Identification and Differentiation of Non-Hemolytic Listeria monocytogenes from Food Processing Environments Using MALDI-TOF MS
Abstract
1. Introduction
2. Results and Discussion
2.1. Identification of L. monocytogenes Reference Strains and Environmental L. monocytogenes Isolates
2.2. Analysis of Protein Profiles in Terms of the Isolation Environment
2.3. Characterization of MALDI-TOF MS Peptide Profiles of L. monocytogenes Reference Strains and Environmental L. monocytogenes Isolates
2.4. Correlation of Peptide Profiles with Phenotypic Traits
2.5. Differentiating Between Reference and Environmental Strains
2.6. Determination of the Ability of Isolates for Hemolysis
2.7. Determination of the Susceptibility of L. monocytogenes to Antibiotics
3. Materials and Methods
3.1. Reference L. monocytogenes Strains
3.2. Environmental Isolates
3.3. Motility, Capability for β-Hemolysis, and Sensitivity to Antibiotics
3.4. Sample Storage and Preparation for MALDI-TOF MS
3.5. Sample Preparation for MALDI-TOF MS
3.6. MALDI-TOF MS Analysis
3.7. Statistical Data Analysis
4. Conclusions
5. Patents
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO. World Health Organization 2023. Available online: https://www.who.int/news-room/fact-sheets/detail/listeriosis (accessed on 15 April 2025).
- Guillet, C.; Join-Lambert, O.; Le Monnier, A.; Leclerq, A.; Mechai, F.; Mamzer-Bruneel, M.F.; Bielecka, M.; Scortti, M.; Disson, O.; Berche, P.; et al. Human listeriosis caused by Listeria ivanovii. Emerg. Infect. Dis. 2010, 16, 136. [Google Scholar] [CrossRef] [PubMed]
- Favaro, M.; Sarmati, L.; Sancesario, G.; Fontana, C. First case of Listeria innocua meningitis in a patient on steroids and etanercept. J. Med. Microbiol. Case Rep. 2014, 1, 003103. [Google Scholar] [CrossRef]
- Orsi, R.H.; Carlin, C.R.; Wiedmann, M. Taxonomy, ecology and relevance to food safety of the genus Listeria with a particular consideration of new Listeria species described between 2010 and 2022. mBio 2023, 15, e0093823. [Google Scholar] [CrossRef] [PubMed]
- Szymczak, B. Phenotypic and genotypic characteristics of non-hemolytic Listeria monocytogenes isolated from food and processing environments. Foods 2023, 12, 3630. [Google Scholar] [CrossRef] [PubMed]
- CDC. Listeria Outbreaks, Centers for Disease Control and Prevention. 2024. Available online: https://www.cdc.gov/listeria/causes/index.html (accessed on 2 May 2025).
- Kawacka, I.; Olejnik-Schmidt, A. Genoserotyping of Listeria monocytogenes originating from meat products and meat processing environments. Food. Sci. Tech. Qual. 2022, 29, 34–44. [Google Scholar] [CrossRef]
- ISO 11290-1:2017; Microbiology of the Food Chain—Horizontal Method for the Detection and Enumeration of Listeria monocytogenes and of Listeria spp.—Part 1: Detection Method. ISO International Organization for Standardization: Geneva, Switzerland, 2017. Available online: https://www.iso.org/standard/60313.html (accessed on 1 March 2025).
- Feng, Y.; Yao, H.; Chen, S.; Sun, X.; Yin, Y.; Jiao, X. Rapid detection of hypervirulent serovar 4h Listeria monocytogenes by multiplex PCR. Front. Microbiol. 2020, 11, 1309. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Z.; Li, S.; Boeren, S.; Smid, E.J.; Notebaart, R.A.; Abee, T. Anaerobic growth of Listeria monocytogenes on rhamnose is stimulated by vitamin B12 and bacterial microcompartment—Dependent 1,2—Propanediol utilization. mSphere 2021, 6, e0043421. [Google Scholar] [CrossRef] [PubMed]
- Commission Regulation (EU) 2024/2895 of 20 November 2024 amending Regulation (EC) No 2073/2005 as regards Listeria monocytogenes C/2024/7974. Available online: http://data.europa.eu/eli/reg/2024/2895/oj (accessed on 2 December 2024).
