A Novel Cold-Adapted Lipase from Sorangium cellulosum Strain So0157-2: Gene Cloning, Expression, and Enzymatic Characterization
Abstract
:1. Introduction
2. Results and Discussion
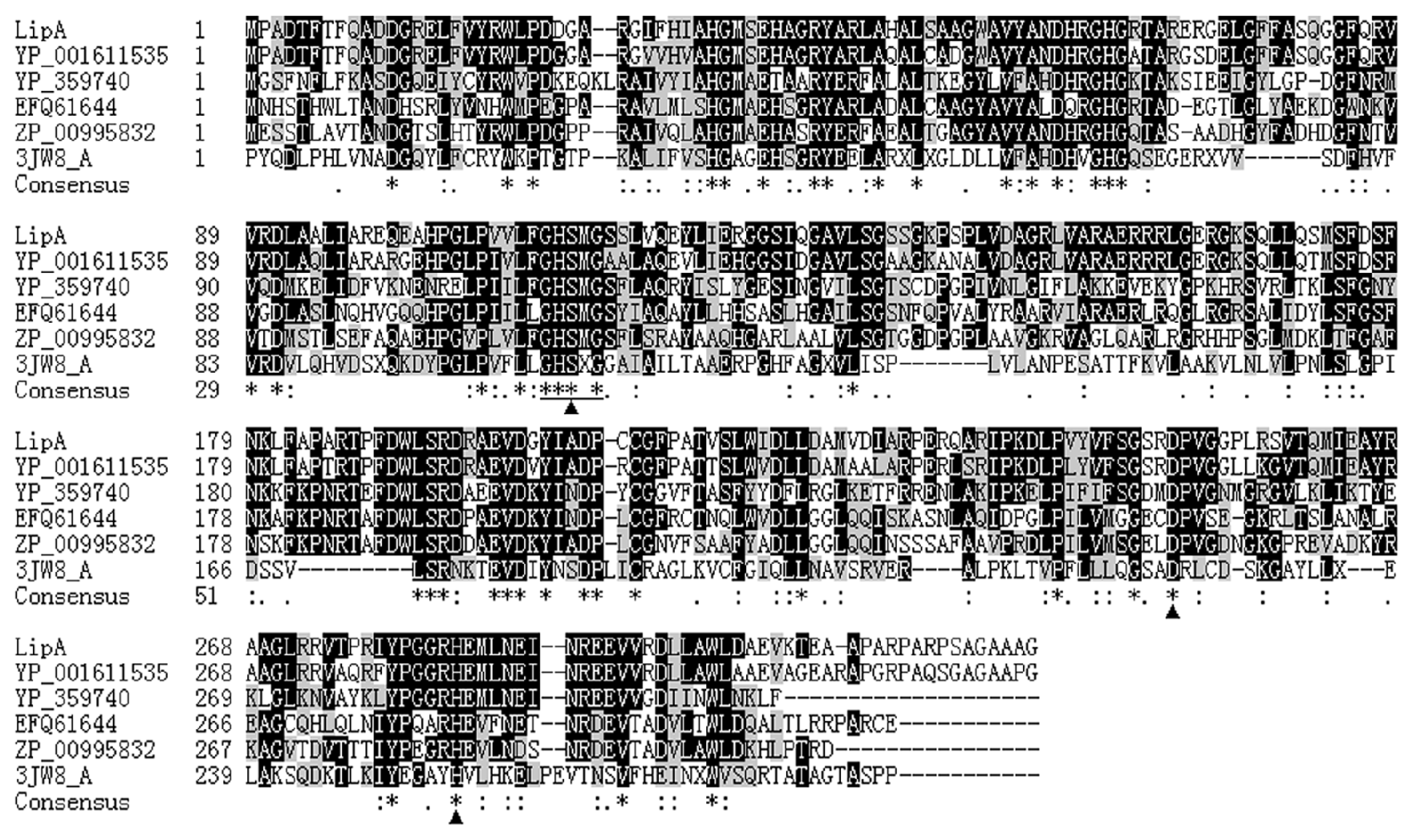
2.1. Gene Cloning and Sequence Analysis
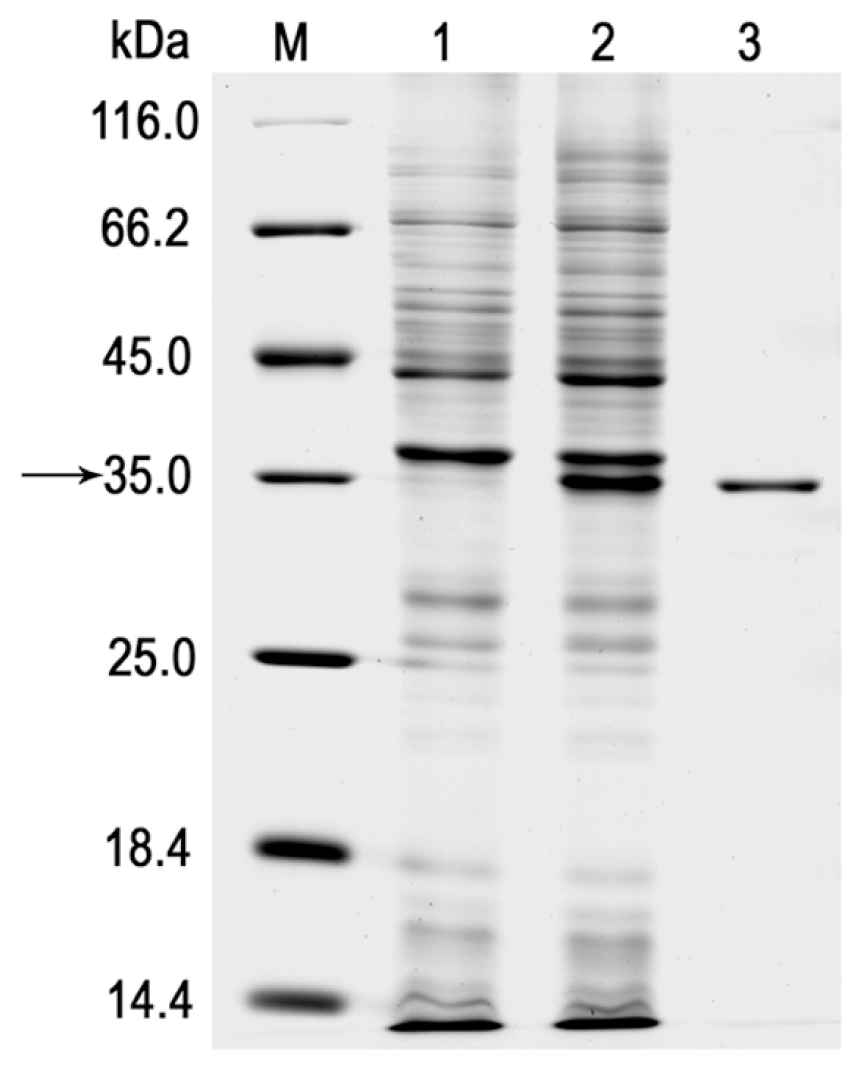
2.2. Expression and Purification of the r-LipA
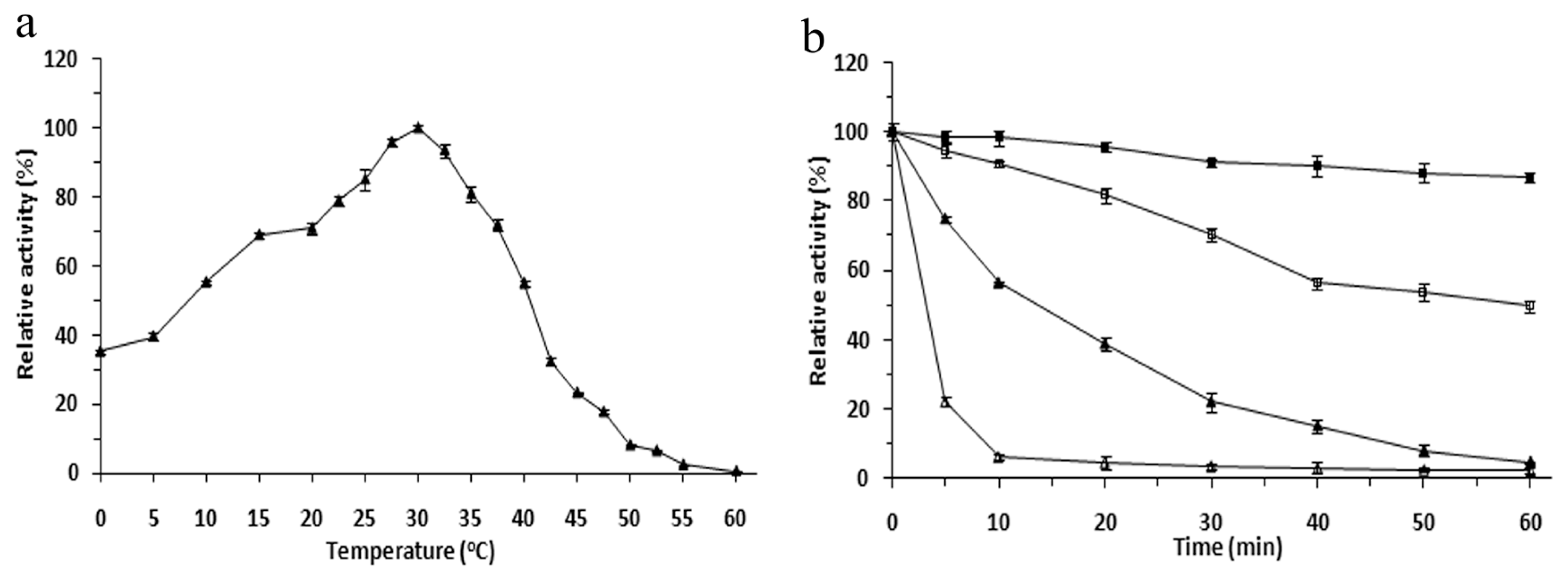
2.3. Effects of Temperature on the r-LipA Avtivity and Stability
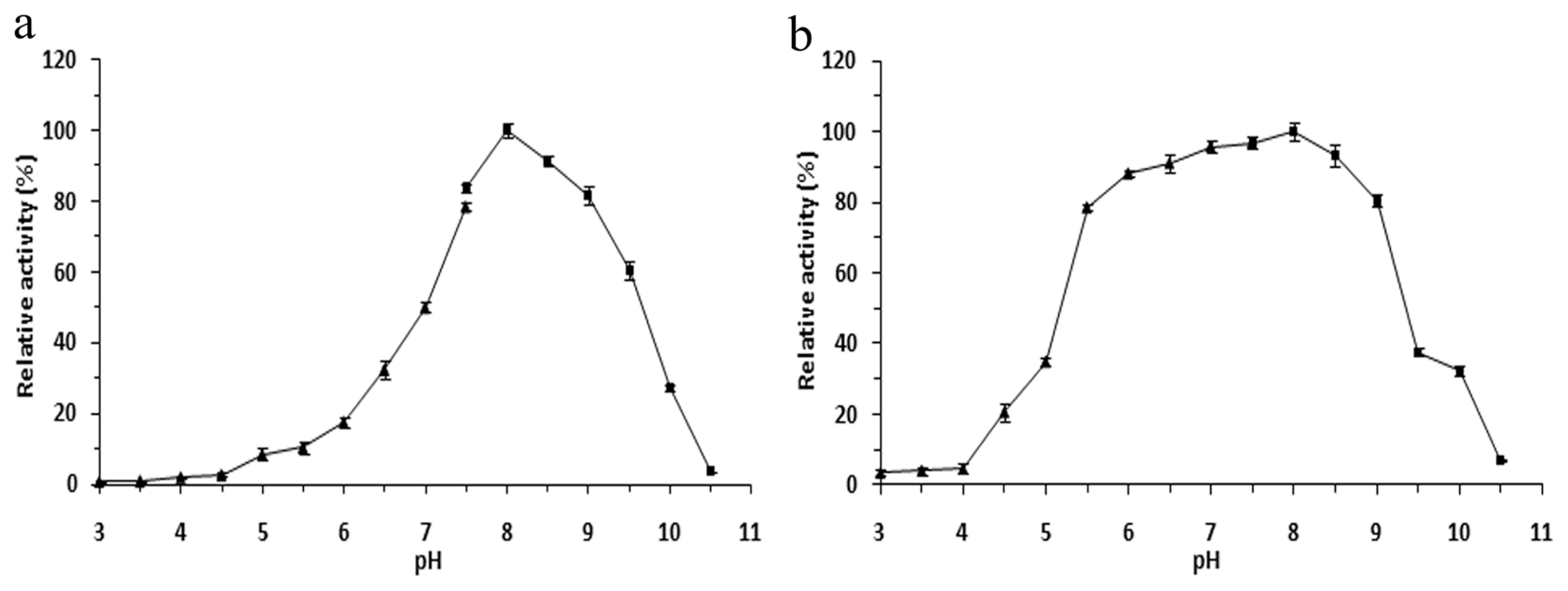
2.4. Effects of pH on the r-LipA Avtivity and Stability
2.5. Effects of Metal Ions, Detergents and Organic Solvents on the r-LipA Activity
2.6. Substrate Specificity of the r-LipA
3. Experimental Section
3.1. Strains, Plasmids and Chemicals
3.2. Cloning and Sequence Analysis
3.3. Construction of Recombinant Plasmid
3.4. Expression and Purification of the r-LipA
3.5. Lipase Activity Assay
3.6. Effects of Temperature on the r-LipA Avtivity and Stability
3.7. Effects of pH on the r-LipA Avtivity and Stability
3.8. Effect of Metal Ions, Detergents and Organic Solvents on the r-LipA Avtivity
3.9. Substrate Specificity of the r-LipA
4. Conclusions
Acknowledgments
References
- Joseph, B; Ramteke, PW; Thomas, G. Cold active microbial lipases: some hot issues and recent developments. Biotechnol. Adv 2008, 26, 457–470. [Google Scholar]
