Synthesis and Antifungal Activity of 1,2,4-Oxadiazole Derivatives
Abstract
1. Introduction
2. Results
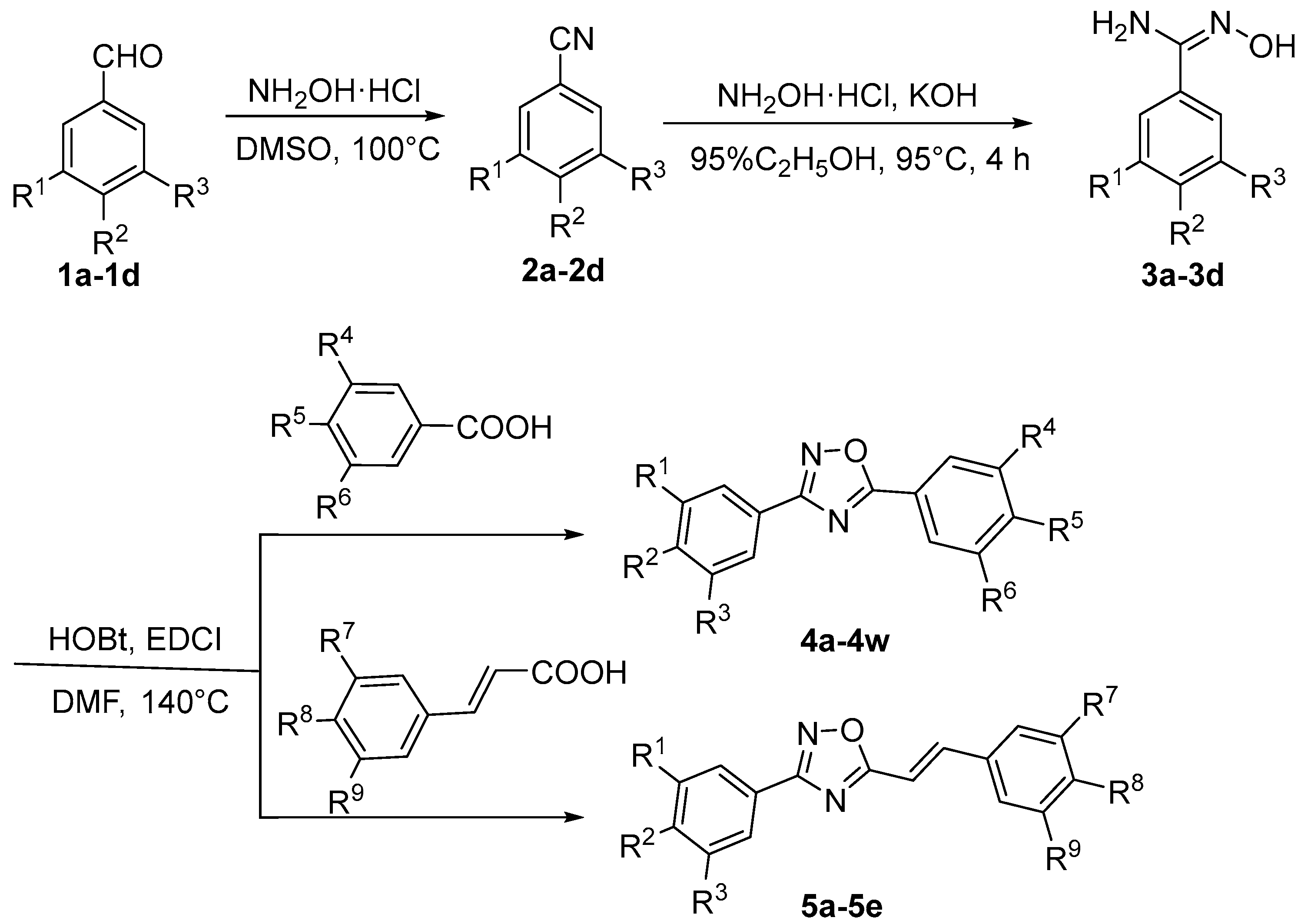
2.1. Synthesis of 1,2,4-Oxadiazole
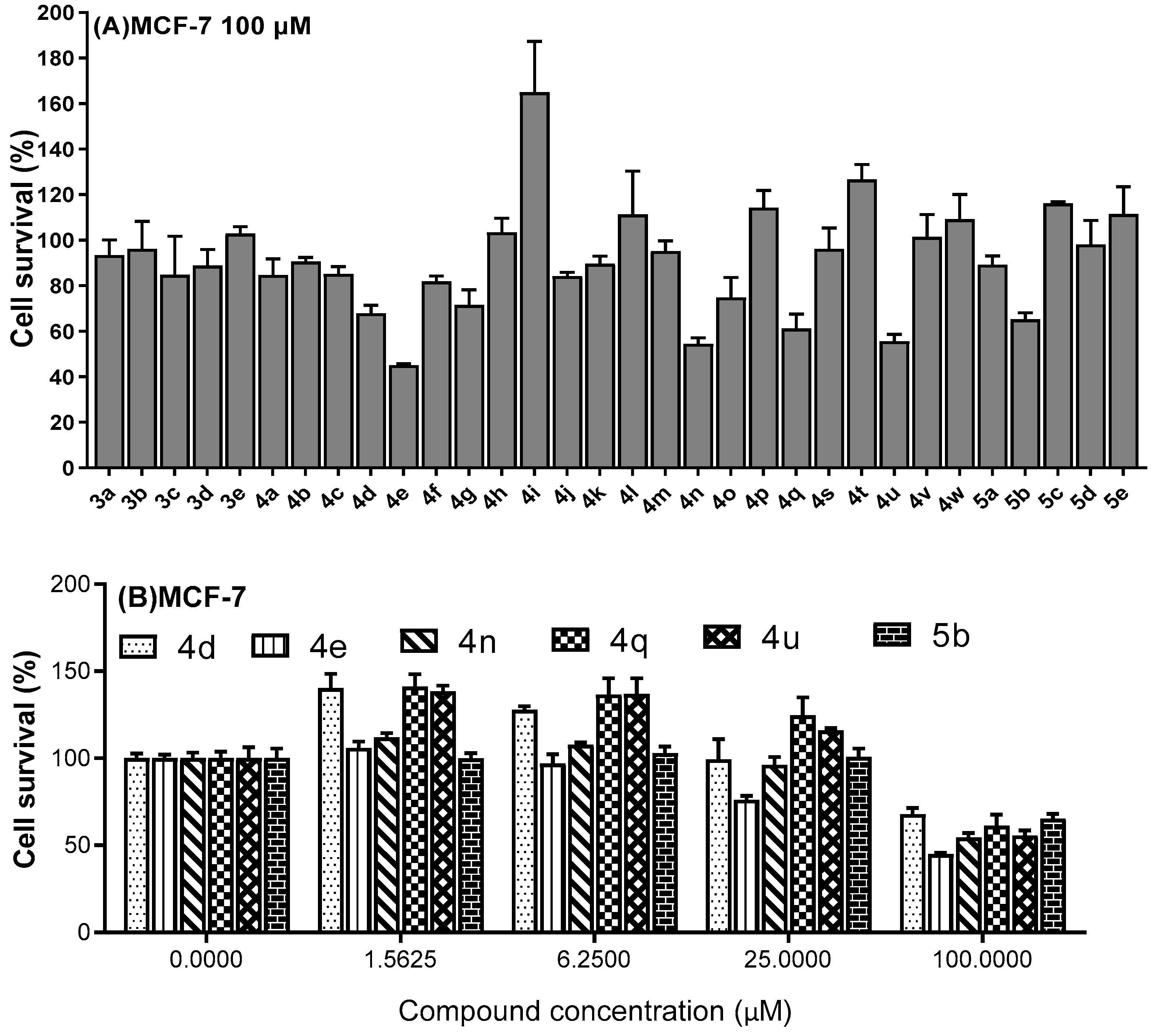
2.2. Cytotoxicity Evaluation
2.3. In Vitro Fungicidal Activities and Structure–Activity Relationship
2.4. EC50 of Compounds Against Plant-Pathogenic Fungi
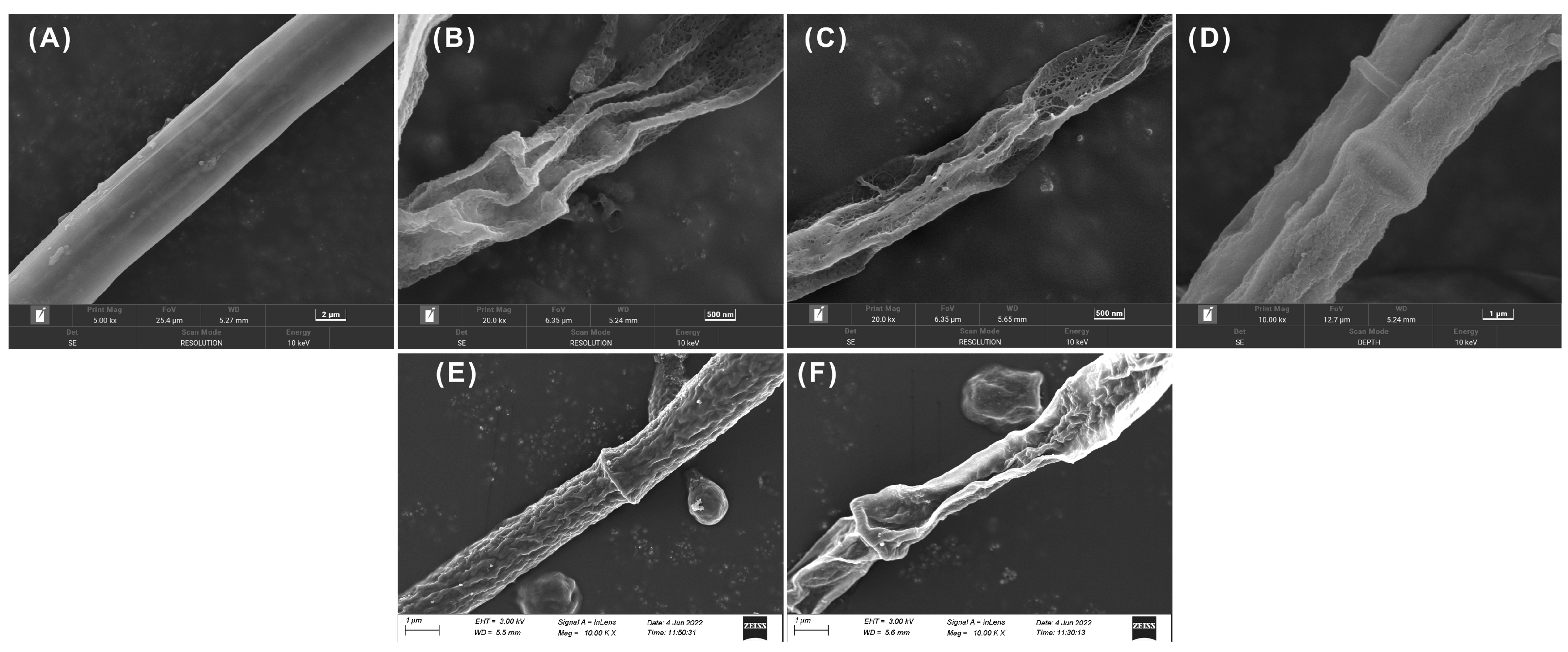
2.5. Effect of Compounds on Plant Pathogen Morphology
2.6. The Effect of Antifungal Activity In Vivo
2.7. Analysis of Molecular Docking
3. Experimental Section
3.1. Chemistry
3.1.1. Synthesis of Benzonitrile
3.1.2. Synthesis of (E)-N′-Hydroxybenzimidamide
- (E)-N′-Hydroxybenzimidamide (3a)
- Prepared from 2a (1.03 g, 10 mmol). Purification procedure: column chromatography (eluent: ethyl acetate in petroleum ether 65%). White solid, yield 85%. m.p.: 67.3–69.3 °C. 1H NMR (400 MHz, DMSO-d6) δ 9.64 (s, 1H, –OH), 7.70–7.65 (m, 2H, Ar-H), 7.39–7.36 (m, 3H, Ar-H), 5.82–5.77 (d, 2H, –NH2). Calculate m/z for C7H8N2O: 136.15, found: 136.09.
- (E)-N′-Hydroxy-4-methylbenzimidamide (3b)
- Prepared from 2b (1.51 g, 10 mmol). Purification procedure: column chromatography (eluent: ethyl acetate in petroleum ether 65%).White solid, yield 87%. m.p.: 143.5–145.1 °C; 1H NMR (400 MHz, DMSO-d6) δ 9.54 (s, 1H, –OH), 7.57–7.55 (d, 2H, Ar-H), 7.19–7.17(d, 2H, Ar-H), 5.76 (s, 2H, –NH2), 2.31(s, 3H, –CH3). Calculate m/z for C8H10N2O: 150.18, found: 150.09.
- (E)-N′-Hydroxy-4-methoxybenzimidamide (3c)
- Prepared from 2c (1.36 g, 10 mmol). Purification procedure: column chromatography (eluent: ethyl acetate in petroleum ether 60%). White solid, yield 82%. m.p.: 119.5–121.2 °C. 1H NMR (400 MHz, DMSO-d6) δ 9.47 (s, 1H, –OH), 7.62–7.60 (d, 2H, Ar-H), 6.94–6.92 (d, 2H, Ar-H), 5.74 (s, 2H, –NH2), 3.77 (s, 3H, –OCH3). Calculate m/z for C8H10N2O2: 166.18, found: 166.09.
- (E)-N′-Hydroxy-3,4-dimethoxybenzimidamide (3d)
