1. Introduction
Interstitial fluids interact with minerals in sediments, undergoing adsorption, dissolution, and precipitation reactions at the mineral/solution interface. The structure of this region and its physical–chemical conditions (temperature, pH, ionic strength, concentration of organic and inorganic ions, complexing agents, redox potential, etc.) determine mineral evolution. Clay minerals play a key role in the transport and retention of solutes, nutrients, and contaminants in soils and sediments, due to their small particle size, high specific surface area, and capacity to adsorb ions and molecules. The decrease in the dissolution rate of silicates with increasing pH is commonly observed under experimental and environmental conditions. Dissolution and adsorption experiments on clay minerals have shown that the reactivity of the surface changes from one face to another [
1,
2,
3,
4,
5,
6,
7]. As a function of the pH, the surface atomic groups undergo protonation or deprotonation reactions. Therefore, dissolution reactions in acidic conditions involve protonation/deprotonation reactions, which trigger the release of structural cations [
6,
8,
9]; the dissolution rate close to neutral pH is the lowest, varying as a function of the nature of the mineral, and as a function of the isomorphic substitutions in consequence. It has been suggested that the rate-limiting step in the dissolution of aluminosilicates was the breaking of bonds to the bridge oxygens, Al–O–Si (e.g., [
2,
6,
10,
11]). Nevertheless, experimental methods cannot be used to fully identify some individual steps of the dissolution reaction mechanism, such as the surface groups where protons were specifically adsorbed to decompose the structure through a protolysis reaction. Xio and Lasaga [
12], in a pioneering ab initio study, identified the proton attack on bridge oxygens as the step controlling the dissolution reaction.
Hendricks [
13] deduced the development of surface charge on the edge face of crystalline silicates, and Schofield and Samson [
14] modeled the clay mineral edge facets and their capacity to exchange protons and hydroxyls. Later, White and Zelazny [
15] reported an exhaustive crystallochemical analysis of the edge structure of dioctahedral phyllosilicates. Bleam et al. [
16] concluded that pyrophyllite {100} or {130} edge faces seemed to be more reactive than {110} or {010} edge faces, which may explain the crystal shape in natural specimens being dominated by {010} and {110} faces. Therefore, the analysis of the structure and facets of phyllosilicates reveals surface reactive sites with oxygen atoms exhibiting differential characteristics. Limiting to dioctahedral minerals, siloxanic oxygen bonds (Si–O–Si) form the basal surface of the tetrahedral sheet. Bridge oxygens connect the tetrahedral and octahedral sheets, bonding a tetrahedral and two octahedral cations (Si–O < Al
2). Finally, the oxygen of the hydroxyl groups is shared by two octahedral cations (Al-OH-Al). At the edge faces, these oxygens may have dangling bonds that relax by protonation and/or deprotonation, forming silanol (Si–OH) and aluminol (Al-OH) groups, and adsorption of water molecules (Al–OH
2) [
17].
Besides experimental procedures, computational methods can also be used to study the structure, properties, reactivity, and mechanisms at the mineral/solution interface, serving as an additional method to complement experimental inferences and theories. Concerning the dissolution processes of phyllosilicates, computational studies enable us to examine reaction steps that are not accessible through experimental methods. Different types of theoretical surface models are often employed in these studies. Some models involve two main approaches: (i) the crystallographic approach, and (ii) the local approach. The first approach treats the surface as a whole by selecting a crystallographic plane, creating a vacuum within the crystal structure, and applying periodic boundary conditions. In contrast, the local approach also selects a crystallographic plane but models only the local structure of that plane as a molecular system. Density Functional Theory (DFT) is mainly used in the physical chemistry of crystals for both surface models [
18,
19,
20]. Both approaches involve different modeling choices and electron DFT parameters. The crystallographic approach must account for the size of the slab layers, the number of layers optimized in the surface and adsorption model, the vacuum height, the cell or supercell, etc., the exchange and correlation functional, the basis set size, the k-point sampling, and the cutoff parameters, among other factors. The local approach also needs to consider the size of the local site, the cluster size, the closing of dangling bonds, the basis set size, the exchange and correlation basis set, solvent environment, etc. (e.g., [
21,
22]). These methods help explain many surface processes at the microscopic level through local atomic interactions. These studies often provide qualitative and sometimes quantitative insights into the physicochemical processes at mineral interfaces. The crystallographic approach has been applied with the DFT method to study water adsorption on the surface and in the interlayer space of Na- [
23] and Ca-Montmorillonite and kaolinite [
24], yielding clear results. Local molecular systems of moderate size, combined with DFT methods, have been applied to clay minerals (e.g., [
25,
26]). Sainz et al. [
27] reported DFT research on the structure of dioctahedral TOT phyllosilicates (O and T refer to octahedral and tetrahedral sheets, respectively) with high, medium, and low interlayer charges. Molecular cluster models have been successfully used in studies of acid-treated montmorillonite, showing that isomorphic substitutions of Al
3+ by Mg
2+ in the octahedral sheet led to effective protonation of test molecules, more so than substitutions of Si
4+ by Al
3+ in the tetrahedral sheet [
28]. Therefore, theoretical surface models of phyllosilicates should provide a comprehensive understanding of protolysis reactions, with results that can help interpret experimental findings, propose reaction mechanisms, and support broader extrapolations.