- Karasu-Yalcin, S.; Soylemez-Milli, N.; Eren, K.; Eryasar-Orer, O. Reducing time in detection of Listeria monocytogenes from food by MALDI-TOF mass spectrometry. J. Food Sci. Technol. 2020, 58, 4102–4109. [Google Scholar] [CrossRef] [PubMed]
- Koudelka, S.; Gelbicova, T.; Prochazkova, M.; Karpiskova, R. Lineage and serotype identification of Listeria monocytogenes by matrix-assisted laser desorption ionization-time of flight mass spectrometry. Czech J. Food Sci. 2018, 36, 452–458. [Google Scholar] [CrossRef]
- Pyz-Łukasik, R.; Gondek, M.; Winiarczyk, D.; Michalak, K.; Paszkiewicz, W.; Piróg-Komorowska, A.; Policht, A.; Ziomek, M. Occurence of Listeria monocytogenes in artisanal cheeses from Poland and its identification by MALDI-TOF MS. Pathogens 2021, 10, 632. [Google Scholar] [CrossRef] [PubMed]
- Wenning, M.; Bretenwieser, F.; Konrad, R.; Huber, I. Identification and differentiation of food-related bacteria: A comparison of FTIR spectroscopy and MALDI TOF mass spectrometry. J. Microbiol. Methods 2014, 103, 44–52. [Google Scholar] [CrossRef] [PubMed]
- Popovic, N.T.; Kazazić, S.P.; Strunjak-Perović, I.; Coz-Rakovac, R. Differentiation of environmental aquatic bacterial isolates by MALDI-TOF MS. Environ. Res. 2017, 152, 7–16. [Google Scholar] [CrossRef] [PubMed]
- Starostin, K.V.; Demidov, E.A.; Bryanskaya, A.V.; Efimov, V.M.; Rozanov, A.S.; Peltek, S.E. Identification of Bacillus strains by MALDI TOF MS using geometric approach. Sci. Rep. 2015, 5, 16989. [Google Scholar] [CrossRef] [PubMed]
- Seuylemezian, A.; Aronson, H.S.; Tan, J.; Lin, M.; Schubert, W.; Vaishampayan, P. Development of a custom MALDI-TOF MS database for species-level identification of bacterial isolates collected from spacecraft and associated surfaces. Front. Microbiol. 2018, 9, 780. [Google Scholar] [CrossRef] [PubMed]
- Kawacka, I.; Olejnik-Schmidt, A.; Schmidt, M. Nonhemolytic Listeria monocytogenes—Prevalence rate, reasons underlying atypical phenotype, and methods for accurate hemolysis assessment. Microorganisms 2022, 10, 483. [Google Scholar] [CrossRef] [PubMed]
- Finn, l.; Onyeaka, H.; O’Neill, S. Listeria monocytogenes biofilms in food-associated environments: A persistent enigma. Foods 2023, 12, 3339. [Google Scholar] [CrossRef] [PubMed]
- Quereda, J.J.; Moron-Garcia, A.; Palacios-Gorba, C.; Dessaux, C.; Garcia-del Portillo, F.; Graciela Pucciarelli, M.; Ortega, A.D. Pathogenicity and virulence of Listeria monocytogenes: A trip from environmental to medical microbiology. Virulence 2021, 12, 2509–2545. [Google Scholar] [CrossRef] [PubMed]
- Akimowicz, M.; Bucka-Kolendo, J. MALDI-TOF MS—Application in food microbiology. Acta Biochim. Pol. 2020, 67, 327–332. [Google Scholar] [CrossRef] [PubMed]
- Jadhav, S.; Gulati, V.; Fox, E.M.; Karpe, A.; Beale, D.J.; Sevior, D.; Bhave, M.; Palombo, E.A. Rapid identification and source-tracking of Listeria monocytogenes using MALDI-TOF MS. Int. J. Food Microbiol. 2015, 202, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Hazen, T.H.; Martinez, R.J.; Chen, Y.; Lafon, P.C.; Garrett, N.M.; Parsons, M.B.; Sobecky, P.A. Rapid identification of Vibrio parahaemolyticus by whole-cell matrix-assisted laser desorption ionization-time of flight mass spectrometry. Appl. Environ. Microbiol. 2009, 75, 6745–6756. [Google Scholar] [CrossRef] [PubMed]
- Niestępski, S.; Harnisz, M.; Korzeniewska, E.; Osińska, A. Isolation of anaerobic bacteria of the Bacteroides fragilis group from environmental samples. In Proceedings of the 11th Conference on Interdisciplinary Problems in Environmental Protection and Engineering EKO-DOK 2019, Polanica-Zdrój, Poland, 8–10 April 2019; E3S Web Conf.; EDP Sciences: Les Ulis, France, 2019; Volume 100, p. 00058. [Google Scholar] [CrossRef]
- Ashfaq, M.Y.; Da’na, D.A.; Al-Ghouti, M.A. Application of MALDI-TOF MS for identification of environmental bacteria: A review. J. Environ. Manag. 2022, 305, 114359. [Google Scholar] [CrossRef] [PubMed]
- Thouvenot, P.; Vales, G.; Bracq-Dieye, H.; Tessaud-Rita, N.; Maury, M.M.; Moura, A.; Lecuit, M.; Leclerq, A. MALDI-TOF mass spectrometry-based identification of Listeria species in surveillance: A prospective study. J. Microbiol. Methods 2018, 144, 29–32. [Google Scholar] [CrossRef] [PubMed]
- Karbiwnyk, C.; Nevins, C.; Wetherby, A.; Geyer, R.; Silverman, M.; Wei, C. Listeria monocytogenes identification by subtyping MALDI-TOF MS Biotyper™ analysis. FDA Lab. Inf. Bull. 2019, 4658. Available online: https://www.fda.gov/media/169755 (accessed on 1 March 2025).