- Prepared from 2d (1.63 g, 10 mmol). Purification procedure: column chromatography (eluent: ethyl acetate in petroleum ether 60%). White solid, yield 88%. m.p.: 163.2–164.5 °C. 1H NMR (400 MHz, DMSO-d6) δ 9.47 (s, 1H, –OH), 7.25–7.22 (t, 2H, Ar-H), 6.95–6.93 (d, 1H, Ar-H), 5.765.74(s, 2H, –NH2), 3.77(s, 6H, –OCH3). Calculate m/z for C9H12N2O3: 196.20, found: 196.01.
- (E)-N′-Hydroxy-3,4,5-trimethoxybenzimidamide (3e)
- Prepared from 2e (1.93g, 10 mmol). Purification procedure: column chromatography (eluent: ethyl acetate in petroleum ether 60%). White solid, yield 81%. m.p.: 168.2–169.1 °C. 1H NMR (400 MHz, DMSO-d6) δ 9.57 (s, 1H, –OH), 6.99 (s, 2H, Ar-H), 5.84 (s, 2H, –NH2), 3.79 (s, 6H, –OCH3), 3.67(s, 3H, –OCH3). Calculate m/z for C10H14N2O4: 226.23, found: 226.09.
3.1.3. Synthesis of 1,2,4-Oxadiazoles
- 5-(4-Chlorophenyl)-3-phenyl-1,2,4-oxadiazole (4a)
- Prepared from 2a (0.20 g, 1.47 mmol), 4-chlorobenzoic acid (0.18 g, 1.15 mmol), EDCI (0.25 g, 1.30 mmol), HOBt (0.17 g, 1.26 mmol). Purification procedure: column chromatography (eluent: ethyl acetate in petroleum ether 14.3% with 1% TEA). White solid, yield 65%. m.p.: 120.3–123.4 °C. 1H NMR (400 MHz, CDCl3) δ 8.20–8.18 (m, 4H, Ar-H), 7.58–7.52 (m, 5H, Ar-H). 13C NMR (101 MHz, CDCl3) δ 174.78, 169.03, 139.19, 131.33, 129.53, 129.46, 129.43, 128.90, 127.52, 127.47, 126.73, 122.73. Calculate m/z for C14H9ClN2O: 256.04, found: 256.09.
- 5-(3,4-Dimethoxyphenyl)-3-phenyl-1,2,4-oxadiazole (4b)
- Prepared from 2a (0.25 g, 1.84 mmol), 3,4-dimethoxybenzoic acid (0.16 g, 0.88 mmol), EDCI (0.34 g, 1.77 mmol), HOBt (0.24 g, 1.78 mmol). Purification procedure: column chromatography (eluent: ethyl acetate in petroleum ether 50% with 1% TEA). Light yellow solid, yield 60%. m.p.: 104.9–106.7 °C. 1H NMR (400 MHz, CDCl3) δ 8.21–8.17 (m, 2H, Ar-H), 7.88–7.86 (q, 1H, Ar-H), 7.72 (d, 1H, Ar-H) 7.56–7.46 (m, 3H, Ar-H), 7.04–7.02 (d, 1H, Ar-H), 4.04–4.01 (d, 6H, –OCH3) 13C NMR (101 MHz, CDCl3) δ 175.60, 168.85, 152.79, 149.23, 131.13, 128.84, 128.82, 128.62, 127.51, 127.17, 127.06, 122.07, 116.85, 111.05, 110.39, 56.17, 56.11. Calculate m/z for C16H14N2O3: 282.10, found: 282.19.