- Pleiss, J; Fischer, M; Peiker, M; Thiele, C; Schmid, RD. Lipase engineering database: understanding and exploiting sequence-structure-function relationships. J. Mol. Catal. B Enzym 2000, 10, 491–508. [Google Scholar]
- Arpigny, JL; Jaeger, KE. Bacterial lipolytic enzymes: classification and properties. J. Biochem 1999, 343, 177–183. [Google Scholar]
- Ruiz, C; Pastor, FRJ; Diaz, P. Isolation and characterization of Bacillus sp. BP-6 LipA, a ubiquitous lipase among mesophilic Bacillus species. Lett. Appl. Microbiol 2003, 37, 354–359. [Google Scholar]
- Fan, ZX; Yue, CW; Tang, Y; Zhang, YZ. Cloning, sequence analysis and expression of bacterial lipase-coding DNA fragments from environment in Escherichia coli. Mol. Biol. Rep 2009, 36, 1515–1519. [Google Scholar]
- Jaeger, KE; Dijkstra, BW; Reetz, MT. Bacterial biocatalysis: Molecular biology, three-dimensional structures, and biotechnological applications of lipases. Annu. Rev. Microbiol 1999, 53, 315–351. [Google Scholar]
- Pandey, A; Benjamin, S; Soccol, CR; Nigam, P; Krieger, N; Soccol, VT. The realm of microbial lipases in biotechnology. Biotechnol. Appl. Biochem 1999, 29, 119–131. [Google Scholar]