- Dos Reis, C.M.F.; Ramos, G.L.P.A.; Pereira, R.C.L.; Vallim, D.C.; Costa, L.E.O. Evaluation of VITEK 2 and MALDI TOF/MS automated methodologies in the identification of atypical Listeria spp. isolated from food in different regions of Brazil. J. Microbiol. Methods 2022, 194, 106434. [Google Scholar] [CrossRef] [PubMed]
- Rychert, J.; Burnham, C.A.; Bythrow, M.; Garner, O.B.; Ginocchio, C.C.; Jennemann, R.; Lewinski, M.A.; Manji, R.; Mochon, A.B.; Procop, G.W.; et al. Multicenter evaluation of the Vitek MS matrix-assisted laser desorption ionization-time of flight mass spectrometry system for identification of Gram-positive aerobic bacteria. J. Clin. Microbiol. 2013, 51, 2225–2231. [Google Scholar] [CrossRef] [PubMed]
- Farfour, E.; Leto, J.; Barritault, M.; Barberis, C.; Meyer, J.; Dauphin, B.; Le Guern, A.S.; Leflèche, A.; Badell, E.; Guiso, N.; et al. Evaluation of the Andromas matrix-assisted laser desorption ionization–time of flight mass spectrometry system for identification of aerobically growing Gram-positive bacilli. J. Clin. Microbiol. 2012, 50, 2702–2707. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Gan, Z.; Zhou, X.; Chen, Z. Accurate classification of Listeria species by MALDI-TOF mass spectrometry incorporating denoising autoencoder and machine learning. J. Microbiol. Methods 2022, 192, 106378. [Google Scholar] [CrossRef] [PubMed]
- Barbuddhe, S.B.; Maier, T.; Schwarz, G.; Kostrzewa, M.; Hof, H.; Domann, E.; Chakraborty, T.; Hain, T. Rapid identification and typing of Listeria species by matrix-assisted laser desorption ionization-time of flight mass spectrometry. Appl. Environ. Microbiol. 2008, 74, 5402–5407. [Google Scholar] [CrossRef] [PubMed]
- Abdelhamed, H.; Nho, S.W.; Kim, S.W.; Reddy, J.S.; Park, S.B.; Sung, J.T.; Lawrence, M.L. Serotype-identifying ions in Listeria monocytogenes using matrix-assisted laser desorption ionization-time of flight mass spectrometry. Heliyon 2022, 8, e11769. [Google Scholar] [CrossRef] [PubMed]
- Mandal, G.; Moráň, L.; Pečinka, L.; Vaňhara, P.; Havel, J. Matrix enrichment by black phosphorus improves ionization and reproducibility of mass spectrometry of intact cells, peptides, and amino acids. Sci. Rep. 2022, 12, 1175. [Google Scholar] [CrossRef] [PubMed]
- Tracy, M.B.; Chen, H.; Weaver, D.M.; Malyarenko, D.I.; Sasinowski, M.; Cazares, L.H.; Drake, R.R.; Semmes, O.J.; Tracy, E.R.; Cooke, W.E. Precision enhancement of MALDI-TOF MS using high-resolution peak detection and label-free alignment. Proteomics 2008, 8, 1530–1538. [Google Scholar] [CrossRef] [PubMed]
- Clark, C.M.; Costa, M.S.; Sanchez, L.M.; Murphy, B.T. Coupling MALDI-TOF mass spectrometry protein and specialized metabolite analyses to rapidly discriminate bacterial function. Proc. Natl. Acad. Sci. USA 2018, 115, 4981–4986. [Google Scholar] [CrossRef] [PubMed]
- Nomura, F. Proteome-based bacterial identification using MALDI-TOF MS: A revolutionary shift in clinical diagnostic microbiology. Biochim. Biophys. Acta Proteins Proteomics 2015, 1854, 528–537. [Google Scholar] [CrossRef] [PubMed]
- Sauget, M.; Valot, B.; Bertrand, X.; Hocquet, D. Can MALDI-TOF mass spectrometry reasonably type bacteria? Trends Microbiol. 2017, 25, 447–455. [Google Scholar] [CrossRef] [PubMed]
- Hsueh, P.R.; Lee, T.F.; Du, S.H.; Teng, S.H.; Liao, C.H.; Sheng, W.H. Bruker biotyper matrix-assisted laser desorption ionization-time of flight mass spectrometry system for identification of Nocardia, Rhodococcus, Kocuria, Gordonia, Tsukamurella, and Listeria species. J. Clin. Microbiol. 2014, 52, 2371–2379. [Google Scholar] [CrossRef] [PubMed]
- Samad, R.A.; Al Disi, Z.; Ashfaq, M.Y.M.; Wahib, S.M.; Zouari, N. The use of principal component analysis and MALDI-TOF MS for the differentiation of mineral forming Virgibacillus and Bacillus species isolated from sabkhas. RSC Adv. 2020, 10, 14606–14616. [Google Scholar] [CrossRef] [PubMed]
- Rychli, K.; Müller, A.; Zaiser, A.; Schoder, D.; Allerberger, F.; Wagner, M.; Schmitz-Esser, S. Genome sequencing of Listeria monocytogenes “Quargel” listeriosis outbreak strains reveals two different strains with distinct in vitro virulence potential. PLoS ONE 2014, 9, e89964. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Zhang, Y.; Zhang, T.; Xu, M.; Wang, H.; Zhang, S.; Zhang, T.; Zhou, W. Establishing a MALDI TOF MS method for rapid identification of three common Gram-positive bacteria (Bacillus cereus, Listeria monocytogenes, and Micrococcus luteus) associated with foodborne diseases. Food Sci. Technol. 2022, 42, e117021. [Google Scholar] [CrossRef]
- Luciani, M.; Krasteva, I.; Schirone, M.; D’Onofrio, F.; Iannetti, L.; Torresi, M.; Di Pancrazio, C.; Perletta, F.; Valeninuzzi, S.; Tittarelli, M.; et al. Adaptive strategies of Listeria monocytogenes: An in-depth analysis of the virulent strain involved in an outbreak in Italy through quantitative proteomics. Int. J. Food Microbiol. 2025, 427, 110951. [Google Scholar] [CrossRef] [PubMed]
- Maury, M.M.; Chenal-Francisque, V.; Bracq-Dieye, H.; Han, L.; Leclercq, A.; Vales, G.; Moura, A.; Gouin, E.; Scortti, M.; Disson, O.; et al. Spontaneous loss of virulence in natural populations of Listeria monocytogenes. Infect. Immun. 2017, 85, e00541-17. [Google Scholar] [CrossRef] [PubMed]
- Abril, A.G.; Carrera, M.; Böhme, K.; Barros-Velázquez, J.; Calo-Mata, P.; Sánchez-Pérez, A.; Villa, T.G. Proteomic characterization of antibiotic resistance in Listeria and production of antimicrobial and virulence factors. Int. J. Mol. Sci. 2021, 22, 8141. [Google Scholar] [CrossRef] [PubMed]
- Ojima-Kato, T.; Yamamoto, N.; Takahashi, H.; Tamura, H. Desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) can precisely discriminate the lineages of Listeria monocytogenes and species of Listeria. PLoS ONE 2016, 11, e0159730. [Google Scholar] [CrossRef] [PubMed]
- Griffin, P.M.; Price, G.R.; Schooneveldt, J.M.; Schlebusch, S.; Tilse, M.H.; Urbanski, T.; Venter, D. Use of MALDI-TOF MS to identify vancomycin-resistant enterococci and investigate the epidemiology of an outbreak. J. Clin. Microbiol. 2012, 50, 2918–2931. [Google Scholar] [CrossRef] [PubMed]
- Florio, W.; Tavanti, A.; Barnini, S.; Ghelardi, E.; Lupetti, A. Recent advances and ongoing challenges in the use of MALDI-TOF mass spectrometry for diagnostic microbiology. Expert Rev. Proteom. 2020, 17, 593–607. [Google Scholar] [CrossRef]
- Johansson, A.; Nagy, E.; Soki, J. Detection of carbapenemase activities of Bacteroides fragilis strains with MALDI-TOF MS. Anaerobe 2014, 26, 49–52. [Google Scholar] [CrossRef] [PubMed]
- Feucherolles, M.; Cauchie, H.M.; Penny, C. MALDI-TOF mass spectrometry and specific biomarkers: Potential new key for swift identification of antimicrobial resistance in foodborne pathogens. Microorganisms 2019, 7, 593. [Google Scholar] [CrossRef] [PubMed]
- EUCAST. European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters. 2022. Version 12.0. Available online: http://www.eucast.org (accessed on 15 April 2025).