- 5-(4-(tert-Butyl)phenyl)-3-phenyl-1,2,4-oxadiazole (4c)
- Prepared from 2a (0.50 g, 3.67 mmol), 4-(tert-butyl) benzoic acid (0.49 g, 2.75 mmol), EDCI (0.64 g, 3.34 mmol), HOBt (0.45 g, 3.33 mmol). Purification procedure: column chromatography (eluent: ethyl acetate in petroleum ether 14.3% with 1% TEA). Light yellow solid, yield 45%. m.p.: 50.5–51.8 °C. 1H NMR (400 MHz, CDCl3) δ 8.22–8.14 (m, 4H, Ar-H), 7.61–7.51 (m, 5H, Ar-H), 1.40–1.37 (d, 9H, –C(CH3)3). 13C NMR (101 MHz, CDCl3) δ 175.79, 168.90, 156.49, 131.13, 128.84, 128.03, 127.54, 127.09, 126.11, 121.52, 35.23, 31.12. Calculate m/z for C18H18N2O: 278.14, found: 278.09.
- 3-(3-Phenyl-1,2,4-oxadiazol-5-yl) phenol (4d)
- Prepared from 2a (0.50 g, 3.67 mmol), 3-hydroxybenzoic acid (0.43 g, 3.12 mmol), EDCI (0.70 g, 3.65 mmol), HOBt (0.50 g, 3.70 mmol). Purification procedure: column chromatography (eluent: ethyl acetate in petroleum ether 25%). White solid, yield 78%. m.p.: 184.3–186.5°C. 1H NMR (400 MHz, DMSO-d6) δ 10.13(s, 1H, –OH), 8.12–8.09 (m, 2H, Ar-H), 7.64–7.57 (m, 5H, Ar-H), 7.50–7.46 (t, 1H, Ar-H), 7.14–7.11 (q, 1H, Ar-H). 13C NMR (101 MHz, DMSO-d6) δ 176.01, 168.65, 158.49, 132.10, 131.31, 129.73, 128.65, 127.91, 127.54, 126.64, 124.81, 120.95, 119.04, 114.57. Calculate m/z for C14H10N2O2: 238.07, found: 237.99.
- 5-(3-Phenyl-1,2,4-oxadiazol-5-yl) benzene-1,3-diol (4e)
- Prepared from 2a (0.20 g, 1.47 mmol), 3,5-dihydroxybenzoic acid (0.19 g, 1.23 mmol), EDCI (0.28 g, 1.46 mmol), HOBt (0.20 g, 1.47 mmol). Purification procedure: column chromatography (eluent: ethyl acetate in petroleum ether 25%). Light yellow solid, yield 33%. m.p.: 265.0–267.2 °C. 1H NMR (400 MHz, DMSO-d6) δ 9.96 (s, 2H, –OH), 8.09–8.07 (m, 2H, Ar-H), 7.64–7.58 (m, 3H, Ar-H), 7.05–7.04 (d, 2H, Ar-H), 6.54–6.53 (t, 1H, Ar-H). 13C NMR (101 MHz, DMSO-d6) δ 176.07, 168.60, 159.71, 132.09, 129.73, 128.65, 127.90, 127.53, 126.67, 125.06, 107.75, 106.18. Calculate m/z for C14H10N2O3: 254.07, found: 254.11.
- 4-(3-Phenyl-1,2,4-oxadiazol-5-yl) benzene-1,2-diol (4f)
- Prepared from 2a (0.24 g, 1.76 mmol), 3,4-dihydroxybenzoic acid (0.19 g, 1.25 mmol), EDCI (0.28 g, 1.46 mmol), HOBt (0.20 g, 1.48 mmol). Purification procedure: column chromatography (eluent: ethyl acetate in petroleum ether 50%). Light yellow solid, yield 42%. m.p.: 237.5–239.2 °C. 1H NMR (400 MHz, DMSO-d6) δ 9.89 (s, 3H, –OH), 8.09–8.06 (m, 2H, Ar-H), 7.62–7.52 (m, 5H, Ar-H), 6.98–6.96 (d, 1H, Ar-H). 13C NMR (101 MHz, DMSO-d6) δ 176.12, 168.43, 151.25, 146.44, 131.92, 129.66, 127.48, 126.92, 120.98, 116.77, 115.08, 114.63. Calculate m/z for C14H10N2O3: 254.07, found: 253.99.
- 5-(3-Phenyl-1,2,4-oxadiazol-5-yl) benzene-1,2,3-triol (4g)
- Prepared from 2a (0.40 g, 2.94 mmol), 3,4,5-trihydroxybenzoic acid (0.21 g, 1.24 mmol), EDCI (0.28 g, 1.46 mmol), HOBt (0.20 g, 1.48 mmol). Purification procedure: column chromatography (eluent: 11% Methanol, 44% CH2Cl2 in petroleum ether with 2% HAc). Grey solid, yield 37%. m.p.: 259.3–262.2 °C. 1H NMR (400 MHz, DMSO-d6) δ 9.50 (s, 3H, –OH), 8.07–8.05 (m, 2H, Ar-H), 7.61–7.59 (d, 3H, Ar-H), 7.15 (s, 2H, Ar-H) 13C NMR (101 MHz, DMSO-d6) δ 176.30, 168.42, 146.94, 139.20, 131.94, 129.69, 127.47, 126.93, 113.42, 107.46. Calculate m/z for C14H10N2O4: 270.06, found: 269.99.
- 3-Phenyl-5-(4-(trifluoromethyl)phenyl)-1,2,4-oxadiazole (4h)
- Prepared from 2a (0.22 g, 1.62 mmol), 4-(trifluoromethyl) benzoic acid (0.26 g, 1.37 mmol), EDCI (0.35 g, 1.83 mmol), HOBt (0.25 g, 1.85 mmol). Purification procedure: column chromatography (eluent: ethyl acetate in petroleum ether 50%). White solid, yield 55%. m.p.: 101.1–101.8 °C. 1H NMR (400 MHz, CDCl3) δ 8.39–8.37 (d, 2H, Ar-H), 8.22–8.19 (q, 2H, Ar-H), 7.87–7.85 (d, 2H, Ar-H), 7.57–7.55 (m, 3H, Ar-H). 13C NMR (101 MHz, DMSO-d6) δ 174.69, 168.89, 133.27, 132.95, 132.30, 129.79, 129.33, 127.59, 127.49, 126.98, 126.94, 126.32. Calculate m/z for C15H9F3N2O: 290.07, found: 291.07.
- 5-(4-Nitrophenyl)-3-phenyl-1,2,4-oxadiazole (4i)
- Prepared from 2a (0.23 g, 1.70 mmol), 4-nitrobenzoic acid (0.21 g, 1.26 mmol), EDCI (0.28 g, 1.46 mmol), HOBt (0.21 g, 1.55 mmol). Purification procedure: column chromatography (eluent: ethyl acetate in petroleum ether 18%). White solid, yield 77%. m.p.: 223.0–225.2 °C. 1H NMR (400 MHz, CDCl3) δ 8.45 (s, 4H, -Ar-H), 8.22–8.20(q, 2H, -Ar-H), 7.59–7.55 (m, 3H, -Ar-H). 13C NMR (101 MHz, CDCl3) δ 173.64, 169.41, 150.17, 131.62, 129.54, 129.23, 129.01, 127.56, 126.31, 124.39. Calculate m/z for C14H9N3O3: 267.06, found: 268.07.
- 5-(4-Chlorophenyl)-3-(4-methoxyphenyl)-1,2,4-oxadiazole (4j)
- Prepared from 2b (0.50 g, 3.01 mmol), 4-chlorobenzoic acid (0.39 g, 2.50 mmol), EDCI (0.57 g, 2.97 mmol), HOBt (0.41 g, 3.03 mmol). Purification procedure: column chromatography (eluent: ethyl acetate in petroleum ether 14% with 1% TEA). White solid, yield 55%. m.p.: 134.5–134.9 °C. 1H NMR (400 MHz, CDCl3) δ 8.19–8.11 (q, 4H, Ar-H), 7.57–7.55 (d, 2H, Ar-H), 7.05–7.03 (d, 2H, Ar-H), 3.91 (s, 3H, –OCH3). 13C NMR (101 MHz, CDCl3) δ 174.50, 168.72, 161.99, 139.06, 129.48, 129.43, 129.39, 129.13, 129.09, 122.83, 119.17, 114.27, 114.22, 55.42. Calculate m/z for C15H11ClN2O2: 286.05, found: 286.09.
