006, 30, 3–16. [Google Scholar]
- Serganov, A. The long and the short of riboswitches. Curr. Opin. Struct. Biol 2009, 19, 251–259. [Google Scholar]
- Browning, DF; Busby, SJW. The regulation of bacterial transcription initiation. Nature Rev. Microbiol 2004, 2, 1–9. [Google Scholar]


| Plasmid fusion | LysR motif | ANU843 (wt) | ANU851 (nodD−) | ||
|---|---|---|---|---|---|
| Clover root exudates (μM) | |||||
| 0 | 10 | 0 | 10 | ||
| pEP1 | + | 2630 ± 288 | 3613 ± 352 | 2682 ± 253 | 2789 ± 245 |
| pEP9 | − | 1778 ± 185 | 1866 ± 191 | 1709 ± 167 | 1811 ± 177 |
| Strain or plasmid | Relevant characteristics | Reference |
|---|---|---|
| Rhizobium leguminosarum bv. trifolii | ||
| Rt24.2 | Wild type, Rifr, Nxr | [36] |
| Rt2472 | Rt24.2 derivative carrying mini-Tn5 between 151–152 bp position of rosR, Rifr, Kmr | [41] |
| ANU843 | Wild type, Rifr | [57] |
| ANU851 | ANU843 derivative carrying Tn5::nodD, Kmr | [57] |
| E. coli | ||
| VH1000 | MG1655 derivative, lacZ, lacI, pyrE+ | [58] |
| LG01 | MG1655 derivative, phoB519::Tn5, Kmr | [59] |
| Plasmids | ||
| pUC19 | Cloning and sequencing vector, Apr | [60] |
| pMP220 | IncP, mob, promoterless lacZ, Tcr | [61] |
| pMJ221 | pUC19 containing 126-bp EcoRI-XbaI fragment of the rosR upstream region (based on primers: pROS2/pREW2 and pRBAM1/pREW3) | This work |
| pMJ222 | pUC19 containing 126-bp EcoRI-XbaI fragment of the rosR upstream region (based on primers: pROS2/pREW4 and pRBAM1/pREW3) | This work |
| pMJ223 | pUC19 containing 126-bp EcoRI-XbaI fragment of the rosR upstream region (based on primers: pROS2/pREW1) | This work |
| pBR1 | pBBR1MCS-2 containing 1100-bp EcoRI-BamHI fragment with rosR of Rt24.2 | [41] |
| pEP1 | pMP220 carrying the −403 to +243 bp fragment of the rosR coding region | [36] |
| pEP9 | pMP220 carrying the −403 to −185 bp fragment of the rosR upstream region | [36] |
| pEP10 | pMP220 carrying the −403 to −232 bp fragment of the rosR upstream region | [36] |
| pEP13 | pMP220 carrying the −357 to −232 bp fragment of the rosR upstream region | [42] |
| pEP14 | pMP220 carrying the −357 to −268 bp fragment of the rosR upstream region | [42] |
| pEP15 | pMP220 carrying the −339 to −268 bp fragment of the rosR upstream region | [42] |
| pEP21 | pMP220 containing 126-bp EcoRI-XbaI fragment from pMJ221 | This work |
| pEP22 | pMP220 containing 126-bp EcoRI-XbaI fragment from pMJ222 | This work |
| pEP23 | pMP220 containing 126-bp EcoRI-XbaI fragment from pMJ223 | This work |
| Oligonucleotide primers | Sequence (5′→3′) * | |
| pROS2 | GAGCCCCTGAATTCTTCATCTGTCA | This work |
| pREW1 | AGGGATCTAGAAGCAGTACGCTAGACATTCAC | This work |
| pREW2 | GTAATTCGGATCCAGAACTCTACTTGCA | This work |
| pREW3 | GAAATCAAAACTGAGGGATCTAGAAGCA | This work |
| pREW4 | GTAATTCGGATCCCGAACTCTACTTGCA | This work |
| pRBAM1 | AAATGCAAGTAGAGTTCTGGATCCGAATTA | This work |
© 2011 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Janczarek, M.; Skorupska, A. Modulation of rosR Expression and Exopolysaccharide Production in Rhizobium leguminosarum bv. trifolii by Phosphate and Clover Root Exudates. Int. J. Mol. Sci. 2011, 12, 4132-4155. https://doi.org/10.3390/ijms12064132
Janczarek M, Skorupska A. Modulation of rosR Expression and Exopolysaccharide Production in Rhizobium leguminosarum bv. trifolii by Phosphate and Clover Root Exudates. International Journal of Molecular Sciences. 2011; 12(6):4132-4155. https://doi.org/10.3390/ijms12064132
Chicago/Turabian StyleJanczarek, Monika, and Anna Skorupska. 2011. "Modulation of rosR Expression and Exopolysaccharide Production in Rhizobium leguminosarum bv. trifolii by Phosphate and Clover Root Exudates" International Journal of Molecular Sciences 12, no. 6: 4132-4155. https://doi.org/10.3390/ijms12064132
APA StyleJanczarek, M., & Skorupska, A. (2011). Modulation of rosR Expression and Exopolysaccharide Production in Rhizobium leguminosarum bv. trifolii by Phosphate and Clover Root Exudates. International Journal of Molecular Sciences, 12(6), 4132-4155. https://doi.org/10.3390/ijms12064132