- Reetz, MT. Lipases as practical biocatalysts. Curr. Opin. Chem. Biol 2002, 6, 145–150. [Google Scholar]
- Schmid, RD; Verger, R. Lipases: interfacial enzymes with attractive applications. Angew. Chem. Int. Ed 1998, 37, 1608–1633. [Google Scholar]
- Choo, DW; Kurihara, T; Suzuki, T; Soda, K; Esaki, N. A cold-adapted lipase of an Alaskan psychrotroph, Pseudomonas sp. strain B11-1: Gene cloning and enzyme purification and characterization. Appl. Environ. Microbiol 1998, 64, 486–491. [Google Scholar]
- Park, IH; Kim, SH; Lee, YS; Lee, SC; Zhou, Y; Kim, CM; Ahn, SC; Choi, YL. Gene cloning, purification, and characterization of a cold-adapted lipase produced by Acinetobacter baumannii BD5. J. Microbiol. Biotechnol 2009, 19, 128–135. [Google Scholar]
- Suzuki, T; Nakayama, T; Kurihara, T; Nishino, T; Esaki, N. Cold-active lipolytic activity of psychrotrophic Acinetobacter sp. strain No. 6. J. Biosci. Bioeng 2001, 92, 144–148. [Google Scholar]
- Zheng, XM; Chu, XY; Zhang, W; Wu, NF; Fan, YL. A novel cold-adapted lipase from Acinetobacter sp. XMZ-26: Gene cloning and characterisation. Appl. Microbiol. Biotechnol 2011, 90, 971–980. [Google Scholar]