- 5-(3,4-Dimethoxyphenyl)-3-(4-methoxyphenyl)-1,2,4-oxadiazole (4k)
- Prepared from 2b (0.50 g, 3.01 mmol), 3,4-dimethoxybenzoic acid (0.46 g, 2.53 mmol), EDCI (0.57 g, 2.97 mmol), HOBt(0.41 g, 3.03 mmol). Purification procedure: column chromatography (eluent: ethyl acetate in petroleum ether 50% with 1% TEA). Light red solid, yield 44%. m.p.: 123.3–124.5 °C. 1H NMR (400 MHz, CDCl3) δ 8.14–8.12 (m, 2H, Ar-H), 7.87–7.84 (q, 1H, Ar-H), 7.71 (d, 1H, Ar-H), 7.05–7.01 (m, 3H, Ar-H), 4.04–4.00 (d, 6H, –OCH3), 3.91 (s, 3H, –OCH3). 13C NMR (101 MHz, CDCl3) δ 175.31, 168.53, 161.84, 152.71, 149.19, 129.10, 122.01, 119.51, 116.95, 114.19, 111.02, 110.38, 56.16, 56.09, 55.39. Calculate m/z for C17H16N2O4: 312.11, found: 312.19.
- 5-(4-(tert-Butyl)phenyl)-3-(4-methoxyphenyl)-1,2,4-oxadiazole (4l)
- Prepared from 2b (0.50 g, 3.01 mmol), 4-(tert-butyl) benzoic acid (0.45 g, 2.53 mmol), EDCI (0.57 g, 2.97 mmol), HOBt (0.41 g, 3.03 mmol). Purification procedure: column chromatography (eluent: ethyl acetate in petroleum ether 20% with 1% TEA). Light yellow solid, yield 44%. m.p.: 77.5–81.0 °C. 1H NMR (400 MHz, CDCl3) δ 8.17–8.13 (m, 4H, Ar-H), 7.60–7.58 (d, 2H, Ar-H), 7.05–7.03 (m, 2H, Ar-H), 3.91 (s, 3H, –OCH3), 1.40 (s, 9H, –C(CH3)3). 13C NMR (101 MHz, CDCl3) δ 175.51, 168.59, 161.86, 156.36, 129.13, 127.99, 126.07, 121.60, 119.54, 114.21, 55.40, 35.21, 31.12. Calculate m/z for C19H20N2O2: 308.15, found: 308.19.
- 5-(4-Chlorophenyl)-3-(3,4-dimethoxyphenyl)-1,2,4-oxadiazole (4m)
- Prepared from 2c (0.30 g, 1.53 mmol), 4-chlorobenzoic acid (0.20 g, 1.28 mmol), EDCI (0.28 g, 1.46 mmol), HOBt (0.20 g, 1.48 mmol). Purification procedure: column chromatography (eluent: ethyl acetate in petroleum ether 25% with 1% TEA). White solid, yield 52%. m.p.: 153.4–154.3 °C. 1H NMR (400 MHz, CDCl3) δ 8.19–8.17 (d, 2H, Ar-H), 7.81–7.79 (q, 1H, Ar-H), 7.67 (d, 1H, Ar-H), 7.58–7.54 (m, 2H, Ar-H), 7.02–6.96 (q, 1H, Ar-H), 4.02–3.97 (t, 6H, –OCH3). 13C NMR (101 MHz, CDCl3) δ 174.55, 168.79, 151.56, 149.14, 139.12, 129.49, 129.44, 122.76, 120.98, 119.25, 110.99, 109.82, 56.07, 55.99. Calculate m/z for C16H13ClN2O3: 316.06, found: 316.09.
- 5-(4-(tert-Butyl)phenyl)-3-(3,4-dimethoxyphenyl)-1,2,4-oxadiazole (4n)
- Prepared from 2c (0.20 g, 1.02 mmol), 4-(tert-butyl)benzoic acid (0.15 g, 0.84 mmol), EDCI (0.20 g, 1.04 mmol), HOBt (0.14 g, 1.04 mmol). Purification procedure: column chromatography (eluent: ethyl acetate in petroleum ether 50% with 1% TEA). Light yellow solid, yield 52.2%. m.p.: 98.1–101.5 °C. 1H NMR (400 MHz, CDCl3) δ 8.18–8.16 (d, 2H, -Ar-H), 7.84–7.81 (q, 1H, -Ar-H), 7.70–7.69 (d, 1H, -Ar-H), 7.60–7.58 (d, 3H, -Ar-H), 7.02–7.00 (d, 1H, -Ar-H), 4.03 (s, 3H, –OCH3), 3.99 (s, 3H, –OCH3), 1.40 (s, 9H, –C(CH3)3). 13C NMR (101 MHz, CDCl3) δ 175.58, 168.66, 156.44, 151.40, 149.09, 128.01, 126.08, 121.52, 120.96, 119.62, 110.96, 109.87, 56.08, 56.03, 55.99, 35.21, 31.11. Calculate m/z for C20H22N2O3: 338.16, found: 338.19.
- 3-(3-(3,4-Dimethoxyphenyl)-1,2,4-oxadiazol-5-yl)phenol (4o)
- Prepared from 2c (0.3 g, 1.53 mmol), 3-hydroxybenzoic acid (0.18 g, 1.17 mmol), EDCI (0.28 g, 1.46 mmol), HOBt (0.21 g, 1.55 mmol). Purification procedure: column chromatography (eluent: 33% ethyl acetate and 33% CH2Cl2 in petroleum ether). White solid, yield 34%. m.p.: 177.0–178.0 °C. 1H NMR (400 MHz, CDCl3) δ 7.82–7.78 (m, 3H, Ar-H), 7.66 (d, 1H, Ar-H), 7.46–7.42 (q, 1H, Ar-H), 7.13–7.11 (m, 1H, Ar-H), 7.00–6.98 (d, 1H, Ar-H), 4.00–3.97 (d, 6H, Ar-H). 13C NMR (101 MHz, DMSO-d6) δ 175.57, 168.47, 158.46, 151.95, 149.43, 131.27, 124.87, 120.96, 120.84, 119.01, 118.85, 114.54, 112.29, 110.01, 56.06, 55.99. Calculate m/z for C16H14N2O4: 298.10, found: 298.21.
- 5-(3-(3,4-Dimethoxyphenyl)-1,2,4-oxadiazol-5-yl)benzene-1, 3-diol (4p)
- Prepared from 2c (0.30 g, 1.53 mmol), 3,5-dihydroxybenzoic acid (0.20 g, 1.30 mmol), EDCI (0.28 g, 1.46 mmol), HOBt (0.21 g, 1.55 mmol). Purification procedure: column chromatography (eluent: 33% ethyl acetate in petroleum ether). Light yellow solid, yield 51%. m.p.: 140.4–151.0 °C. 1H NMR (400 MHz, DMSO-d6) δ 9.95 (s, 2H, –OH), 7.68–7.66 (q, 1H, Ar-H), 7.54 (d, 1H, Ar-H), 7.18–7.16 (d, 1H, Ar-H), 7.04–7.03 (d, 2H, Ar-H), 6.52–6.51 (t, 1H, Ar-H), 3.87 (d, 6H,-OCH3). 13C NMR (101 MHz, DMSO-d6) δ 175.73, 168.43, 159.70, 151.95, 149.45, 125.13, 120.93, 118.89, 112.34, 110.01, 107.65, 106.14, 56.09, 56.00. Calculate m/z for C16H14N2O5: 314.09, found: 314.11.