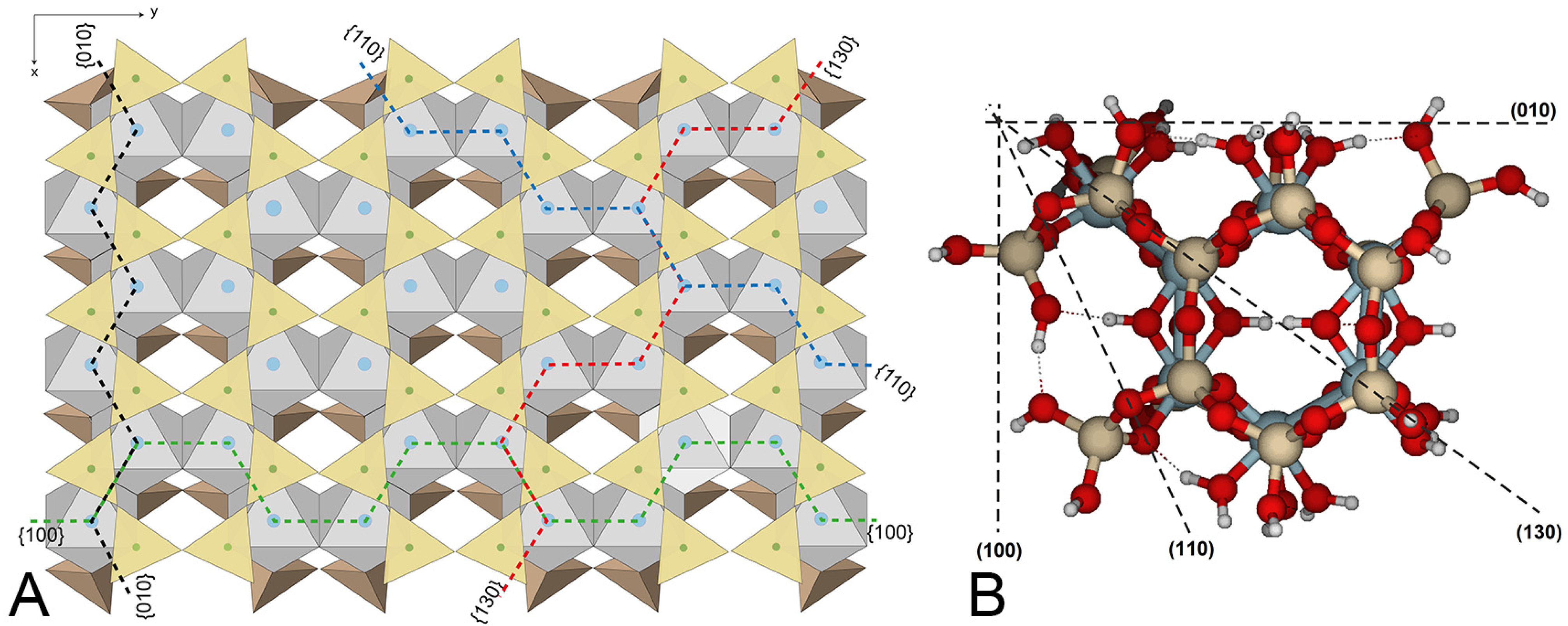
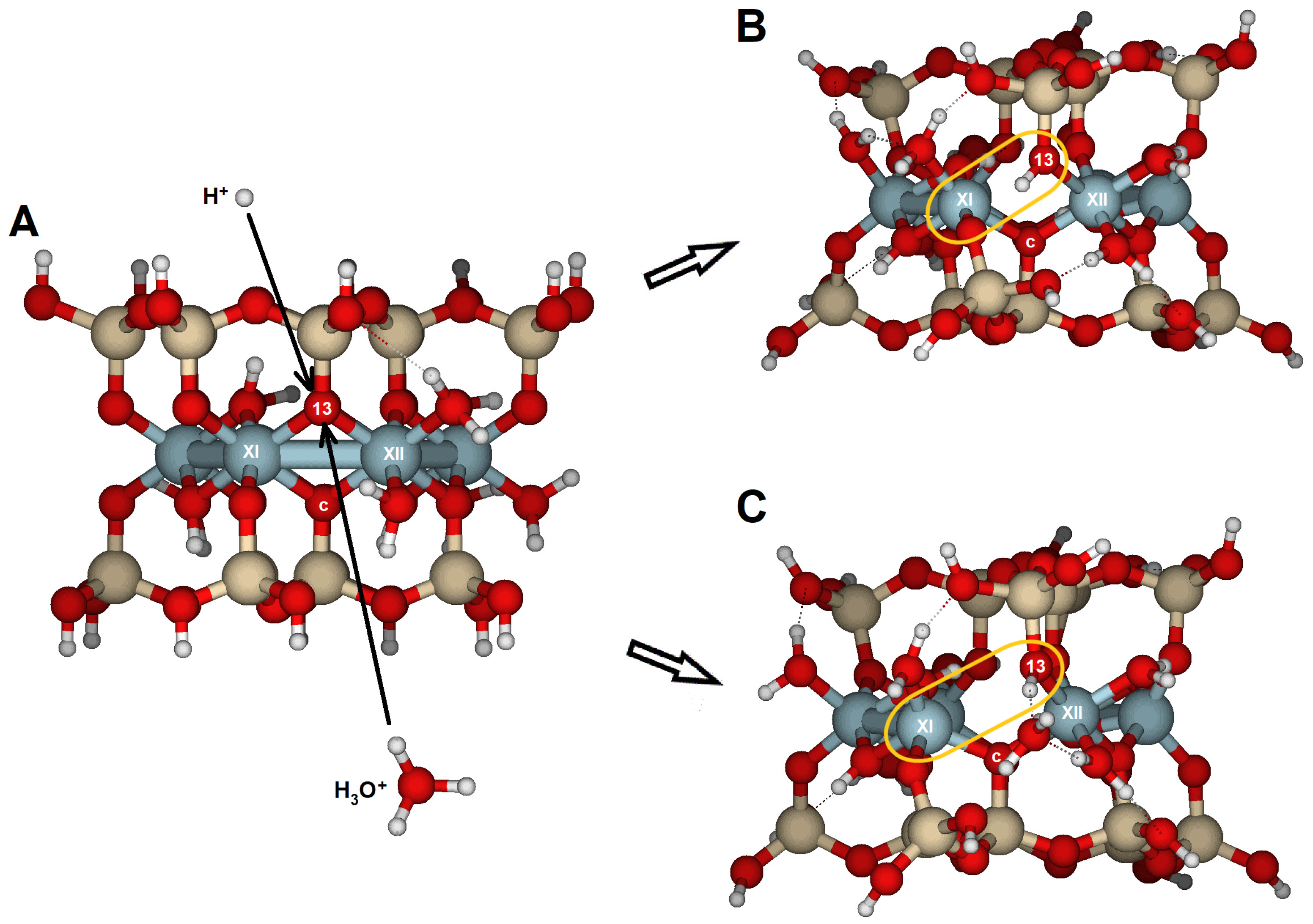
Pyrophyllite is a 2:1 dioctahedral phyllosilicate with no isomorphic substitutions and does not possess any structural charge (
Figure 1A). Thus, it is the simplest model for a large variety of clay mineral studies. There are many studies related to the basal surfaces of these minerals, although the basal plane is much less reactive than the edges of the clay particles [
29]. Few studies have modeled the edge faces of phyllosilicates.
Churakov [
31] calculated the composition and relative stability of the edge faces of pyrophyllite and later provided results on the proton exchange process on the hydrated lateral facets. He found that the Si–O–Al groups had the highest proton affinity on {110}, {100}, and {130} edges. However, in the {010} edge, the proton affinity was associated with the Al–OH groups.
Systematic quantum mechanical calculations of pyrophyllite edge surfaces revealed that, although the basal {001} surface is hydrophobic, the edge surfaces display hydrophilic properties due to hydroxyl groups and dangling atoms [
32]. Ulian and Valdrè [
33] thoroughly characterized pyrophyllite thermochemical and thermo-physical properties using DFT, providing data on heat capacity and thermal expansion coefficients across a broad temperature range (0–900 K).