- Ryu, HS; Kim, HK; Choi, WC; Kim, MH; Park, SY; Han, NS; Oh, TK; Lee, JK. New cold-adapted lipase from Photobacterium lipolyticum sp. nov. that is closely related to filamentous fungal lipases. Appl. Microbiol. Biotechnol 2006, 70, 321–326. [Google Scholar]
- Reichenbach, H; Höfle, G. Myxobacteria as Producers of Secondary Metabolites. In Drug Discovery from Nature; Grabley, S, Thiericke, R, Eds.; Spinger: Berlin, Germany, 1999; Volume Chapter 9, pp. 149–179. [Google Scholar]
- Shimkets, LJ. Social and developmental biology of the myxobacteria. Microbiol. Rev 1990, 54, 473–501. [Google Scholar]
- Gerth, K; Pradella, S; Perlova, O; Beyer, S; Müller, R. Myxobacteria: proficient producers of novel natural products with various biological activities-past and future biotechnological aspects with the focus on the genus Sorangium. J. Biotechnol 2003, 106, 233–253. [Google Scholar]
- Schneiker, S; Perlova, O; Kaiser, O; Gerth, K; Alici, A; Altmeyer, MO; Bartels, D; Bekel, T; Beyer, S; Bode, E; et al. Complete genome sequence of the myxobacterium Sorangium cellulosum. Nat. Biotechnol 2007, 25, 1281–1289. [Google Scholar]
- SignalP Server. SignalP 3.0; Center for Biological Sequence Analysis (CBS): Lyngby, Denmark, 2011. Available online: http://www.cbs.dtu.dk/services/SignalP/ accessed on 11 July 2011.