- 4-(3-(3,4-Dimethoxyphenyl)-1,2,4-oxadiazol-5-yl)benzene-1,2-diol (4q)
- Prepared from 2c (0.30 g, 1.53 mmol), 3,4-dihydroxybenzoic acid (0.20 g, 1.30 mmol), EDCI (0.28 g, 1.46 mmol), HOBt (0.21 g, 1.55 mmol). Purification procedure: column chromatography (eluent: 50% ethyl acetate in petroleum ether). Grey solid, yield 34%. m.p.: 209.9–210.2 °C. 1H NMR (400 MHz, DMSO-d6) δ 7.67–7.65 (q, 1H, Ar-H), 7.55–7.51 (m, 3H, Ar-H), 7.17–7.14 (d, 1H, Ar-H), 6.96–6.94 (d, 1H, Ar-H), 3.87–3.85 (d, 6H, –OCH3) 13C NMR (101 MHz, DMSO-d6) δ 175.77, 168.26, 151.83, 151.05, 149.40, 146.38, 120.92, 120.86, 119.16, 116.75, 115.08, 114.75, 112.29, 110.01, 56.07, 55.99. Calculate m/z for C16H14N2O5: 314.09, found: 314.11
- 5-(4-Chlorophenyl)-3-(3,4,5-trimethoxyphenyl)-1,2,4-oxadiazole (4s)
- Prepared from 2d (0.10 g, 0.44 mmol), 4-chlorobenzoic acid (0.06 g, 0.38 mmol), EDCI (0.08 g, 0.42 mmol), HOBt (0.06 g, 0.44 mmol). Purification procedure: column chromatography (eluent: ethyl acetate in petroleum ether 67% with 1% TEA). White solid, yield 65%. m.p.: 178.7–181.0 °C. 1H NMR (400 MHz, CDCl3) δ 8.21–8.19 (d,2H, Ar-H), 7.58–7.56 (d, 2H, Ar-H), 7.43 (s, 2H, Ar-H), 4.00–3.91 (d, 9H, –OCH3). 13C NMR (101 MHz, CDCl3) δ 174.76, 168.86, 153.56, 140.54, 139.26, 129.54, 129.49, 122.65, 121.93, 104.55, 61.01, 56.31. Calculate m/z for C17H15ClN2O4: 346.07, found: 346.01.
- 5-(3,4-Dimethoxyphenyl)-3-(3,4,5-trimethoxyphenyl)-1,2,4-oxadiazole (4t)
- Prepared from 2d (0.10 g, 0.44 mmol), 3,4-dimethoxybenzoic acid (0.06 g, 0.33 mmol), EDCI (0.08 g, 0.42 mmol), HOBt (0.06 g, 0.44 mmol). Purification procedure: column chromatography (eluent: ethyl acetate in petroleum ether 33% with 1% TEA). White solid, yield 59%. m.p.: 158.6–158.6 °C. 1H NMR (400 MHz, CDCl3) δ 7.89–7.86 (q, 1H, Ar-H), 7.72–7.71 (d, 1H, Ar-H), 7.44 (s, 2H, Ar-H), 7.05–7.02 (d, 1H, Ar-H), 4.05 (s, 3H, –OCH3), 4.01 (d, 9H, –OCH3), 3.95 (s, 3H, –OCH3). 13C NMR (101 MHz, CDCl3) δ 175.59, 168.68, 153.52, 152.86, 149.24, 140.38, 122.29, 122.15, 116.75, 111.05, 110.40, 104.55, 61.00, 56.33, 56.22, 56.22, 56.13. Calculate m/z for C19H20N2O6: 372.13, found: 372.19.
- 5-(4-(tert-Butyl)phenyl)-3-(3,4,5-trimethoxyphenyl)-1,2,4-oxadiazole (4u)
- Prepared from 2d (0.50 g, 2.21 mmol), 4-(tert-butyl)benzoic acid (0.33 g, 1.85 mmol), EDCI (0.42 g, 2.19 mmol), HOBt (0.30 g, 2.22 mmol). Purification procedure: column chromatography (eluent: ethyl acetate in petroleum ether 50% with 1% TEA). White solid, yield 53%. m.p.: 101.5–102.3 °C. 1H NMR (400 MHz, CDCl3) δ 8.18–8.16 (d, 2H, Ar-H), 7.60–7.58 (d, 2H, Ar-H), 7.45 (s, 2H, Ar-H), 4.00 (s, 6H, –OCH3), 3.95 (s, 3H, –OCH3). 13C NMR (101 MHz, CDCl3) δ 175.76, 168.72, 156.56, 153.52, 140.39, 128.04, 126.11, 122.31, 121.42, 104.57, 61.00, 56.31, 35.23, 31.11. Calculate m/z for C21H24N2O4: 368.17, found: 368.29.
- 3-(3-(3,4,5-Trimethoxyphenyl)-1,2,4-oxadiazol-5-yl) phenol (4v)
- Prepared from 2d (0.30 g, 1.33 mmol), 3-hydroxybenzoic acid (0.16 g, 1.16 mmol), EDCI (0.25 g, 1.30 mmol), HOBt (0.18 g, 1.33 mmol). Purification procedure: column chromatography (eluent: 78% ethyl acetate in petroleum ether). White solid, yield 48%. m.p.: 182.0–183.5 °C. 1H NMR (400 MHz, CDCl3) δ 7.85–7.80 (m, 2H, Ar-H), 7.49–7.41 (m, 3H, Ar-H), 7.15–7.12 (m, 1H, Ar-H), 4.00–3.96 (d, 9H, –OCH3). 13C NMR (101 MHz, DMSO-d6) δ 175.81, 168.51, 158.48, 153.84, 140.60, 131.32, 124.74, 121.87, 120.96, 119.07, 114.57, 104.71, 60.64, 56.48. Calculate m/z for C17H16N2O5: 328.11, found: 328.01.
- 3-(3,4-Dimethoxyphenyl)-5-(4-methoxyphenyl)-1,2,4-oxadiazole (4w)
- Prepared from 2c (0.30 g, 1.53 mmol), (E)-3-(4-chlorophenyl) acrylic acid (0.19 g, 1.25 mmol), EDCI (0.29 g, 1.51 mmol), HOBt (0.20 g, 1.48 mmol). Purification procedure: column chromatography (eluent: 33% ethyl acetate in petroleum ether with 1% TEA). White solid, yield 64%. m.p.: 157.0–158.0 °C. 1H NMR (400 MHz, CDCl3) δ 8.20–8.16 (m, 2H, -Ar-H), 7.82–7.80 (q, 1H, -Ar-H), 7.69–7.68 (d, 1H, -Ar-H), 7.08–7.05 (m, 2H, -Ar-H), 7.02–6.99 (d, 1H, -Ar-H), 4.02 (s, 3H, –OCH3), 3.99 (s, 3H, –OCH3), 3.93 (s, 3H, –OCH3). 13C NMR (101 MHz, CDCl3) δ 175.37, 168.57, 163.10, 151.37, 149.08, 130.07, 126.49, 120.90, 119.67, 116.88, 114.47, 113.85, 110.95, 109.84, 56.07, 55.99, 55.54. Calculate m/z for C17H16N2O4: 312.11, found: 312.19.
- (E)-3-Phenyl-5-styryl-1,2,4-oxadiazole (5a)
- Prepared from 2a (0.25 g, 1.84 mmol), cinnamic acid (0.22 g, 1.21 mmol), EDCI (0.35 g, 1.83 mmol), HOBt (0.25 g, 1.85 mmol). Purification procedure: column chromatography (eluent: 14% CH2Cl2 in petroleum ether with 2% TEA). White solid, yield 40%. m.p.: 92.6–93.0 °C. 1H NMR (400 MHz, CDCl3) δ 8.17–8.14 (m, 2H, Ar-H), 7.94–7.88 (m, 1H, –HC=CH–), 7.66–7.63 (m, 2H, Ar-H), 7.56–7.50 (m, 3H, Ar-H) 7.49–7.44 (m, 3H, Ar-H), 7.13–7.06 (m, 1H, –HC=CH–). 13C NMR (101 MHz, CDCl3) δ 175.24, 175.20, 168.72, 142.73, 134.40, 131.19, 130.57, 129.10, 128.88, 127.96, 127.46, 126.93, 110.23. Calculate m/z for C16H12N2O: 248.09, found: 248.19.
- (E)-3-Phenyl-5-(3,4,5-trimethoxystyryl)-1,2,4-oxadiazole (5b)
- Prepared from 2a (0.25 g, 1.84 mmol), (E)-3-(3,4,5-trimethoxyphenyl)acrylic acid (0.40 g, 1.68 mmol), EDCI (0.35 g, 1.83 mmol), HOBt (0.25 g, 1.85 mmol). Purification procedure: column chromatography (eluent: 17% ethyl acetate and 17% CH2Cl2 in petroleum ether with 2% TEA). Yellow solid, yield 56%. m.p.: 126.8–128.2 °C. 1H NMR (400 MHz, CDCl3) δ 8.16–8.12 (m, 2H, Ar-H), 7.86–7.80 (q, 1H, –CH=CH–), 7.55–7.51 (m, 3H, Ar-H), 7.04–6.98 (q, 1H, –CH=CH–), 6.87–6.85 (d, 2H, Ar-H), 3.96–3.91 (t, 9H, –OCH3). 13C NMR (101 MHz, CDCl3) δ 175.20, 168.67, 153.54, 142.62, 140.25, 131.20, 129.91, 128.88, 127.41, 126.90, 109.46, 105.00, 61.05, 56.18. Calculate m/z for C19H18N2O4: 338.13, found: 338.19.