The most recent studies on molecular dynamics of dissolution at the {110} edge facets of pyrophyllite revealed that dissolution in pure water is a complex process with multiple steps, characterized by a series of simultaneous direct and reversible elementary reactions [
34]. These reactions result in a gradual reduction in surface site density, ultimately leading to the release of silanol and aluminol complexes into the solution.
It is experimentally clear [
35] that protons are involved in the dissolution mechanism of silicates; however, the precise mechanism is not yet clearly understood. Therefore, this study aims to enhance our understanding of the dissolution mechanisms of dioctahedral phyllosilicates in acidic conditions by providing atomistic evidence, given their importance in geological and environmental processes. To achieve this, DFT calculations will be used to model the protolysis reaction at the edge faces. Molecular models for four edge faces—{100}, {010}, {110}, and {130}—will be developed. The reactivity of each face during the protolysis process will be assessed based on its Proton Reaction Energy (PRE) and the distortion of its structure after protonation.
3. Discussion
A systematic study of the initial steps of the dissolution reaction was performed by modeling the protolysis of the {100}, {010}, {110}, and {130} surfaces in molecular cluster models of pyrophyllite. These surfaces are attacked by either H+ or H3O+, which can be considered the first steps of hydrolysis in acidic media. Before reaction, the dangling atoms produced by cutting the structure were relaxed with H or H2O. The reactive oxygens corresponded to those in the octahedral sheet, either as bridge oxygens (Si–O < Al2), bridge aluminol oxygens (Si–O–Al), or hydroxyl groups (Al–OH–Al).