- Bertrand, T; Augé, F; Houtmann, J; Rak, A; Vallée, F; Mikol, V; Berne, PF; Michot, N; Cheuret, D; Hoornaert, C; Mathieu, M. Structural basis for human monoglyceride lipase inhibition. J. Mol. Biol 2010, 396, 663–673. [Google Scholar]
- Finn, RD; Tate, J; Mistry, J; Coggill, PC; Sammut, SJ; Hotz, HR; Ceric, G; Forslund, K; Eddy, SR; Sonnhammer, ELL; et al. The Pfam protein families database. Nucleic Acids Res 2008, 36, D281–D288. [Google Scholar]
- Olchowy, J; Kur, K; Sachadyn, P; Milewski, S. Construction, purification, and functional characterization of His-tagged Candida albicans glucosamine-6-phosphate synthase expressed in Escherichia coli. Protein Expr. Purif 2006, 46, 309–315. [Google Scholar]
- Schlieben, NH; Niefind, K; Schomburg, D. Expression, purification, and aggregation studies of His-tagged thermoalkalophilic lipase from Bacillus thermocatenulatus. Protein Expr. Purif 2004, 34, 103–110. [Google Scholar]
- Panteghini, M; Bonora, R; Pagani, F. Measurement of pancreatic lipase activity in serum by a kinetic colorimetric assay using a new chromogenic substrate. Ann. Clin. Biochem 2001, 38, 365–370. [Google Scholar]
- ClustalW2-Multiple Sequence Alignment; European Bioinformatic Institute (EBI): Cambridge, UK, 2011. Available online: http://www.ebi.ac.uk/Tools/msa/clustalw2/ accessed on 11 July 2011.