- (E)-5-(4-Chlorostyryl)-3-phenyl-1,2,4-oxadiazole (5c)
- Prepared from 2a (0.20 g, 1.47 mmol), (E)-3-(4-chlorophenyl)acrylic acid (0.23 g, 1.26 mmol), EDCI (28 g, 1.46 mmol), HOBt (0.20 g, 1.47 mmol). Purification procedure: column chromatography (eluent: 25% ethyl acetate and 25% CH2Cl2 in petroleum ether). Yellow solid, yield 57%. m.p.: 162.5–164.0 °C. 1H NMR (400 MHz, CDCl3) δ 8.17–8.14 (m, 2H, Ar-H), 7.89–7.85 (d, 1H, –CH=CH–), 7.59–7.51 (m, 5H, Ar-H), 7.45–7.43 (d, 2H, Ar-H), 7.09–7.05 (d, 1H, –CH–CH–). 13C NMR (101 MHz, DMSO-d6) δ 175.74, 168.51, 141.94, 135.65, 133.70, 132.08, 130.59, 129.75, 129.53, 127.49, 126.69, 111.49. Calculate m/z for C16H11ClN2O: 282.06, found: 282.09.
- (E)-3-(4-Methoxyphenyl)-5-styryl-1,2,4-oxadiazole (5d)
- Prepared from 2b (0.28 g, 1.69 mmol), cinnamic acid (0.21 g, 1.42 mmol), EDCI (0.32 g, 1.67 mmol), HOBt (0.23 g, 1.7 mmol). Purification procedure: column chromatography (eluent: 67% CH2Cl2 in petroleum ether with 1% TEA). Light yellow solid, yield 38%. m.p.: 124.2–125.5 °C. 1H NMR (400 MHz, CDCl3) δ 8.12–8.07 (m, 2H, Ar-H), 7.92–7.88 (d, 1H, –HC=CH–), 7.65–7.63 (m, 2H, Ar-H), 7.50–7.44 (m, 3H, Ar-H), 7.11–7.07 (d, 1H, –HC=CH–), 7.05–7.02 (d, 2H, Ar-H), 3.90 (s, 3H, –OCH3). 13C NMR (101 MHz, DMSO-d6) δ 175.54, 168.17, 162.15, 143.08, 134.73, 131.07, 129.45, 129.16, 128.83, 119.01, 115.09, 110.75, 55.85. Calculate m/z for C17H14N2O2: 278.11, found: 278.19.
- (E)-3-(3,4-Dimethoxyphenyl)-5-styryl-1,2,4-oxadiazole (5e)
- Prepared from 2c (0.30 g, 1.53 mmol), cinnamic acid (0.19 g, 1.28 mmol), EDCI (0.28 g, 1.46 mmol), HOBt (0.20 g, 1.47 mmol). Purification procedure: column chromatography (eluent: 25% ethyl acetate and 25% CH2Cl2 in petroleum ether). Light yellow solid, yield 51%. m.p.: 115.4–116.5 °C. 1H NMR (400 MHz, CDCl3) δ 7.93–7.89 (d, 1H, –HC=CH–), 7.78–7.76 (q, 1H, Ar-H), 7.65–7.63 (m, 3H, Ar-H), 7.48–7.45 (m, 3H, Ar-H), 7.11–7.07 (d, 1H, –HC=CH–), 7.01–6.99 (d, 1H, -Ar-H), 4.01 (s, 3H, –OCH3), 3.98(s, 3H, –OCH3). 13C NMR (101 MHz, CDCl3) δ 175.00, 168.47, 151.43, 149.11, 142.60, 134.41, 130.53, 129.08, 127.92, 120.84, 119.46, 110.98, 110.23, 109.76, 56.06, 55.98. Calculate m/z for C18H16N2O3: 308.12, found: 308.19.
3.2. Cell Culture and Cytotoxicity Evaluation
3.3. Fungicidal Activity Testing
3.3.1. Plant Pathogens
3.3.2. In Vitro Antifungal Assay
- A: The average diameter of the blank medium colony.
- B: Diameter of mushroom cake (3 mm).
- C: The average diameter of the colonies in the medium containing compound.
3.3.3. Determination of EC50
3.3.4. Study on the State of Fungi
3.3.5. Determination of the Antifungal Activity In Vivo
- A: The average diameter of the blank medium colony.
- B: The average diameter of the colonies in the medium containing compound.
- C: Diameter of mushroom cake (3 mm/4 mm).
3.4. Molecular Docking
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- United Nations. 2019 Revision of World Population Prospects. Available online: https://population.un.org/wpp/ (accessed on 10 April 2025).
- Giovanni, B.; Roman, P.; Riccardo, P.; Loredana, C.; Giuseppe, S.; Dennis, F.; Stefania, S.; Stefano, D.; Angelo, C.; Filippo, M. The essential oil from industrial hemp (Cannabis sativa L.) by-products as an effective tool for insect pest management in organic crops. Ind. Crops Prod. 2018, 122, 308–315. [Google Scholar]
- Lykogianni, M.; Bempelou, E.; Karamaouna, F.; Aliferis, K.A. Do pesticides promote or hinder sustainability in agriculture? The challenge of sustainable use of pesticides in modern agriculture. Sci. Total Environ. 2021, 795, 148625. [Google Scholar] [CrossRef]
- Epstein, L. Fifty Years Since Silent Spring. Annu. Rev. Phytopathol. 2014, 52, 377–402. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.; Waterhouse, S.; Paveley, N. Evidence For Reduced Sexual Reproduction of Zymoseptoria Tritici Following Treatment with Fluxapyroxad and Implications for Initial Infection of Wheat Crops. Commun. Agric. Appl. Biol. Sci. 2014, 79, 385–395. [Google Scholar] [PubMed]
- Neils, A.L.; Brisco-McCann, E.I.; Harlan, B.R.; Hausbeck, M.K. Management strategies for Alternaria leaf blight on American ginseng. Crop Prot. 2021, 139, 105302. [Google Scholar] [CrossRef]
- Matheron, M.E.; Porchas, M. Activity of Boscalid, Fenhexamid, Fluazinam, Fludioxonil, and Vinclozolin on Growth of Sclerotinia minor and S. sclerotiorum and Development of Lettuce Drop. Plant Dis. 2004, 88, 665–668. [Google Scholar] [CrossRef]
- Gutiérrez-Alonso, O.; Hawkins, N.J.; Cools, H.J.; Shaw, M.W.; Fraaije, B.A. Dose-dependent selection drives lineage replacement during the experimental evolution of SDHI fungicide resistance in Zymoseptoria tritici. Evol. Appl. 2017, 10, 1055–1066. [Google Scholar] [CrossRef]
- Fernández-Ortuño, D.; Pérez-García, A.; Chamorro, M.; de la Peña, E.; de Vicente, A.; Torés, J.A. Resistance to the SDHI Fungicides Boscalid, Fluopyram, Fluxapyroxad, and Penthiopyrad in Botrytis cinerea from Commercial Strawberry Fields in Spain. Plant Dis. 2017, 101, 1306–1313. [Google Scholar] [CrossRef]
- Schmeling, B.V.; Kulka, M. Systemic Fungicidal Activity of 1,4-Oxathiin Derivatives. Science 1966, 152, 659–660. [Google Scholar] [CrossRef]
- Keinath, A.P. Differential Sensitivity to Boscalid in Conidia and Ascospores of Didymella bryoniae and Frequency of Boscalid-Insensitive Isolates in South Carolina. Plant Dis. 2012, 96, 228–234. [Google Scholar] [CrossRef]
- Zeun, R.; Scalliet, G.; Oostendorp, M. Biological activity of sedaxane—A novel broad-spectrum fungicide for seed treatment. Pest Manag. Sci. 2013, 69, 527–534. [Google Scholar] [CrossRef] [PubMed]
- Mills, E.L.; Kelly, B.; O’Neill, L.A.J. Mitochondria are the powerhouses of immunity. Nat. Immunol. 2017, 18, 488–498. [Google Scholar] [CrossRef] [PubMed]