- WHO. Critically Important Antimicrobials for Human Medicine: Categorization for the Development of Risk Management Strategies to Contain Antimicrobial Resistance Due to Non-Human Antimicrobial Use Report of the Second WHO Expert Meeting, 2007, Copenhagen. pp. 29–31. Available online: https://iris.who.int/handle/10665/43765 (accessed on 15 January 2025).
- Szymczak, B. Method of Modifying the Revival of Atypical Listeria monocytogenes Bacteria and Isolation of their DNA. Patent No. Pat.245727, 16 July 2024. (In Polish). [Google Scholar]
- Freiwald, A.; Sauer, S. Phylogenetic classification and identification of bacteria by mass spectrometry. Nat. Protoc. 2009, 4, 732–742. [Google Scholar] [CrossRef] [PubMed]
- Szymczak, B. Method for Determining the Sensitivity of Listeria Monocytogenes Bacteria, Especially Atypical Strains, to Antibiotics. P.450342, 25 November 2024. [Google Scholar]
- Szymczak, B. Method for Identifying Typical and Atypical Isolates of Listeria Monocytogenes Bacteria Capable of Hemolysis. P.449917, 30 September 2024. [Google Scholar]
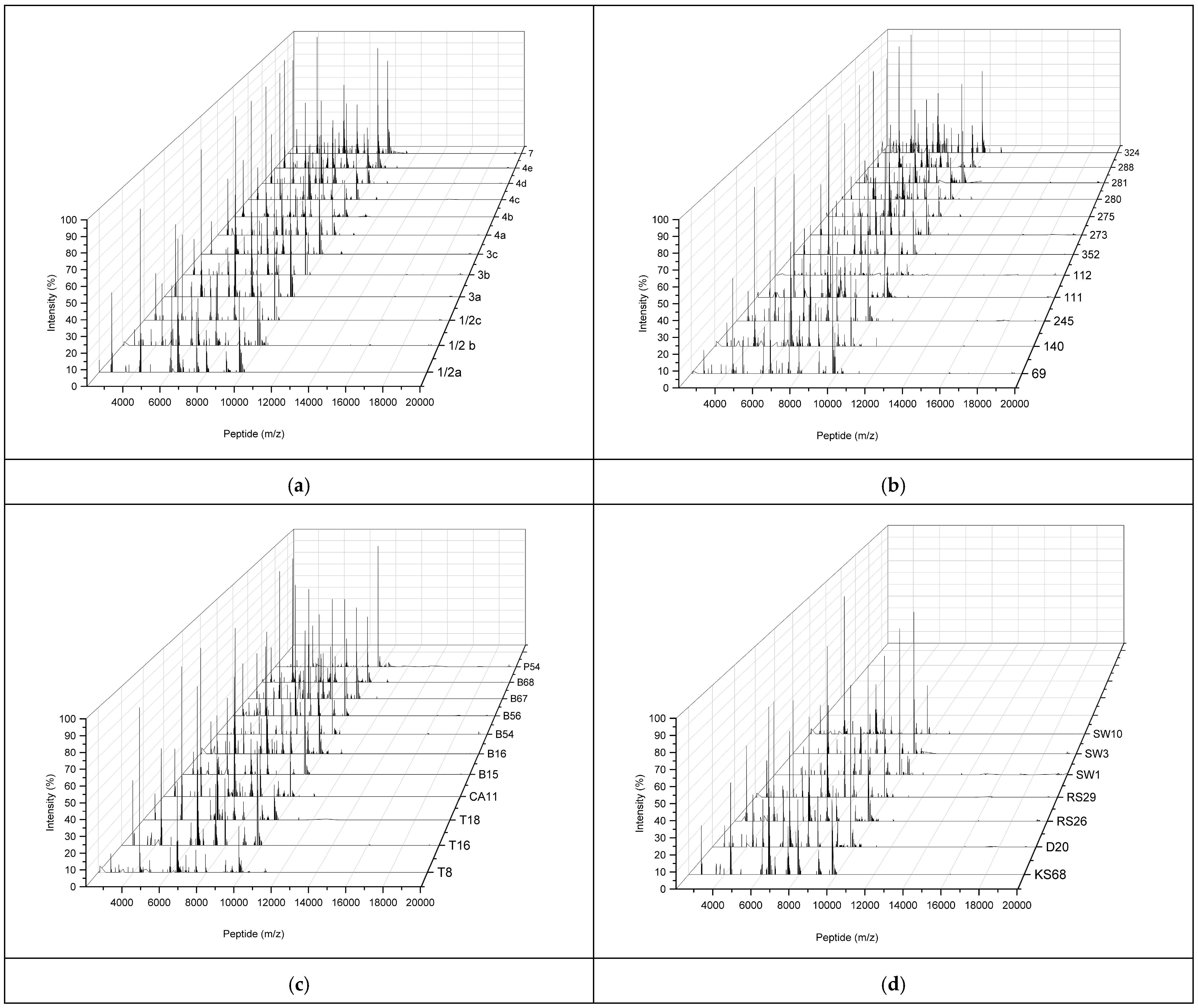
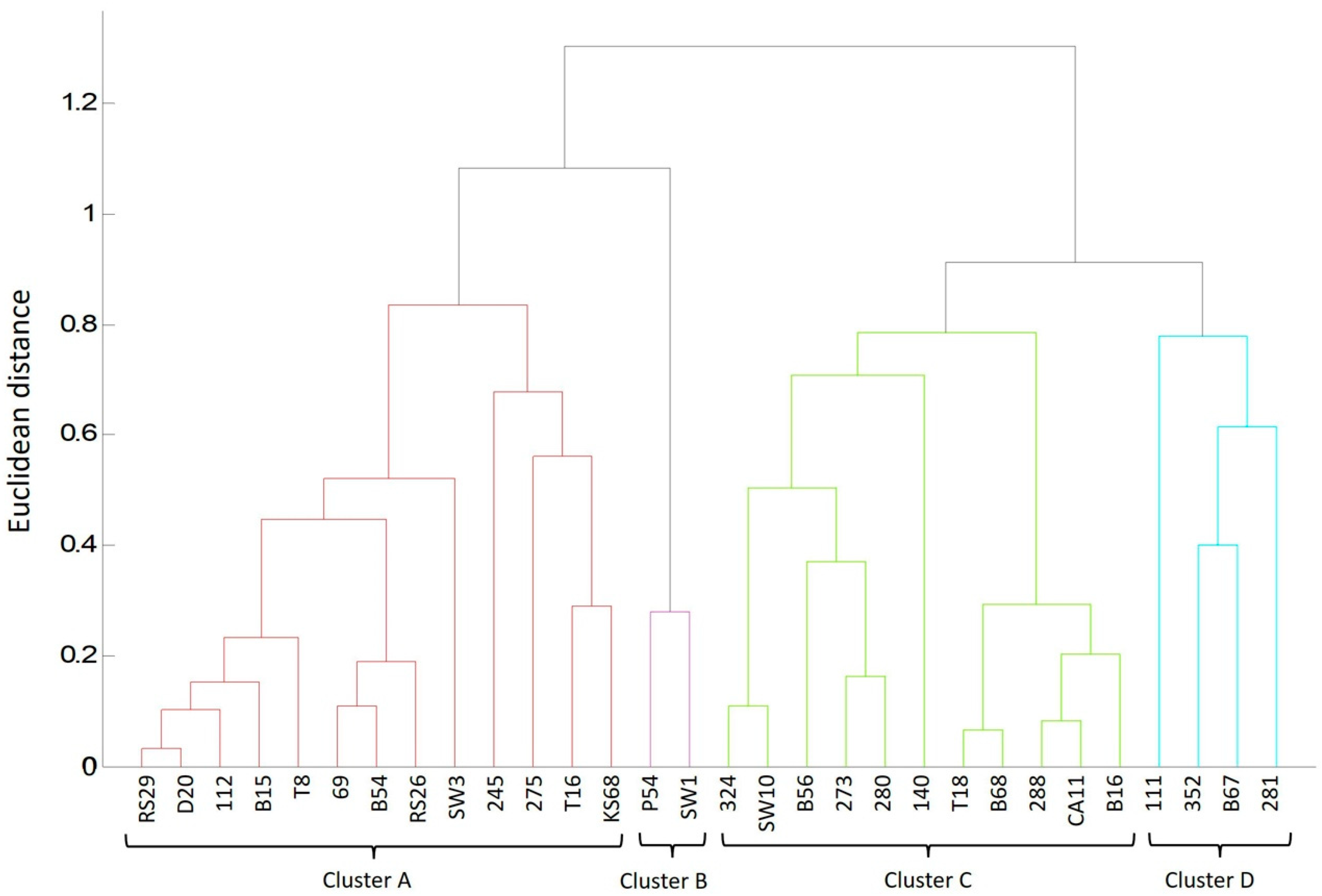
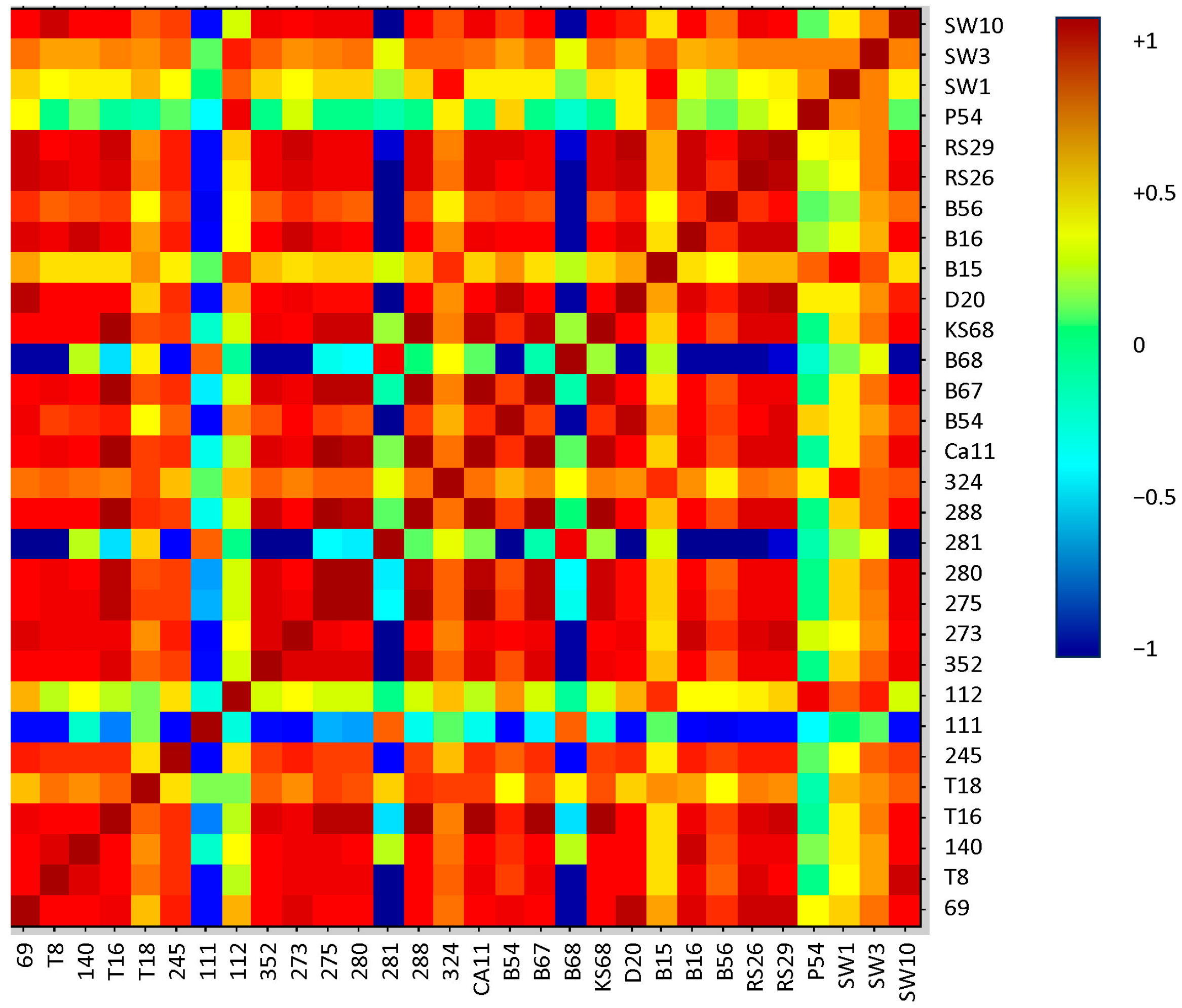
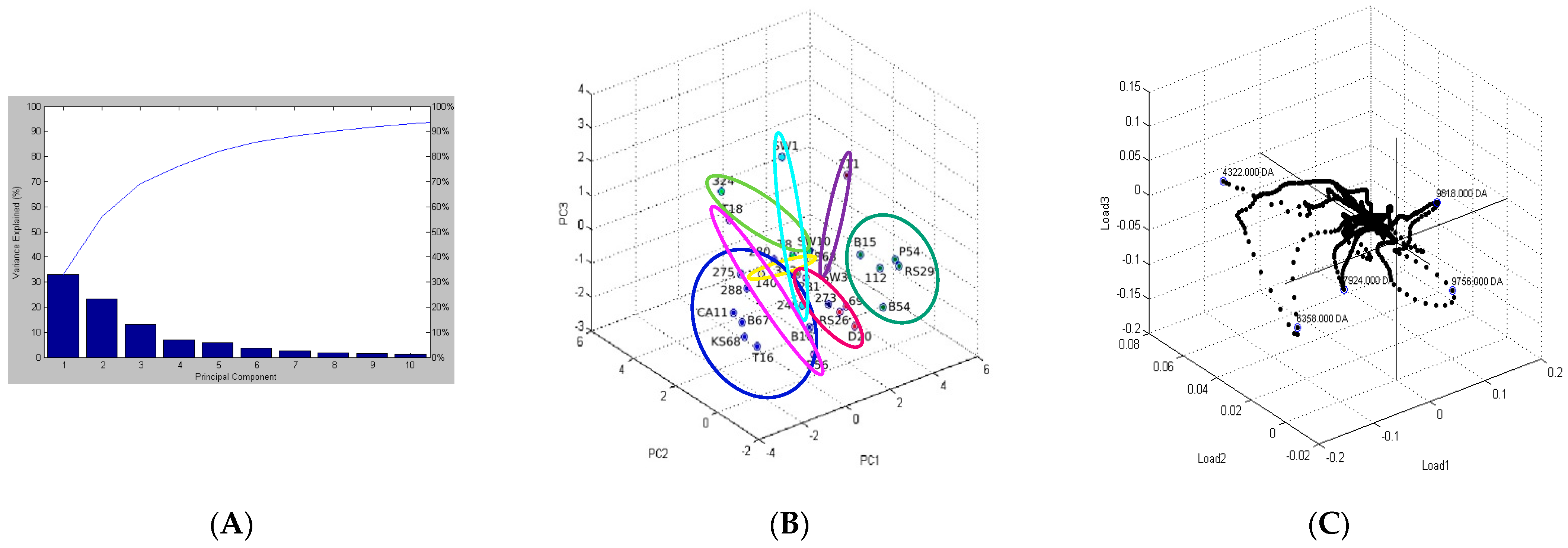
| Isolate IDs. | Bruker Database | In-House Database | ||||
|---|---|---|---|---|---|---|
| Score Value (Best Match) | Score Value (Second Best Batch) | Species ID According to MALDI Biotyper 3.1 | Score Value (Best Match) | Score Value (Second Best Batch) | Species ID According to MALDI Biotyper 3.1 | |
| 1/2a | 1.708 | 1.694 | L. innocua | 2.732 | 2.354 | L. monocytogenes ATCC 19111 |
| 1/2b | 2.263 | 2.214 | L. monocytogenes | 2.732 | 2.285 | L. monocytogenes CIP 7832 |
| 1/2c | 1.880 | 1.865 | L. monocytogenes | 2.655 | 2.324 | L. monocytogenes ATTC 10112 |
| 3a | 1.737 | 1.735 | L. monocytogenes | 2.685 | 2.374 | L. monocytogenes ATCC 19113 |