- Feller, G; Narinx, E; Arpigny, JL; Aittaleb, M; Baise, E; Geniot, S; Gerday, C. Enzymes from Psychrophilic organisms. FEMS Microbiol 1996, 18, 189–202. [Google Scholar]
- Zhang, JW; Lin, S; Zeng, R. Cloning, expression, and characterization of a cold-adapted lipase gene from an antarctic deep-sea psychrotrophic bacterium, Psychrobacter sp. 7195. J. Microbiol. Biotechnol 2007, 17, 604–610. [Google Scholar]
- Jeon, JH; Kim, JT; Kim, YJ; Kim, HK; Lee, HS; Kang, SG; Kim, SJ; Lee, JH. Cloning and characterization of a new cold-active lipase from a deep-sea sediment metagenome. Appl. Microbiol. Biotechnol 2009, 81, 865–874. [Google Scholar]
- Zhang, JW; Zeng, RY. Molecular cloning and expression of a cold-adapted lipase gene from an antarctic deep sea psychrotrophic bacterium Pseudomonas sp. 7323. Mar. Biotechnol 2008, 10, 612–621. [Google Scholar]
- Shimkets, LJ; Dworkin, M; Reichenbach, H. The Myxobacteria. In The Prokaryotes; Dworkin, M, Falkow, S, Rosenberg, E, Schleifer, K-H, Stackebrandt, E, Eds.; Springer: Berlin, Germany, 2006; pp. 31–115. [Google Scholar]
- Joseph, B; Ramteke, PW; Thomas, G; Shrivastava, N. Standard review cold-active microbial lipases: a versatile tool for industrial applications. Biotechnol. Mol. Biol. Rev 2007, 2, 39–48. [Google Scholar]
- Laane, C; Boeren, S; Vos, K; Veeder, C. Rules for optimization of biocatalysis in organic solvents. Biotechnol. Bioeng 1987, 30, 81–87. [Google Scholar]
- Gupta, MN. Enzyme function in organic solvents. Eur. J. Biochem 1992, 203, 25–32. [Google Scholar]
- Zaks, A; Klibanov, AM. Enzyme-catalyzed processes in organic solvents. Proc. Natl. Acad. Sci. USA 1985, 82, 3192–3196. [Google Scholar]
- Inoue, A; Yamamoto, M; Horikoshi, K. Pseudomonas putida which can grow in the presence of toluene. Appl. Environ. Microbiol 1991, 57, 1560–1562. [Google Scholar]
- Cai, YJ; Wang, L; Liao, XR; Ding, YR; Sun, J. Purification and partial characterization of two new cold-adapted lipases from mesophilic Geotrichum sp. SYBC WU-3. Process Biochem 2009, 44, 786–790. [Google Scholar]
- Lan, DM; Yang, N; Wang, WK; Shen, YF; Yang, B; Wang, YH. A Novel cold-active lipase from Candida albicans: cloning, expression and characterization of the recombinant enzyme. Int. J. Mol. Sci 2011, 12, 3950–3965. [Google Scholar]
- Zhao, L; Li, PF; Lu, CH; Li, SG; Shen, YM; Li, YZ. Glycosylation and production characteristics of epothilones in alkali-tolerant Sorangium cellulosum strain So0157-2. J. Microbiol 2010, 48, 438–444. [Google Scholar]
- Sambrook, J; Russel, DW. Molecular Cloning: A Laboratory Manual, 3rd ed; Cold Spring Harbor Laboratory: New York, NY, USA, 2001. [Google Scholar]
- NCBI/BLAST-Basic Logic Alignment Search Tool; Natinal Center for Biotechnology Information (NCBI): Bethesda, MD, USA, 2011. Available online: http://blast.ncbi.nlm.nih.gov/Blast.cgi accessed on 11 July 2011.