We should remember that the molecular clusters are calculated in a vacuum. Surface structure calculations were done without an explicit bulk water phase, which could influence the relative stability of surface proton configurations. Under natural conditions, dangling atoms at the surface interface relax through interactions with solvent molecules that may be adsorbed at specific sites. Oxygen atoms on the surface can carry a local excess of negative charge, although the entire surface remains neutral in non-protonated or non-hydronium structures. Martins et al. [
32] reported strong interactions between water molecules and edge surfaces, with some water molecules being trapped at particular Al sites—especially those with low coordination—which aligns with our observations in the molecular models.
Furthermore, bridge aluminol oxygens act as Brønsted bases due to two lone electron pairs and an excess negative charge (Si–O
δ-–Al). These correspond to bridge oxygens on the outermost face where one Al atom is missing; in aqueous media, relaxation of these sites may lead to protonation of the oxygens, which have only one lone electron pair and carry an excess positive charge (Si–O
δ+H–Al). Such reactive groups function as amphoteric surface sites in water [
17,
36,
37].
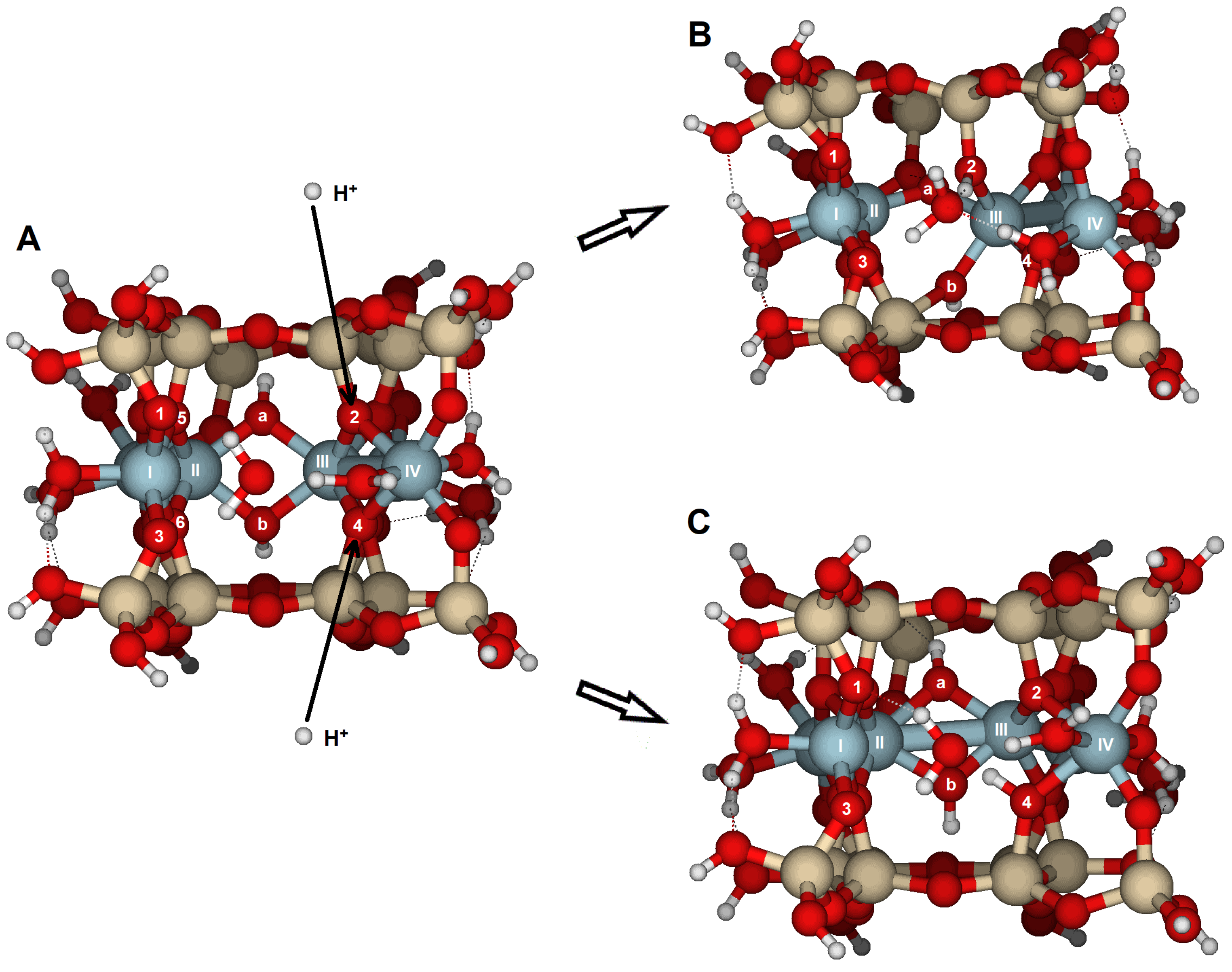
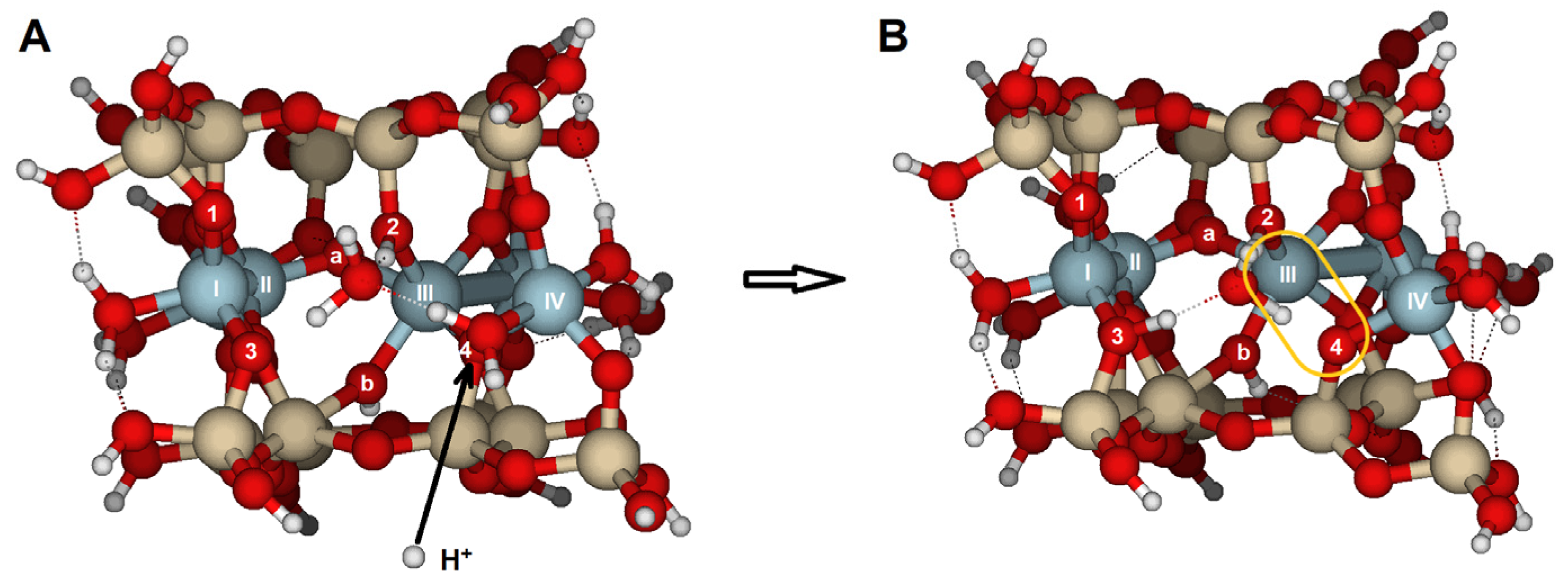
The most reactive oxygens were the bridge oxygens, whose coordination sphere is completed with one Si and two Al, especially those located at the inner part of the edge. These oxygens have only a lone electron pair that a proton can attack. That is the case with O2 and O4 on the {100} face, or O5 on the {010} face. This protonation renders the oxygen atom unstable, causing bond breaking (
Figure 4 and
Figure 8). The results from modeling the four selected faces showed them as the most reactive oxygens vulnerable to proton attack, as well as highlighting the reactivity differences among the faces.
The third type of oxygen, the hydroxyl groups (Al–OH–Al), such as O
a, O
b, and O