- Zuniga, A.I.; Oliveira, M.S.; Rebello, S.S.; Peres, N.A. Baseline Sensitivity of Botrytis cinerea Isolates from Strawberry to Isofetamid Compared to other SDHIs. Plant Dis. 2020, 104, 1224–1230. [Google Scholar] [CrossRef]
- Pei, Q.H.; Li, Y.; Ge, X.Z.; Tian, P.F. Multipath effects of berberine on peach Brown rot fungus Monilinia fructicola. Crop Prot. 2019, 116, 92–100. [Google Scholar] [CrossRef]
- Avenot, H.F.; Michailides, T.J. Progress in understanding molecular mechanisms and evolution of resistance to succinate dehydrogenase inhibiting (SDHI) fungicides in phytopathogenic fungi. Crop Prot. 2010, 29, 643–651. [Google Scholar] [CrossRef]
- Giornal, F.; Pazenok, S.; Rodefeld, L.; Lui, N.; Vors, J.-P.; Leroux, F.R. Synthesis of diversely fluorinated pyrazoles as novel active agrochemical ingredients. J. Fluor. Chem. 2013, 152, 2–11. [Google Scholar] [CrossRef]
- Lamberth, C. Small ring chemistry in crop protection. Tetrahedron 2019, 75, 4365–4383. [Google Scholar] [CrossRef]
- Verma, S.K.; Verma, R.; Kumar, K.S.S.; Banjare, L.; Shaik, A.B.; Bhandare, R.R.; Rakesh, K.P.; Rangappa, K.S. A key review on oxadiazole analogs as potential methicillin-resistant Staphylococcus aureus (MRSA) activity: Structure-activity relationship studies. Eur. J. Med. Chem. 2021, 219, 113442. [Google Scholar] [CrossRef]
- Dhonnar, S.L.; More, R.A.; Adole, V.A.; Jagdale, B.S.; Sadgir, N.V.; Chobe, S.S. Synthesis, spectral analysis, antibacterial, antifungal, antioxidant and hemolytic activity studies of some new 2,5-disubstituted-1,3,4-oxadiazoles. J. Mol. Struct. 2022, 1253, 132216. [Google Scholar] [CrossRef]
- Izgi, S.; Sengul, I.F.; Şahin, E.; Koca, M.S.; Cebeci, F.; Kandemir, H. Synthesis of 7-azaindole based carbohydrazides and 1,3,4-oxadiazoles; Antioxidant activity, α-glucosidase inhibition properties and docking study. J. Mol. Struct. 2022, 1247, 131343. [Google Scholar] [CrossRef]
- Bezerra, N.M.M.; De Oliveira, S.P.; Srivastava, R.M.; Da Silva, J.R. Synthesis of 3-aryl-5-decapentyl-1,2,4-oxadiazoles possessing antiinflammatory and antitumor properties. Il Farm. 2005, 60, 955–960. [Google Scholar] [CrossRef]
- Srivastava, R.M.; de Almeida Lima, A.; Viana, O.S.; da Costa Silva, M.J.; Catanho, M.T.J.A.; de Morais, J.O.F. Antiinflammatory Property of 3-Aryl-5-(n-propyl)-1,2,4-oxadiazoles and Antimicrobial Property of 3-Aryl-5-(n-propyl)-4,5-dihydro-1,2,4-oxadiazoles: Their Syntheses and Spectroscopic Studies. Bioorganic Med. Chem. 2003, 11, 1821–1827. [Google Scholar] [CrossRef] [PubMed]
- Janardhanan, J.; Chang, M.; Mobashery, S. The oxadiazole antibacterials. Curr. Opin. Microbiol. 2016, 33, 13–17. [Google Scholar] [CrossRef]
- Fei, Q.; Liu, C.; Luo, Y.; Chen, H.; Ma, F.; Xu, S.; Wu, W.J.M.D. Rational design, synthesis, and antimicrobial evaluation of novel 1,2,4-trizaole-substituted 1,3,4-oxadiazole derivatives with a dual thioether moiety. Mol. Divers. 2025, 29, 255–267. [Google Scholar] [CrossRef]
- Verma, S.K.; Verma, R.; Verma, S.; Vaishnav, Y.; Tiwari, S.P.; Rakesh, K.P. Anti-tuberculosis activity and its structure-activity relationship (SAR) studies of oxadiazole derivatives: A key review. Eur. J. Med. Chem. 2021, 209, 112886. [Google Scholar] [CrossRef] [PubMed]
- De, S.S.; Khambete, M.P.; Degani, M.S. Oxadiazole scaffolds in anti-tuberculosis drug discovery. Bioorganic Med. Chem. Lett. 2019, 29, 1999–2007. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.; Tian, W.Y.; Zhang, Q.; Liu, J.Z.; Liu, Z.Y.; Jin, J.; Guo, Y.; Bai, L.P. Novel 1,3,4-thiadiazole/oxadiazole-linked honokiol derivatives suppress cancer via inducing PI3K/Akt/mTOR-dependent autophagy. Bioorganic Chem. 2021, 115, 105257. [Google Scholar] [CrossRef]
- Ayoup, M.S.; Abu-Serie, M.M.; Abdel-Hamid, H.; Teleb, M. Beyond direct Nrf2 activation; reinvestigating 1,2,4-oxadiazole scaffold as a master key unlocking the antioxidant cellular machinery for cancer therapy. Eur. J. Med. Chem. 2021, 220, 113475. [Google Scholar] [CrossRef]
- Manjunath, R.; Anantharaju, P.G.; Madhunapantulas, S.V.; Gaonkar, S.L. Design, synthesis, and biological evaluation of 2-butyl-4-chloroimidazole-derived 1,3,4-oxadiazoles: ACE inhibition, anticancer, and antitubercular activities. J. Mol. Struct. 2025, 1322, 140630. [Google Scholar]
- Blokhina, S.V.; Sharapova, A.V.; Ol’khovich, M.V.; Doroshenko, I.A.; Levshin, I.B.; Perlovich, G.L. Synthesis and antifungal activity of new hybrids thiazolo[4,5-d]pyrimidines with (1H-1,2,4)triazole. Bioorganic Med. Chem. Lett. 2021, 40, 127944. [Google Scholar] [CrossRef]
- Wang, X.B.; Chai, J.Q.; Kong, X.Y.; Jin, F.; Chen, M.; Yang, C.L.; Xue, W. Expedient discovery for novel antifungal leads: 1,3,4-Oxadiazole derivatives bearing a quinazolin-4(3H)-one fragment. Bioorganic Med. Chem. 2021, 45, 116330. [Google Scholar] [CrossRef] [PubMed]
- Çevik, U.A.; Celik, I.; Işık, A.; Pillai, R.R.; Tallei, T.E.; Yadav, R.; Özkay, Y.; Kaplancıklı, Z.A. Synthesis, molecular modeling, quantum mechanical calculations and ADME estimation studies of benzimidazole-oxadiazole derivatives as potent antifungal agents. J. Mol. Struct. 2022, 1252, 132095. [Google Scholar] [CrossRef]
- Çavuşoğlu, B.K.; Yurttaş, L.; Cantürk, Z. The synthesis, antifungal and apoptotic effects of triazole-oxadiazoles against Candida species. Eur. J. Med. Chem. 2018, 144, 255–261. [Google Scholar] [CrossRef]
- Faria, J.V.; Vegi, P.F.; Miguita, A.G.C.; dos Santos, M.S.; Boechat, N.; Bernardino, A.M.R. Recently reported biological activities of pyrazole compounds. Bioorganic Med. Chem. 2017, 25, 5891–5903. [Google Scholar] [CrossRef]