| 3b | 1.860 | 1.821 | L. monocytogenes | 2.724 | 2.412 | L. monocytogenes CIP 7835 |
| 3c | 1.902 | 1.788 | L. monocytogenes | 2.778 | 2.271 | L. monocytogenes CIP 7836 |
| 4a | 1.841 | 1.817 | L. monocytogenes | 2.732 | 2.328 | L. monocytogenes ATCC 19114 |
| 4b | 1.801 | 1.769 | L. monocytogenes | 2.602 | 2.232 | L. monocytogenes ATCC 13932 |
| 4c | 1.805 | 1.734 | L. monocytogenes | 2.644 | 2.355 | L. monocytogenes ATCC 19116 |
| 4d | 1.867 | 1.865 | L. monocytogenes | 2.623 | 2.352 | L. monocytogenes ATCC 19117 |
| 4e | 1.841 | 1.817 | L. innocua | 2.670 | 2.258 | L. monocytogenes ATCC 19118 |
| 7 | 1.818 | 1.808 | L. monocytogenes | 2.708 | 2.364 | L. monocytogenes NCTC 10890 |
| Average | 1.860 | 1.827 | 2.690 | 2.326 | ||
| Isolate IDs. | Bruker Database | In-House Database | ||||
|---|---|---|---|---|---|---|
| Score Value (Best Match) | Score Value (Second Best Batch) | Species ID According to MALDI Biotyper 3.1 | Score Value (Best Match) | Score Value (Second Best Batch) | Species ID According to MALDI Biotyper 3.1 | |
| 69 | 2.245 | 2.117 | L. innocua | 2.419 | 2.255 | L. monocytogenes |
| 140 | 2.172 | 2.110 | L. innocua | 2.307 | 2.289 | L. monocytogenes |
| 245 | 1.961 | 1.911 | L. innocua/L. monocytogenes | 2.377 | 2.356 | L. monocytogenes |
| 111 | 1.465 | 1.359 | Not a reliable identification | 1.686 | 1.362 | Not a reliable identification |
| 112 | 2.133 | 2.126 | L. innocua | 2.187 | 2.053 | L. monocytogenes |
| 352 | 1.808 | 1.606 | L. monocytogenes/not a reliable identification | 2.373 | 2.359 | L. monocytogenes |
| 273 | 1.974 | 1.970 | L. innocua | 2.273 | 2.242 | L. monocytogenes |
| 275 | 1.895 | 1.825 | L. innocua | 2.265 | 2.088 | L. monocytogenes |
| 280 | 2.003 | 1.889 | L. innocua | 2.374 | 2.326 | L. monocytogenes |
| 281 | 1.750 | 1.686 | L. innocua/not a reliable identification | 2.109 | 2.106 | L. monocytogenes |
| 288 | 1.739 | 1.734 | L. innocua/L. seeligeri | 2.133 | 2.122 | L. monocytogenes |