c, do not protonate in our model calculations. Hydroxyl groups have a passive role in protolysis. After protonation of a bridge oxygen (e.g., O2,
Figure 4B), the hydroxyl groups help accommodate the structural distortions by reorganizing the Al–O bonds. This reorganization facilitates the dissociation of the tetrahedral and octahedral sheets, allowing the structure to open and leading to the breaking/hydrolysis of the structure.
The optimized configurations resulting from protonation reactions on the four studied edge faces led to protolysis at three of the edges: {100}, {010}, and {130}. However, the {110} face accommodated protons with minimal structural distortions, and no bonds were broken. The average values of the PRE (∆E
i) and the overall structural distortion (RMS calculated for all configurations in a series) for each face after one or two protons were added are highlighted. These results emphasize the unique behavior of the {110} face. The oxygens on this edge exhibited the highest PRE values of all the faces, but some of the lowest overall RMS of structural changes. These findings identify the {110} face as the least reactive among the four edges, which, according to Wulff’s rule, should be the predominant face in equilibrium crystals. This conclusion aligns with the morphology of idiomorphic pyrophyllite crystals (e.g., [
31]).
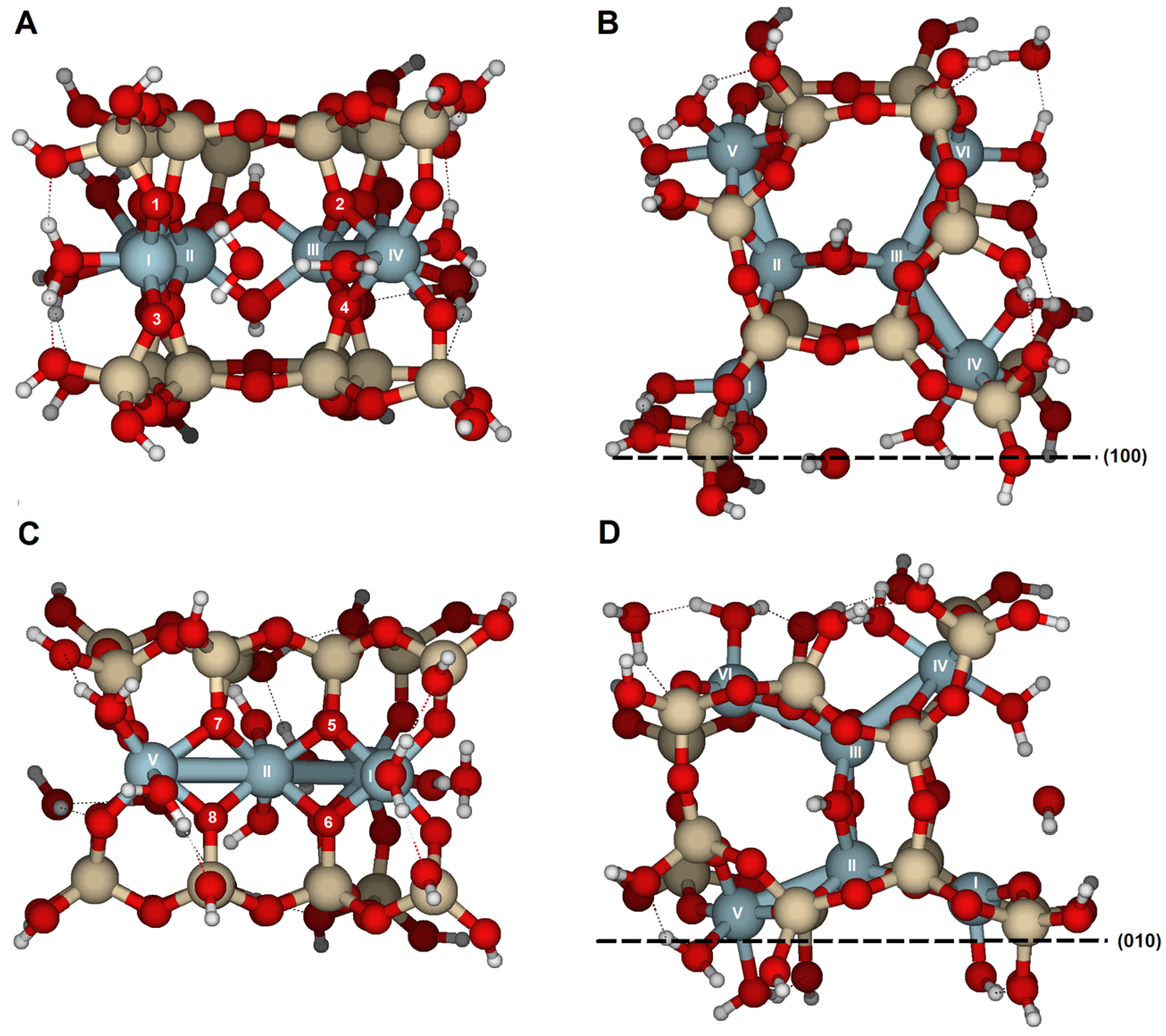
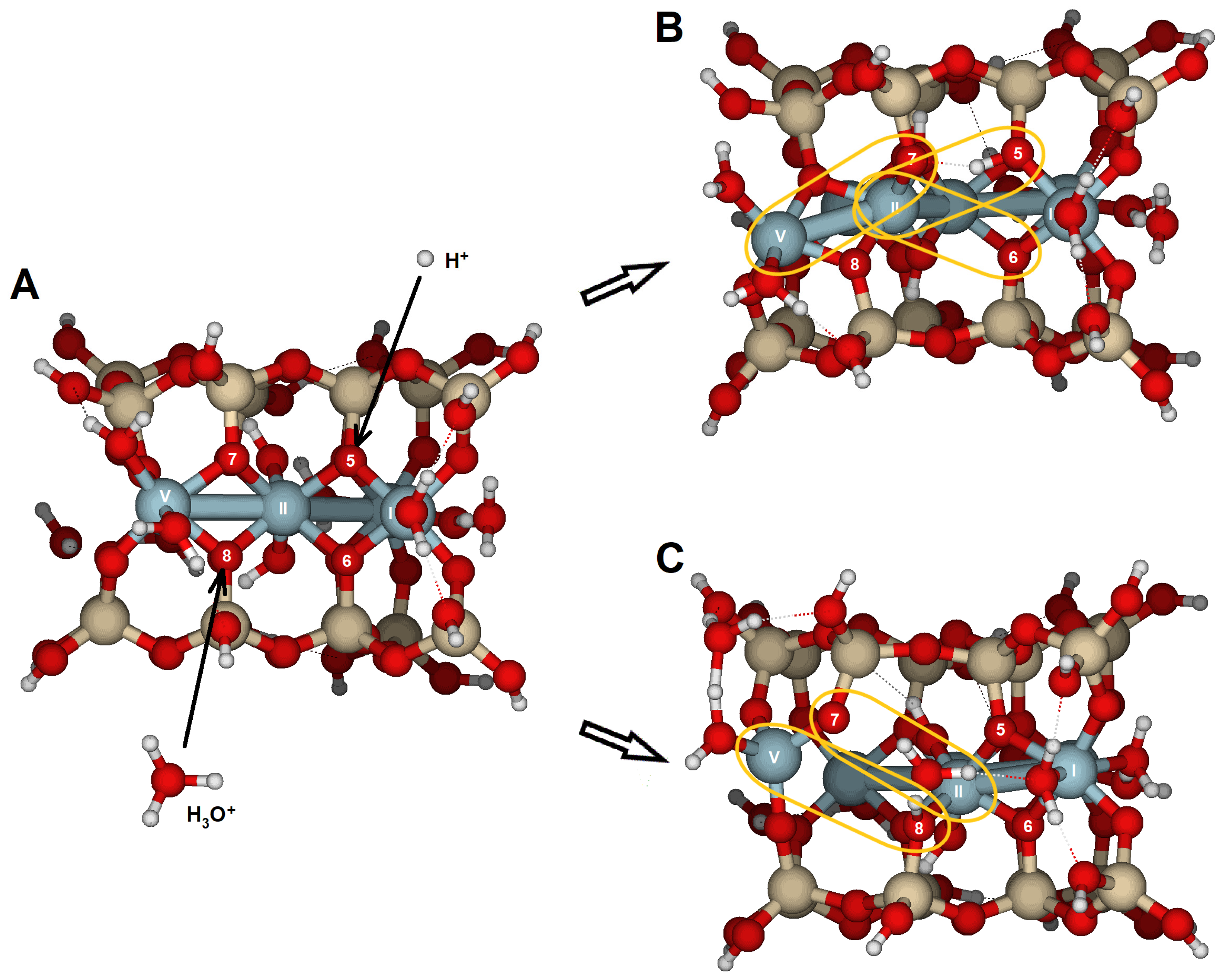
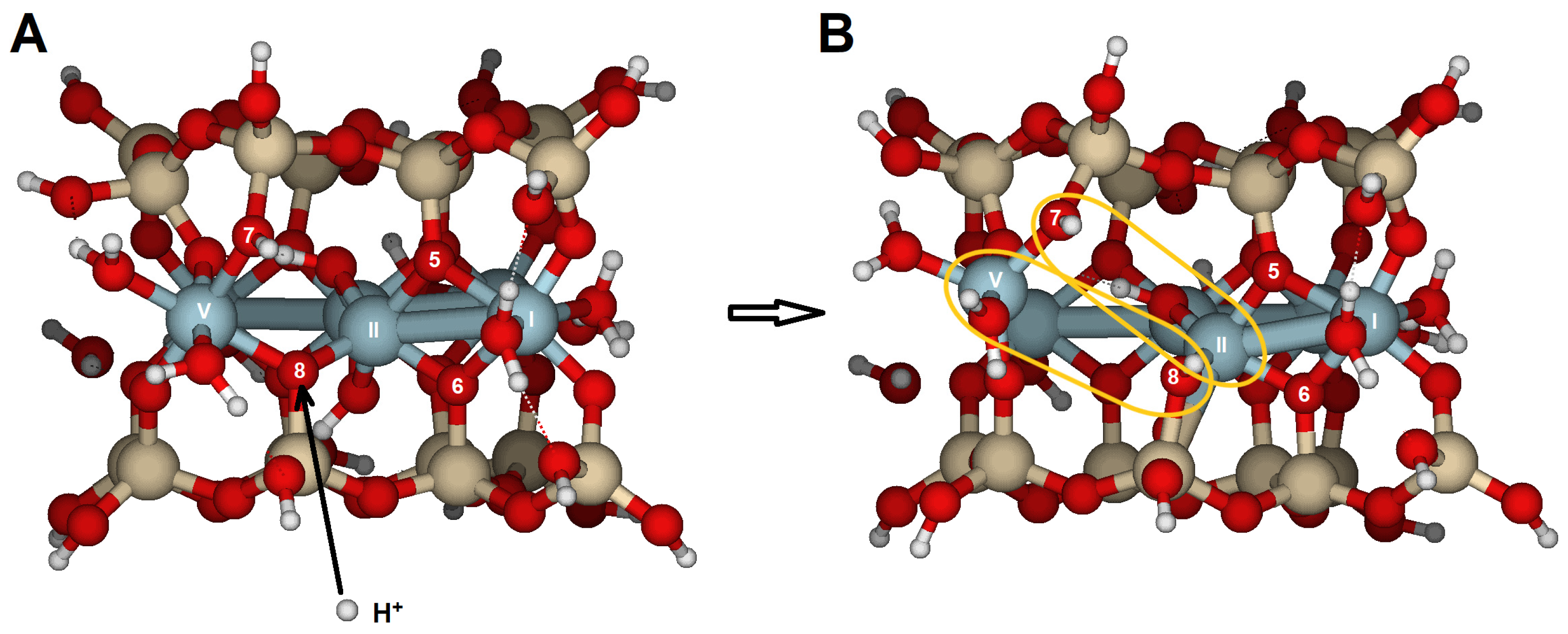
The {110} edge consists of a chain of octahedra (Al) sandwiched between two chains of tetrahedra (Si), arranged in a zig-zag pattern (
Figure 1A). The three types of oxygen are exposed (
Figure 3A,B). The optimized cluster configurations show their high PRE and the edge’s capacity to protonate with minimal structural distortions (
Table 6 and
Table 7;