- Zandonella, G; Haalck, L; Spener, F; Faber, K; Paltauf, F; Hermetter, A. Enantiomeric perylene-glycerolipids as fluorogenic substrates for a dual wavelength assay of lipase activity and stereoselectivity. Chirality 1996, 8, 481–489. [Google Scholar]
| Metal ions | Relative remaining activity (%) of r-LipA
| |
|---|---|---|
| 1 mM | 10 mM | |
| Control | 100.0 ± 3.0 | 100.0 ± 1.6 |
| Li+ | 96.5 ± 1.2 | 93.3 ± 1.5 |
| Na+ | 97.1 ± 3.8 | 105.9 ± 5.5 |
| K+ | 94.3 ± 0.6 | 96.0 ± 4.7 |
| Mg2+ | 105.1 ± 3.8 | 126.1 ± 2.5 |
| Ca2+ | 112.6 ± 1.5 | 140.2 ± 0.3 |
| Fe2+ | 140.8 ± 2.4 | 77.1 ± 2.4 |
| Mn2+ | 120.1 ± 1.9 | 103.4 ± 0.9 |
| Cr3+ | 110.2 ± 3.0 | 59.5 ± 1.9 |
| Co2+ | 104.1 ± 1.8 | 99.2 ± 1.0 |
| Ni2+ | 97.6 ± 1.9 | 63.3 ± 2.6 |
| Cu2+ | 57.8 ± 1.5 | 4.0 ± 0.8 |
| Zn2+ | 56.8 ± 0.9 | 42.9 ± 4.8 |
| Hg2+ | 88.4 ± 2.2 | 40.1 ± 1.5 |
| EDTA | 109.8 ± 1.5 | 99.8 ± 1.4 |
| Detergents | Relative remaining activity (%) of r-LipA
| |
|---|---|---|
| 0.1% | 1% | |
| Control | 100.0 ± 2.9 | 100.0 ± 2.6 |
| SDS | 10.2 ± 0.8 | 9.0 ± 0.7 |
| CTAB | 203.7 ± 3.1 | 95.4 ± 1.4 |
| Tween 20 | 139.2 ± 0.8 | 113.0 ± 5.5 |
| Tween 80 | 132.4 ± 2.2 | 119.4 ± 2.2 |
| Triton X-100 | 146.8 ± 0.4 | 196.3 ± 2.1 |
| Organic solvents | log P | Relative remaining activity (%) of r-LipA
| |
|---|---|---|---|
| 30 min incubation | 2 h incubation | ||
| Control | - | 100.0 ± 1.6 | 100.0 ± 2.1 |
| Dimethyl sulfoxide | −1.22 | 97.1 ± 0.3 | 88.1 ± 2.2 |
| Methanol | −0.76 | 97.7 ± 2.5 | 90.2 ± 1.4 |
| Ethanol | −0.24 | 75.4 ± 3.1 | 67.6 ± 1.1 |
| Acetone | −0.23 | 78.8 ± 1.3 | 69.2 ± 2.7 |
| Isopropanol | 0.14 | 57.2 ± 3.1 | 46.6 ± 2.8 |
| n-Propanol | 0.28 | 76.3 ± 1.7 | 50.2 ± 0.7 |
| n-Butanol | 0.8 | 99.2 ± 4.5 | 82.6 ± 1.6 |
| Diethylether | 0.85 | 113.7 ± 2.0 | 118.5 ± 1.8 |
| Chloroform | 2 | 139.0 ± 1.1 | 142.8 ± 3.5 |
| Benzene | 2 | 140.5 ± 5.5 | 160.2 ± 3.3 |
| Toluene | 2.5 | 150.9 ± 0.6 | 159.2 ± 0.9 |
| p-Xylene | 3.1 | 153.7 ± 5.2 | 152.9 ± 2.5 |
| Cyclohexane | 3.2 | 156.0 ± 3.7 | 136.9 ± 0.8 |
| n-Hexane | 3.5 | 153.0±2.8 | 130.8 ± 3.5 |
| n-Heptane | 4 | 159.1 ± 3.4 | 167.1 ± 1.2 |
| Isooctane | 4.7 | 163.7 ± 0.5 | 132.8 ± 0.7 |
| Substrates | Relative remaining activity (%)
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| pH 7.0, 20 °C | pH 7.0, 30 °C | pH 7.0, 40 °C | pH 8.0, 20 °C | pH 8.0, 30 °C | pH 8.0, 40 °C | pH 9.0, 20 °C | pH 9.0, 30 °C | pH 9.0, 40 °C | |
| pNP acetate | 16.6 ± 0.3 | 32.6 ± 1.5 | 43.2 ± 4.5 | 71.6 ± 2.2 | 100.0 ± 2.6 | 60.5 ± 1.6 | 72.5 ± 1.4 | 86.1 ± 1.6 | 60.8 ± 0.8 |
| pNP butyrate | 7.9 ± 3.0 | 18.3 ± 2.3 | 26.2 ± 3.9 | 38.4 ± 1.3 | 63.4 ± 2.3 | 53.3 ± 2.1 | 54.1 ± 3.6 | 83.3 ± 1.0 | 64.6 ± 1.4 |