| 324 | 2.076 | 2.066 | L. innocua/L. monocytogenes | 2.366 | 2.310 | L. monocytogenes |
| Average for soil | 1.954 | 1.867 | 2.221 | 2.156 | ||
| T8 | 2.129 | 2.088 | L. innocua | 2.353 | 2.330 | L. monocytogenes |
| T16 | 1.908 | 1.888 | L. innocua | 1.723 | 1.662 | L. monocytogenes |
| T18 | 1.762 | 1.727 | L. monocytogenes/L. innocua | 2.293 | 2.268 | L. monocytogenes |
| Average for fruits | 1.933 | 1.988 | 2.123 | 2.087 | ||
| CA11 | 2.028 | 1.994 | L. innocua | 2.353 | 2.247 | L. monocytogenes |
| B15 | 1.812 | 1.771 | L. monocytogenes | 1.897 | 1.788 | L. monocytogenes |
| B16 | 2.183 | 2.127 | L. monocytogenes | 2.288 | 2.259 | L. monocytogenes |
| B54 | 1.994 | 1.968 | L. monocytogenes | 2.700 | 2.007 | L. monocytogenes |
| B56 | 1.745 | 1.741 | L. ivanovii/L. innocua | 2.165 | 2.165 | L. monocytogenes |
| B67 | 1.741 | 1.718 | L. seeligeri/L. innocua | 2.272 | 2.191 | L. monocytogenes |
| B68 | 1.937 | 1.929 | L. innocua/L. monocytogenes | 2.369 | 2.315 | L. monocytogenes |
| P54 | 2.077 | 2.071 | L. innocua | 1.688 | 1.575 | L. monocytogenes |
| Average for vegetables | 1.940 | 1.915 | 2.217 | 2.068 | ||
| KS68 | 1.737 | 1.657 | L. innocua/not a reliable identification | 2.169 | 2.135 | L. monocytogenes |
| D20 | 2.047 | 1.967 | L. innocua | 2.008 | 1.874 | L. monocytogenes |
| RS26 | 2.069 | 2.062 | L. innocua | 2.346 | 2.334 | L. monocytogenes |
| RS29 | 2.120 | 2.117 | L. innocua | 2.346 | 2.335 | L. monocytogenes |
| Average for RTE Food | 1.993 | 1.951 | 2.217 | 2.170 | ||
| SW1 | 1.823 | 1.807 | L. innocua | 2.121 | 2.025 | L. monocytogenes |
| SW3 | 1.817 | 1.772 | L. monocytogenes | 2.351 | 2.345 | L. monocytogenes |
| SW10 | 2.040 | 2.028 | L. monocytogenes | 2.442 | 2.382 | L. monocytogenes |
| Average for swabs | 1.893 | 1.869 | 2.305 | 2.251 | ||
| All isolates average | 1.947 | 1.900 | 2.218 | 2.137 | ||