Figure 10). The water molecules released after the dissociation of hydronium ions and the adsorption of protons appear to connect to the edge by completing the Al coordination sphere, while also enabling protons to migrate between oxygens through hydrogen bonds. These findings highlight the active role of water molecules in silicate dissolution by adsorption and as carriers of proton mobility. Likely, breaking bonds would have required prior protonation reactions, increased the positive charge density, and caused more severe structural distortions. Nonetheless, in natural conditions, with a complete set of water molecules in the hydration sphere, spare water molecules can also enter the solvation sphere and have no impact on the system stability.
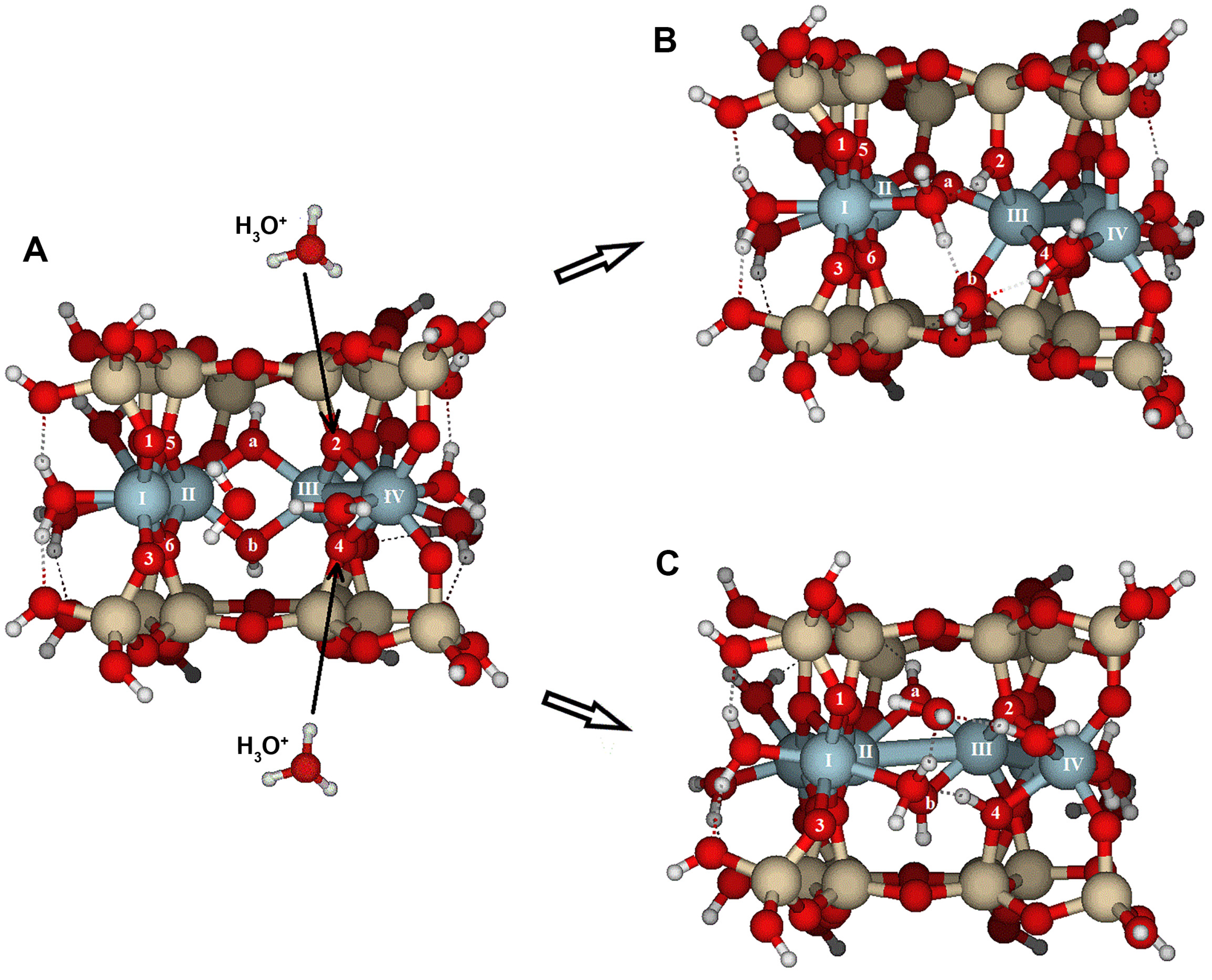
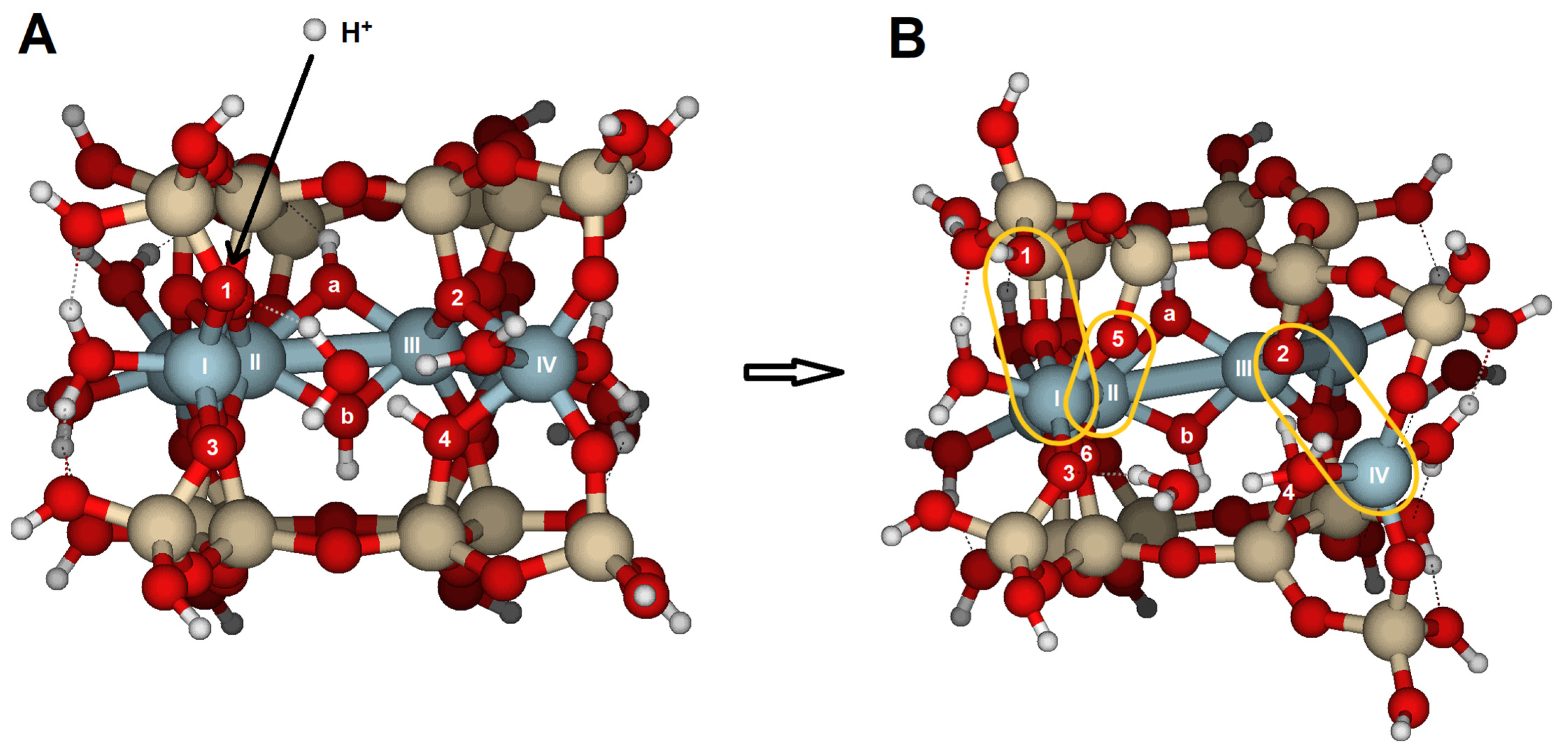
Protonation reactions at the {010} face caused significant structural distortions and protolysis (
Figure 8). The proton attack is most effective at O5, while the hydronium attack is more effective at O8. In both cases, protonation leads to broken bonds, with similar RMS values (
Table 3). However, an attack with hydronium results in Al(V) with 4-fold coordination. The optimized structure is significantly more open than after a single proton attack, with water molecules forming multiple hydrogen bonds. A configuration similar to that obtained after one hydronium attack required two protons, as shown in
Figure 9B and
Table 3 and
Table 4, highlighting the key role of water molecules during hydrolysis.
The {010} and {110} edges are quite similar (
Figure 1A) in terms of atom distribution. In {110} faces, two hydroxyl groups are exposed at the solid/solution interface. They bond with Al(IX) and Al(X) (
Figure 3A,B). However, hydroxyl groups are not exposed on the {010} faces (
Figure 2C,D). As mentioned earlier, hydroxyl groups are not reactive but help accommodate structural distortions caused by protonation reactions. This slight difference may explain the varying reactivity of the two edges. Although the {110} surface has the highest PRE among all the crystal edges, it can still accommodate structural distortions without protolysis. Therefore, the {110} face is