| pNP octanoate | 8.1 ± 1.2 | 12.8 ± 1.8 | 15.7 ± 2.1 | 33.3 ± 0.5 | 52.5 ± 0.3 | 52.1 ± 2.9 | 41.5 ± 2.5 | 68.4 ± 2.3 | 74.2 ± 2.6 |
| pNP decanoate | 1.4 ± 0.2 | 4.9 ± 3.0 | 5.9 ± 0.5 | 22.2 ± 0.7 | 32.1 ± 0.9 | 24.7 ± 3.0 | 24.0 ± 0.2 | 40.4 ± 2.0 | 44.7 ± 2.8 |
| pNP dodecanoate | 1.6 ± 0.3 | 2.5 ± 0.1 | 5.8 ± 1.4 | 13.2 ± 5.2 | 17.0 ± 1.0 | 20.5 ± 1.8 | 23.3 ± 1.8 | 43.9 ± 1.5 | 40.0 ± 1.0 |
| pNP myristate | 1.6 ± 1.0 | 2.2 ± 1.4 | 5.7 ± 2.9 | 5.7 ± 1.3 | 3.9 ± 2.4 | 5.6 ± 2.2 | 5.4 ± 2.7 | 5.2 ± 3.0 | 4.8 ± 3.4 |
| pNP palmitate | 1.6 ± 0.3 | 1.9 ± 1.6 | 4.1 ± 0.6 | 2.0 ± 1.2 | 2.3 ± 0.8 | 1.3 ± 0.8 | 2.6 ± 2.3 | 5.2 ± 0.1 | 1.3 ± 1.3 |
| pNP stearate | 1.3 ± 1.0 | 1.0 ± 0.1 | 2.5 ± 0.6 | 3.4 ± 0.1 | 1.9 ± 1.5 | 0.7 ± 0.8 | 2.5 ± 0.1 | 5.3 ± 2.5 | 1.9 ± 0.4 |
| Substrates | Temperature (°C) | Km (mM) | kcat (s−1) | kcat/Km (s−1·mM−1) |
|---|---|---|---|---|
| pNP acetate (C2) | 4 | 0.037 ± 0.001 | 7.008 ± 0.100 | 186.643 ± 3.201 |
| pNP acetate (C2) | 30 | 0.174 ± 0.006 | 29.225 ± 0.977 | 168.114 ± 2.097 |
| pNP butyrate (C4) | 30 | 0.366 ± 0.211 | 13.617 ± 0.735 | 37.173 ± 0.128 |
| pNP octanoate (C8) | 30 | 0.704 ± 0.026 | 3.294 ± 0.114 | 4.677 ± 0.013 |
| pNP decanoate (C10) | 30 | 1.664 ± 0.047 | 1.565 ± 0.036 | 0.940 ± 0.005 |
| pNP dodecanoate (C12) | 30 | 1.946 ± 0.059 | 1.205 ± 0.032 | 0.619 ± 0.002 |
© 2011 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Cheng, Y.-Y.; Qian, Y.-K.; Li, Z.-F.; Wu, Z.-H.; Liu, H.; Li, Y.-Z. A Novel Cold-Adapted Lipase from Sorangium cellulosum Strain So0157-2: Gene Cloning, Expression, and Enzymatic Characterization. Int. J. Mol. Sci. 2011, 12, 6765-6780. https://doi.org/10.3390/ijms12106765
Cheng Y-Y, Qian Y-K, Li Z-F, Wu Z-H, Liu H, Li Y-Z. A Novel Cold-Adapted Lipase from Sorangium cellulosum Strain So0157-2: Gene Cloning, Expression, and Enzymatic Characterization. International Journal of Molecular Sciences. 2011; 12(10):6765-6780. https://doi.org/10.3390/ijms12106765
Chicago/Turabian StyleCheng, Yuan-Yuan, Yun-Kai Qian, Zhi-Feng Li, Zhi-Hong Wu, Hong Liu, and Yue-Zhong Li. 2011. "A Novel Cold-Adapted Lipase from Sorangium cellulosum Strain So0157-2: Gene Cloning, Expression, and Enzymatic Characterization" International Journal of Molecular Sciences 12, no. 10: 6765-6780. https://doi.org/10.3390/ijms12106765
APA StyleCheng, Y.-Y., Qian, Y.-K., Li, Z.-F., Wu, Z.-H., Liu, H., & Li, Y.-Z. (2011). A Novel Cold-Adapted Lipase from Sorangium cellulosum Strain So0157-2: Gene Cloning, Expression, and Enzymatic Characterization. International Journal of Molecular Sciences, 12(10), 6765-6780. https://doi.org/10.3390/ijms12106765