| Environment | Sampling Locations | Isolate Number | Motility | β-Hemolysis |
|---|---|---|---|---|
| Soil (S) | Arable soil with natural fertilization (S1) | 245 | - | - |
| 280 | - | - | ||
| 281 | - | - | ||
| 288 | - | - | ||
| Garden plots from (S3) | 112 | - | - | |
| 324 | - | - | ||
| Intensive cattle grazing | 273 | - | - | |
| 275 | - | - | ||
| Forest from (S4) | 352 | - | - | |
| 69 | - | - | ||
| The area around the meat processing plant | 111 | - | - | |
| 140 | - | - | ||
| Fruits | Strawberry from S1 | T8 | - | - |
| Strawberry from S3 | T16 | - | - | |
| T18 | - | - | ||
| Vegetables | Beetroot from S1 | B15 | - | - |
| B16 | - | - | ||
| B67 | - | + | ||
| B68 | - | + | ||
| Beetroot from S3 | B54 | - | - | |
| B56 | - | - | ||
| Carrot from S1 | CA11 | - | + | |
| Potato from S3 | P54 | - | + | |
| RTE food | Dumplings | D20 | - | - |
| Kale sprouts | KS68 | - | + | |
| Radish sprouts | RS26 | - | - | |
| RS29 | - | - | ||
| Swabs | Seal in a hall door | SW10 | - | - |
| Pasteurizer belt | SW1 | - | - | |
| Pasteurizer rollers | SW3 | - | - |
| Biomarkers of Environmental L. monocytogenes Isolates | Biomarkers of Hemolysis Ability | ||
|---|---|---|---|
| Peptide Mass (Da) | R2 and p-Value | Peptide Mass (Da) | R2 and p-Value |
| 2755.36 | −0.6728 p = 0.000 | 2738.09 | +0.4194 p = 0.006 |
| 2776.58 | −0.5118 p = 0.001 | 2755.36 | +0.4225 p = 0.005 |
| 2782.37 | −0.4783 p = 0.001 | 2776.58 | +0.5162 p = 0.000 |
| 2793.32 | −0.6524 p = 0.000 | 2793.32 | +0.4553 p = 0.002 |
| 4361.57 | −0.5411 p = 0.000 | 4361.60 | +0.4021 p = 0.008 |
| 6360.25 | +0.4042 p = 0.008 | 9036.76 | +0.4357 p = 0.004 |
| 6388.02 | −0.5305 p = 0.000 | 9390.72 | +0.3964 p = 0.009 |
| 7420.81 | −0.5282 p = 0.000 | 9750.40 | +0.5555 p = 0.000 |
| Name of Antibiotic | Mass of Peptide (Da) | Correlation Strength (R2) | Zone Diameter Formula of Bacterial Growth Inhibition (mm) |
|---|---|---|---|