de facto the least reactive edge.
The {100} face appears to be more reactive than the {010} face, with slightly higher average PRE and lower overall RMS values for one- and two-proton attacks. The RMS values indicate that structural distortions are significant but more limited than at {010}. The zig-zag arrangement of Al atoms makes the edge relatively rigid. However, the {100} edge is formed by a succession of hemihexagonal octahedral cavities (
Figure 1A), which makes the edge more flexible and capable of accommodating distortions. The {100} face contains four bridge oxygens (e.g., O2 and O4) and two hydroxyls (O
a, O
b) within the hexagonal cavity, along with four outer bridge aluminol oxygens (e.g., O1, O3). Protonation of O1 and O3 alone is not enough to trigger hydrolysis. Protolysis becomes effective after protonation of bridge oxygens (Si–O < Al
2), although all protonations contribute to some extent. It is difficult to explain why O2 has a higher PRE than O4 (
Table 1). The optimized Al–O distances before protonation are slightly longer for O2 than for O4. Attack at O2 results in greater distortion, involving the breaking of the Al(IV)–O2 bond and the rearrangement of additional bonds, including the hydroxyl O
b. In contrast, the attack at O4 only results in the breaking of the Al(III)–O4 bond and a slightly less distorted structure. The second protonation increases distortion, leading to a more open hemihexagonal octahedral cavity. The most favorable combinations include a bridge oxygen (O2, O4) and a bridge aluminol oxygen (O1, O3). Clearly, the attack by a proton (or hydronium ion) triggers the hydrolysis of pyrophyllite. Additionally, bond breaking helps to relax the structural stress, resulting in a lower RMS value for the {100} edge compared to the {010} edge. Therefore, the {100} edge is a more reactive face than the {010}.