- Li, S.K.; Li, D.D.; Xiao, T.F.; Zhang, S.S.; Song, Z.H.; Ma, H.Y. Design, Synthesis, Fungicidal Activity, and Unexpected Docking Model of the First Chiral Boscalid Analogues Containing Oxazolines. J. Agric. Food Chem. 2016, 64, 8927–8934. [Google Scholar] [CrossRef]
- Wang, H.Y.; Gao, X.H.; Zhang, X.X.; Jin, H.; Tao, K.; Hou, T.P. Design, synthesis and antifungal activity of novel fenfuram-diarylamine hybrids. Bioorganic Med. Chem. Lett. 2017, 27, 90–93. [Google Scholar] [CrossRef] [PubMed]
- Liang, P.B.; Shen, S.Q.; Xu, Q.B.; Wang, S.M.; Jin, S.H.; Lu, H.Z.; Dong, Y.H.; Zhang, J.J. Design, synthesis biological activity, and docking of novel fluopyram derivatives containing guanidine group. Bioorganic Med. Chem. 2021, 29, 115846. [Google Scholar] [CrossRef]
- Srivastava, R.M.; Pereira, M.C.; Faustino, W.W.M.; Coutinho, K.; dos Anjos, J.V.; de Melo, S.J. Synthesis, mechanism of formation, and molecular orbital calculations of arylamidoximes. Monatshefte Chem. Chem. Mon. 2009, 140, 1319. [Google Scholar] [CrossRef]
- Tang, Z.L.; Li, X.X.; Yao, Y.; Qi, Y.C.; Wang, M.; Dai, N.N.; Wen, Y.H.; Wan, Y.C.; Peng, L.F. Design, synthesis, fungicidal activity and molecular docking studies of novel 2-((2-hydroxyphenyl)methylamino)acetamide derivatives. Bioorganic Med. Chem. 2019, 27, 2572–2578. [Google Scholar] [CrossRef]
- Shang, X.F.; Zhao, Z.M.; Li, J.C.; Yang, G.Z.; Liu, Y.Q.; Dai, L.X.; Zhang, Z.J.; Yang, Z.G.; Miao, X.L.; Yang, C.J.; et al. Insecticidal and antifungal activities of Rheum palmatum L. anthraquinones and structurally related compounds. Ind. Crops Prod. 2019, 137, 508–520. [Google Scholar] [CrossRef]
- Perveen, K.; Bukhari, N.A.; Al Masoudi, L.M.; Alqahtani, A.N.; Alruways, M.W.; Alkhattaf, F.S. Antifungal potential, chemical composition of Chlorella vulgaris and SEM analysis of morphological changes in Fusarium oxysporum. Saudi J. Biol. Sci. 2021, 29, 2501–2505. [Google Scholar] [CrossRef] [PubMed]



| Compound | The Inhibition Rate (%) | ||||
|---|---|---|---|---|---|
| R. solani | F. graminearum | E. turcicum | B. cinerea | C. capsica | |
| 4a | <10 | 19.06 ± 7.15 | 17.54 ± 2.68 | <10 | - |
| 4b | 18.68 ± 5.82 | 23.75 ± 7.22 | 60.39 ± 1.68 | <10 | <10 |
| 4c | 10.74 ± 3.54 | 33.47 ± 3.88 | 30.07 ± 1.95 | <10 | <10 |
| 4d | 18.60 ± 2.71 | 40.22 ± 1.50 | 46.11 ± 0.74 | 36.77 ± 9.53 | 22.28 ± 2.97 |
| 4e | 16.61 ± 1.47 | 30.07 ± 4.51 | 26.47 ± 3.09 | <10 | 18.84 ± 3.10 |
| 4f | 80.87 ± 2.40 | 100 | 85.71 ± 1.97 | 38.85 ± 5.35 | 86.85 ± 3.11 |
| 4g | 39.91 ± 3.44 | 56.86 ± 6.74 | 38.67 ± 1.00 | <10 | 47.65 ± 1.05 |
| 4h | <10 | 28.98 ± 8.54 | <10 | <10 | <10 |
| 4i | <10 | <10 | <10 | <10 | - |
| 4j | <10 | <10 | <10 | <10 | <10 |
| 4k | 22.59 ± 8.85 | 21.67 ± 2.53 | <10 | <10 | <10 |
| 4l | <10 | 27.58 ± 1.78 | 24.16 ± 3.77 | <10 | <10 |
| 4m | 11.73 ± 5.11 | <10 | 15.16 ± 0.77 | - | - |
| 4n | <10 | <10 | 29.79 ± 1.26 | <10 | <10 |
| 4o | 24.43 ± 1.65 | 55.10 ± 2.99 | 28.09 ± 1.65 | <10 | 20.82 ± 0.59 |
| 4p | <10 | <10 | 11.25 ± 1.91 | <10 | 0 |
| 4q | 47.76 ± 8.13 | 43.48 ± 3.70 | 43.54 ± 0.99 | 32.84 ± 12.16 | 55.54 ± 1.34 |
| 4s | <10 | <10 | <10 | <10 | <10 |
| 4t | <10 | 28.27 ± 2.19 | <10 | <10 | - |
| 4u | <10 | <10 | <10 | <10 | <10 |
| 4v | <10 | 26.07 ± 9.47 | 15.46 ± 3.53 | <10 | <10 |
| 4w | <10 | <10 | <10 | <10 | <10 |
| 5a | 14.52 ± 6.32 | <10 | 21.15 ± 3.85 | <10 | - |
| 5b | 21.17 ± 2.78 | 36.09 ± 1.67 | 31.81 ± 1.44 | 30.16 ± 6.40 | 20.62 ± 4.48 |
| 5c | <10 | <10 | <10 | <10 | - |
| 5d | <10 | 19.02 ± 7.40 | 19.22 ± 2.39 | 21.71 ± 6.82 | - |
| 5e | 23.89 ± 0.59 | 28.74 ± 5.14 | 29.42 ± 2.05 | 30.56 ± 1.43 | <10 |
| Plant Pathogen | Compound | The Regression Equation | R2 | EC50 (μg/mL) | 95% CI (μg/mL) |
|---|---|---|---|---|---|
| R. solani | 4q | y = 2.077x + 1.697 | 0.9163 | 38.88 | 24.62–164.03 |
| 4f | y = 4.073x + 0.507 | 0.8752 | 12.68 | 6.02–26.74 | |
| carbendazim | y = 1.790x + 4.924 | 0.9265 | 1.10 | 0.68–1.78 | |
| F. graminearum | 4q | y = 0.969x + 2.894 | 0.9479 | 149.26 | 51.84–429.78 |
| 4g | y = 2.179x + 0.848 | 0.9496 | 80.55 | 45.54–142.46 | |
| 4f | y = 0.992x + 3.536 | 0.9335 | 29.97 | 19.44–46.22 | |
| carbendazim | y = 0.926x + 3.804 | 0.9991 | 19.59 | 18.34–20.91 | |
| E. turcicum | 4q | y = 1.025x + 2.580 | 0.9375 | 228.99 | 82.79–633.38 |
| 4f | y = 1.412x + 2.933 | 0.9893 | 29.14 | 24.70–34.39 | |
| carbendazim | y = 2.246x + 0.419 | 0.9643 | 109.56 | 65.03–184.58 | |
| C. capsica | 4q | y = 1.596x + 2.414 | 0.9947 | 41.67 | 36.61–47.44 |
| 4f | y = 1.184x + 3.882 | 0.9845 | 8.81 | 6.60–11.76 | |
| carbendazim | y = 2.004x + 6.307 | 0.9296 | 0.22 | 0.12–0.43 |
| Treatment | Blank | Carbendazim (5 Days) | 4f | 4q (5 Day) | |||
|---|---|---|---|---|---|---|---|
| Concentration (μg/mL) | - | 500 | 1000 | 500 (5 day) | 1000 (3 day) | 500 | 1000 |
| Control efficacy (%) | 0 | 100 | 100 | 100 | 100 | 50.12 | 76.83 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, L.; Yang, K.; Yao, L.; Wang, N.; Kang, H.; Yao, G.; Li, X.; Qin, B. Synthesis and Antifungal Activity of 1,2,4-Oxadiazole Derivatives. Molecules 2025, 30, 1851. https://doi.org/10.3390/molecules30081851
Yu L, Yang K, Yao L, Wang N, Kang H, Yao G, Li X, Qin B. Synthesis and Antifungal Activity of 1,2,4-Oxadiazole Derivatives. Molecules. 2025; 30(8):1851. https://doi.org/10.3390/molecules30081851
Chicago/Turabian StyleYu, Lili, Kuan Yang, Lin Yao, Nana Wang, Hui Kang, Guangda Yao, Xiaomeng Li, and Bei Qin. 2025. "Synthesis and Antifungal Activity of 1,2,4-Oxadiazole Derivatives" Molecules 30, no. 8: 1851. https://doi.org/10.3390/molecules30081851
APA StyleYu, L., Yang, K., Yao, L., Wang, N., Kang, H., Yao, G., Li, X., & Qin, B. (2025). Synthesis and Antifungal Activity of 1,2,4-Oxadiazole Derivatives. Molecules, 30(8), 1851. https://doi.org/10.3390/molecules30081851