| Gentamycin | 6376.31 | −0.504 | diameter = 25.461 − 0.2833 × PI% |
| Streptomycin | 7926.96 | −0.602 | diameter = 23.017 − 0.8574 × PI% |
| Kanamycin | 10,229.25 | −0.545 | diameter = 24.099 − 11.300 × PI% |
| Chloramphenicol | 7903.42 | −0.512 | diameter = 25.902 − 5.4950 × PI% |
| Rifampicin | 6374.90 | −0.630 | diameter = 25.801 − 0.4315 × PI% |
| Cephalothin | 5944.73 | −0.520 | diameter = 24.511 − 1.6250 × PI% |
| Vancomycin | 9036.76 | +0.612 | diameter = 23.854 + 0.2672 × PI% |
| Clindamycin | 9088.84 | −0.472 | diameter = 15.599 − 13.0800 × PI% |
| Erythromycin | 9999.35 | +0.541 | diameter = 27.606 + 2.3420 × PI% |
| Ampicillin | 7481.42 | +0.529 | diameter = 26.153 + 2.1796 × PI% |
| Mezlocillin | 9010.29 | −0.517 | diameter = 28.195 − 0.2832 × PI% |
| Penicillin | 4341.25 | −0.581 | diameter = 25.000 − 1.4620 × PI% |
| Ciprofloxacin | 9390.72 | −0.438 | diameter = 24.276 − 1.3480 × PI% |
| Tetracycline | 6492.85 | −0.527 | diameter = 22.950 − 9.6020 × PI% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Szymczak, B. Identification and Differentiation of Non-Hemolytic Listeria monocytogenes from Food Processing Environments Using MALDI-TOF MS. Molecules 2025, 30, 3049. https://doi.org/10.3390/molecules30143049
Szymczak B. Identification and Differentiation of Non-Hemolytic Listeria monocytogenes from Food Processing Environments Using MALDI-TOF MS. Molecules. 2025; 30(14):3049. https://doi.org/10.3390/molecules30143049
Chicago/Turabian StyleSzymczak, Barbara. 2025. "Identification and Differentiation of Non-Hemolytic Listeria monocytogenes from Food Processing Environments Using MALDI-TOF MS" Molecules 30, no. 14: 3049. https://doi.org/10.3390/molecules30143049
APA StyleSzymczak, B. (2025). Identification and Differentiation of Non-Hemolytic Listeria monocytogenes from Food Processing Environments Using MALDI-TOF MS. Molecules, 30(14), 3049. https://doi.org/10.3390/molecules30143049