The PRE of the reactive oxygen (O13) at the {130} face is the lowest of the average values (
Table 8). The reaction is efficient, breaking the Al(XI)–O13 bond and opening the area attacked. These values and the optimized cluster configurations indicate that the {130} face is more reactive than the {110} face.
Only two reactive oxygens were identified in the cluster used to model the {130} edge. However, this might be an artifact of the small cluster size, which was limited to keep computational requirements manageable.
Figure 1A shows a similar arrangement of Al atoms on the {130} and {100} faces, featuring a series of hemihexagonal octahedral cavities. Differences result from the relative positions of tetrahedral and octahedral sheets. Although the edges are not identical, the reactive sites on both faces may be similar. Remember that for one-H
+-attack, the overall RMS of {130} is higher than {100}. Therefore, similar reactivity could be expected for faces {100} and {130}. Additionally, the modeling results for both faces may complement each other. The Al(XI) and Al(XII) might correspond to Al(IV) and Al(I), respectively, in clusters modeling the {130} and {100} edges. The Al(II) and Al(III) in {100} could provide insights into the reactivity of oxygens attached to the Al atoms in the innermost hemihexagonal cavity of the {130} face. Based on these points, the reactivity of the {130} edge may resemble that of the {100} edge.
The relative reactivities of the studied faces are approximately {100} ~ {130} > {010} >> {110}. This sequence aligns with Bleam et al.’s [
16] calculation, which indicated that forming {100} and {130} edges required about 16% more energy compared to {010} and {110}. According to Wulff’s rule, the particle morphology is primarily determined by the most stable or less reactive faces, which, in the case of pyrophyllite, are {010} and {110} [
31].
Modeling the attack with a hydronium ion instead of a proton results in the protonation of a reactive oxygen and the release of a water molecule. This water molecule stays nearby and helps relax the local excess or deficit of charge. Protonation caused by one H+ attack leads to notable structural distortions. In contrast, the H3O+ attack is usually more effective because water forms hydrogen bonds, completing the coordination sphere of hydrolyzed aluminum atoms, or acting as a pathway for proton migration between nearby oxygens. The PREs for hydronium attacks are about one-third of those for H+, and the distortion caused is similar, as shown by comparable overall RMS values. The positioning of water molecules near the affected area likely helps stabilize the hydration sphere, bringing it back to the normal surface hydration level. These findings highlight the critical role of water molecules in dissolution reactions.
Generally, the second protonation results in higher RMS values than the first. The attack of two protons (or a hydronium and a proton), whether simultaneous or sequential, begins on a highly reactive bridge oxygen. This increases the reactivity of other oxygens, such as the outer bridge aluminol oxygens, and promotes structural breakdown by splitting the octahedral and tetrahedral sheets. The protolysis mainly affects the octahedral cations, which are gradually surrounded by water molecules before being released into the solution. Fully releasing aluminum into solutions would require additional calculations, including accounting either for water molecules or a continuum solvent method to model the hydration layer surrounding the crystal. These second protonations, which have the highest overall RMS values, suggest that reducing the pH and increasing the temperature would promote pyrophyllite dissolution; highly acidic conditions might cause multiple proton attacks around reactive sites.
Although transition states between reactants and products have been studied, they have not yet been observed. This indicates that protolysis occurs directly, without an intermediate step. Consequently, no energy barrier was detected in any of the protolysis reactions, which is consistent with the molecular dynamics results of Schilemann and Churakov [
34]. Surface protonation reactions are reversible at amphoteric surface sites. Surface protonation shows very little temperature dependence [
37]. They suggested that the slight variations in proton adsorption constants likely stem from changes in the water dissociation constant (K
w) with temperature. This behavior aligns with a very low energy barrier as suggested by DFT calculations.
5. Conclusions
DFT modeling of the reactivity of the four main pyrophyllite edge faces under acidic conditions, represented by proton and/or hydronium attack, allowed the identification of face {110} as the most stable. The reactive oxygens correspond to the bridge aluminol oxygens (Si–O–Al). These oxygens can be protonated and accommodate the positive charge and structural distortions without causing protolysis of nearby bonds. Effective protolysis requires the attack of additional protons/hydroniums. This situation corresponds to more acidic conditions, that is, a lower pH.
In contrast, the other three faces, {100}, {130}, and {010}, exhibited higher reactivity. DFT calculations showed that this increased reactivity was associated with the presence of bridge oxygen groups (Si–O < Al2) linked to Al cations located in the second line. Once oxygen is protonated, the structure destabilizes and evolves by breaking a bond, initiating the separation of the octahedral and tetrahedral sheets, and ultimately leading to the hydrolysis and detachment of the octahedral cations. In addition, it was demonstrated that hydroxyl groups (Al–OH–Al) have a low protonation capacity and are therefore less reactive.
To conclude, we should focus on the primary goal of this study, which is to gain a deeper understanding of the protolysis reaction involved in the dissolution mechanisms of dioctahedral phyllosilicates under acidic conditions and to gather atomistic evidence of the initial steps of these processes. Pyrophyllite is proposed as a simplified model for dioctahedral phyllosilicates. Experimental methods cannot identify individual steps in the dissolution process. Dissolution experiments showed that the most reactive surfaces are the crystal edges, where proton adsorption triggers the process. DFT results are consistent with clay mineral titration experiments that concluded that reactive groups are located on bridge oxygens (Si–O < Al
2), which are much more reactive than bridge aluminol (Si–O–Al) and hydroxyl groups (Al–OH–Al) [
38,
49]. Protonation of these bridge oxygens initiates hydrolysis, confirming that hydrolysis of the bridge oxygens is the rate-limiting step (e.g., [
6,
9]). Protonation of bridge aluminol oxygens on the outermost edge surface induces and enhances the protonation of bridge oxygens, as shown by surface titration experiments [
37,
38]. When hydronium ions react instead of protons, they produce similar or greater structural changes, and water molecules form hydrogen bonds with the resulting structures. During dissolution experiments of dioctahedral phyllosilicates, octahedral cations are preferentially released [
6,
9,
50,
51,
52,
53]. This behavior results from the preferential hydrolysis of the bond linking octahedral cations to the tetrahedral sheets, as demonstrated in the modeling reported here.
Finally, we can see once again that the basic surface properties of the local area can be studied with molecular surface models, providing new insights at the atomistic level on the reactivity of these surfaces. Future research should model the dissolution using extended or periodic surface models, which better approximate the actual mineral/solution interface, including the role of the solvent (water). It will also be essential to explore the contribution of isomorphic substitutions present in most phyllosilicates. The atomistic evidence, primarily obtained from molecular models, will be beneficial in further understanding the role of phyllosilicates in various geological and environmental processes.